SUMMARY
Transcription factor (TF) DNA sequence preferences direct their regulatory activity, but are currently known for only ~1% of all eukaryotic TFs. Broadly sampling DNA-binding domain (DBD) types from multiple eukaryotic clades, we determined DNA sequence preferences for >1,000 TFs encompassing 54 different DBD classes from 131 diverse eukaryotes. We find that closely related DBDs almost always have very similar DNA sequence preferences, enabling inference of motifs for ~34% of the ~170,000 known or predicted eukaryotic TFs. Sequences matching both measured and inferred motifs are enriched in ChIP-seq peaks and upstream of transcription start sites in diverse eukaryotic lineages. SNPs defining expression quantitative trait loci in Arabidopsis promoters are also enriched for predicted TF binding sites. Importantly, our motif “library” (http://cisbp.ccbr.utoronto.ca) can be used to identify specific TFs whose binding may be altered by human disease risk alleles. These data present a powerful resource for mapping transcriptional networks across eukaryotes.
INTRODUCTION
Transcription factor (TF) sequence specificities, typically represented as “motifs”, are the primary mechanism by which cells recognize genomic features and regulate genes. Eukaryotic genomes contain dozens to thousands of TFs encoding at least one of the >80 known types of sequence-specific DNA-binding domains (DBDs) (Weirauch and Hughes, 2011). Yet, even in well-studied organisms, many TFs have unknown DNA sequence preference (de Boer and Hughes, 2012; Zhu et al., 2011), and there are virtually no experimental DNA binding data for TFs in the vast majority of eukaryotes. Moreover, even for the best-studied classes of DBDs, accurate prediction of DNA sequence preferences remains very difficult (Christensen et al., 2012; Persikov and Singh, 2014), despite the fact that identification of “recognition codes” that relate amino acid (AA) sequences to preferred DNA sequences has been a longstanding goal in the study of TFs (De Masi et al., 2011; Desjarlais and Berg, 1992; Seeman et al., 1976). These deficits represent a fundamental limitation in our ability to analyze and interpret the function and evolution of DNA sequences.
The sequence preferences of TFs can be characterized systematically both in vivo (Odom, 2011) and in vitro (Jolma and Taipale, 2011; Stormo and Zhao, 2010). The most prevalent method for in vivo analysis is currently ChIP-seq (Barski and Zhao, 2009; Park, 2009), but ChIP does not inherently measure relative preference of a TF to individual sequences, and may not identify correct TF motifs due to complicating factors such as chromatin structure and partner proteins (Gordan et al., 2009; Li et al., 2011; Liu et al., 2006; Yan et al., 2013). In contrast, it is relatively straightforward to derive motifs from all of the common methods for in vitro analysis of TF sequence specificity, including Protein Binding Microarrays (PBMs), Bacterial 1-hybrid (B1H), and High-Throughput in vitro Selection (HT-SELEX) (Stormo and Zhao, 2010), all of which have been applied to hundreds of proteins (e.g. (Berger et al., 2008; Enuameh et al., 2013; Jolma et al., 2013; Noyes et al., 2008)).
Previous large-scale studies have reported that proteins with similar DBD sequences tend to bind very similar DNA sequences, even when they are from distantly related species (e.g. fly and human). This observation is important because it suggests that the sequence preferences of TFs may be broadly inferred from data for only a small subset of TFs (Alleyne et al., 2009; Berger et al., 2008; Bernard et al., 2012; Noyes et al., 2008). However, these analyses have utilized data for only a handful of DBD classes and species, and they contrast with numerous demonstrations that mutation of one or a few critical DBD AAs can alter the sequence preferences of a TF (e.g. (Aggarwal et al., 2010; Cook et al., 1994; De Masi et al., 2011; Mathias et al., 2001; Noyes et al., 2008)), which suggest that prediction of DNA binding preferences by homology should be highly error-prone. To our knowledge, rigorous and exhaustive analyses of the accuracy and limitations of inference approaches to predicting TF DNA-binding motifs using DBD sequences has not been done.
Here, we determined the DNA sequence preferences for >1,000 carefully-selected TFs from 131 species, representing all major eukaryotic clades, and encompassing 54 DBD classes. We show that, in general, sequence preferences can be accurately inferred by overall DBD AA identity, suggesting that mutations that dramatically impact sequence specificity are relatively rare. By identifying distinct confidence thresholds for each individual DBD class (i.e. levels of protein sequence identity above which motifs can be assumed to be identical between two proteins), we infer sequence preferences for roughly one-third of all known eukaryotic TFs, based on experimental data for fewer than 2% of them. Cross-validation indicates that ~89% of predicted sequence preferences are as accurate as experimental replicates of the same TF. We demonstrate the functional relevance and utility of both known and inferred motifs by showing that they coincide with ChIP-seq binding peak sequences, are enriched in the promoter regions of diverse eukaryotes, and significantly overlap eQTLs in Arabidopsis. We also demonstrate how our data can be used to predict the specific TFs whose binding would be altered by a human disease risk allele. To house the data and the resulting inferences, we have created the Cis-BP database (catalog of inferred sequence binding preferences), freely available at http://cisbp.ccbr.utoronto.ca.
RESULTS
PBM data for >1,000 diverse eukaryotic TFs
We sought to examine the relationship between the DBD AA sequence identity and the DNA sequence specificity of any two proteins, and to simultaneously broadly survey the sequence specificity of eukaryotic TFs. To identify TFs, we first scanned the AA sequences for each of 81 different types of DBDs for which there is an available Pfam model (Weirauch and Hughes, 2011), using known or predicted proteins in 290 sequenced eukaryote genomes. We identified a total of 166,851 putative TFs that fit these criteria. From these, we selected 2,913 individual TFs to analyze, using several different criteria aimed at achieving the goals of our study, including a relatively even balance among DBD classes and species, a survey of different levels of sequence identity among proteins, and a deeper focus on several model organisms and abundant DBD classes (see Experimental Procedures). Figure S1 depicts the overall scheme.
We analyzed each TF using PBM assays, following procedures previously described (Berger et al., 2006; Weirauch et al., 2013). The PBM technique can be summarized as follows: a GST-tagged DNA-binding protein is “hybridized” to a double-stranded DNA microarray, and subsequent addition of a fluorescently tagged antibody reveals the DNA sequences that the protein has bound, and to what degree. Each PBM contains a diverse set of ~41,000 35-mer probes, designed such that all possible 10-mers are present once and only once; thus, all non-palindromic 8-mers are present 32 times, allowing for a robust and unbiased assessment of sequence preference to all possible 8-mers. Values for individual 8-mers are typically given as both E-scores (which represent relative rank of intensities, and range from −0.5 to + 0.5 (Berger et al., 2006)) and Z-scores (which scale approximately with binding affinity (Badis et al., 2009)). PBMs also allow derivation of Position Weight Matrices (PWMs) up to 14 bases wide (Badis et al., 2009; Berger et al., 2006; Mintseris and Eisen, 2006; Weirauch et al., 2013), as well as IUPAC consensus sequences (Ray et al., 2013). PWMs can be represented as sequence logos (Schneider and Stephens, 1990) which are typically taken as synonymous with “motifs”. Here, we report 8-mer scores, PWMs, and consensus sequences, and use whichever is best suited to individual analyses. For example, while motifs are well suited for visualization of sequence preferences, and convenient for scanning longer sequences, they can underestimate reproducibility of experiments due to the introduction of uncertainties in the process of PWM derivation (Weirauch et al., 2013; Zhao and Stormo, 2011).
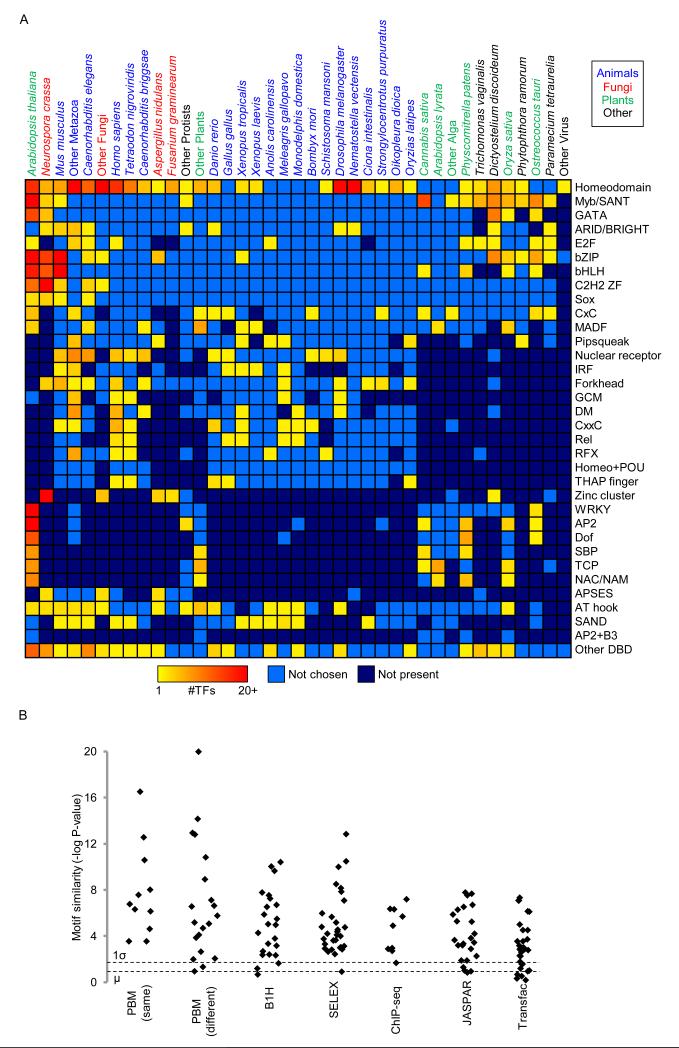
We analyzed each of the 2,913 proteins using two different PBM arrays, designated “ME” and “HK” after their designers (Lam et al., 2011). Of these, 1,032 (encompassing 1,017 different TFs) yielded data that satisfy our stringent success criteria, including E-scores greater than 0.45 on both arrays, and agreement in both 8-mer data and motifs between the two arrays (Berger et al., 2008; Weirauch et al., 2013) (see Experimental Procedures). The distribution of the 1,032 proteins over 131 species and 54 DBD classes is summarized in Figure 1A. Many values in this matrix are zero because the majority of DBD classes are only present in certain subsets of species (e.g. plant-specific TFs) (Weirauch and Hughes, 2011). PBM failures may be due to any of several causes, including protein misfolding, requirement for cofactors, or bona fide lack of sequence-specific DNA binding activity.
Figure 1. Overview of the motif dataset.
(A) TFs characterized in this study, by species and DBD class. TFs with multiple DBD classes are indicated with a “+” (e.g., AP2+B3). DBD classes and species containing fewer than five members are grouped into “Other”. Species are ordered by the total number of TFs with characterized motifs. (B) PBM-derived motifs are similar to previously characterized motifs. We compared new PBM-derived motifs to previously determined motifs for the same TF. P-values were calculated using the TomTom PWM similarity tool (Tanaka et al., 2011), with Euclidean distance and default parameter settings. Dashed lines indicate mean (bottom), and mean plus one standard deviation (top) of P-values obtained from 10,000 randomly selected PWM pairs. ‘PBM (same)’ and ‘PBM (dif)’ indicate PBMs from other studies performed using the same, or different array designs as this study, respectively. See also Figure S1 and Tables S1, S2, and S6.
For 123 of the 1,017 examined TFs, there are previously described motifs. In most cases the new motifs we obtained are highly similar to motifs compiled from the literature (JASPAR and Transfac), derived from PBMs in other studies, derived from other technologies (B1H and HT-SELEX), or derived from ChIP-seq experiments (Figure 1B; sources provided as Table S1; full motif comparisons provided as Table S2). Importantly, a large majority of the proteins we analyzed (894/1,017) had no previous binding data, and among these, roughly half yielded a motif that is highly different from any previously known motif (see Experimental Procedures). For several DBD classes (CG-1, CxC, GRAS, LOB, and Storekeeper) we characterized sequence specificity for the first time. CxC domains, for example, span a wide range of organisms encompassing plants, animals, and protists, and recognize variations of a unique, largely conserved TTTCGAAA motif.
Inference of TF sequence specificity using degree of identity in the DNA-binding domain
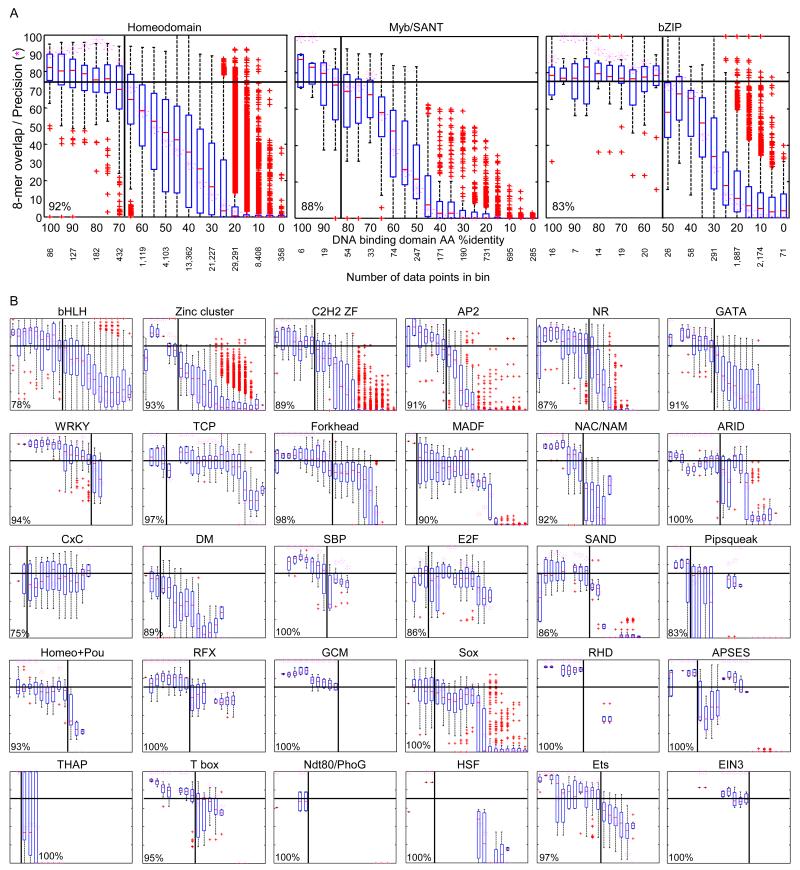
We next used the PBM data to ask how well the percent protein sequence identity in the DBD between two proteins correlates with similarity in their DNA sequence preferences. As a measure of DNA sequence preference similarity, we calculated the overlap in high-scoring 8-mers (e.g., E-score > 0.45) (see Experimental Procedures), as the E-scores are on a uniform scale that facilitates direct comparison. The boxplots shown in Figure 2 and Supplementary Data 1 illustrate that there are different characteristic relationships between DBD identity and 8-mer overlap for different DBD classes. However, virtually all of the plots display a sigmoidal appearance, such that a particular threshold defines the %AA identity over which the majority (75% or more) of protein pairs have very similar DNA binding preferences (i.e., are at least as similar as the 25th percentile of replicates of the same protein – see Experimental Procedures) (Figure 2). For homeodomains, this threshold is 70% AA identity (Figure 2A), almost exactly what we previously reported (65%) (Berger et al., 2008).
Figure 2. Motif inference thresholds by DBD class.
(A) Relationship between similarity in DBD AA sequence and DNA sequence preferences. Boxplots depict the relationship between the %ID of aligned AAs and % of shared 8-mer DNA sequences with E-scores exceeding 0.45, for the three DBD classes with the most PBMs in this study. %ID bins range from 0 to 100, of size 10, in increments of five. Below, number of DBD pairs in each bin. Pink asterisks indicate the precision of the corresponding bin (i.e., the fraction of protein pairs with 8-mer similarity at least as high as the 25th percentile of replicates). Horizontal line indicates the 75% precision line used to choose the inference threshold. Vertical lines indicate AA %ID threshold (i.e., the point before the pink asterisks drop below the horizontal line). Percentage in lower left corner indicates cross validation success rate. (B) Relationship for all DBD classes. Boxplots for all DBD classes for which we could establish an inference threshold, depicted as in (A). DBD classes are ordered by the number of TFs characterized in this study. See also Figures S2 and S6.
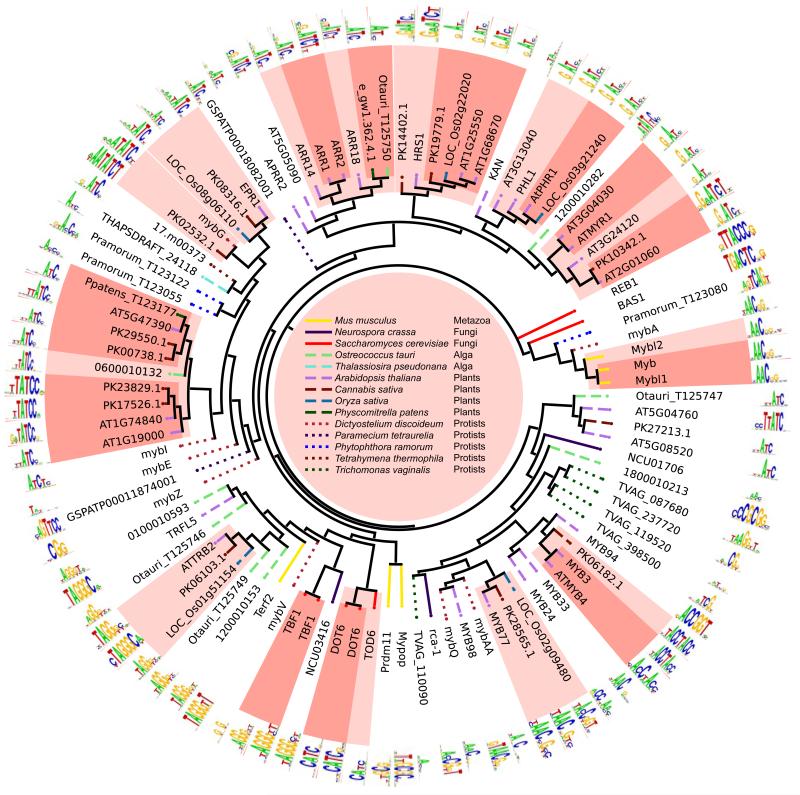
Strikingly, such a threshold can be drawn for virtually every DBD class (Figure 2 and Supplementary Data 1). Moreover, exactly the same trends and thresholds are observed in a leave-one-out cross validation prediction framework, in which the high-scoring 8-mers are predicted to be identical to those of the protein with the closest %AA identity (see Experimental Procedures). Using this framework, we estimate that ~89% of predicted sequence preferences (hereafter, “inferences”) above the thresholds shown in Figure 2 are as accurate as experimental replicates (Supplementary Data 1), with similar precision obtained whether inferences are derived from only orthologs (88%), or only paralogs (90%). The distribution of DBD AA similarities among the 1,017 proteins we analyzed is nearly identical to the distribution among all sequenced eukaryotes (data not shown), so the same numbers can be expected for de novo predictions. Furthermore, comparisons among PWMs (i.e. motifs) confirm these thresholds, despite the potential variation contributed by motif derivation (Weirauch et al., 2013; Zhao and Stormo, 2011). To illustrate this concept, Figure 3 depicts PBM-derived motifs obtained for the Myb/SANT family, grouped according to the relationship of their DBD AA sequences. Darkly shaded regions, which invariably contain nearly indistinguishable motifs, indicate groups of TFs with DBD similarity exceeding the 87.5% threshold for this family.
Figure 3. Overview of Myb/SANT family motifs.
PBM-derived motifs from the Myb/SANT family (84 from this study, 13 from other studies) are shown. Tree reflects the percent of identical AAs after alignment. Dark shading, 87.5% AA identity (standard inference threshold); light shading, > 70% AA identity (relaxed inference threshold). TBF1 and DOT6 each have two motifs because they were examined in two different studies.
We conclude that TFs with DBD AA sequence identity above these thresholds will typically have very similar sequence specificity. As previously proposed, this presents a simple approach for broadly predicting the sequence preferences of TFs (Alleyne et al., 2009; Berger et al., 2008; Bernard et al., 2012; Noyes et al., 2008): the 8-mer data, motifs, and consensus sequences can be directly transferred from the nearest protein above the threshold for which data is available. The data presented here encompass 37 DBD classes with sufficient data (i.e. comparisons in each of the bins in the box plots) to produce thresholds, together representing ~85% of all known eukaryotic TFs. Until more data are available, we propose that a threshold of 70% AA identity (which is the mean, median, and mode across all DBD classes) can be applied to the remaining DBD classes (boxplots in Figure S2 show the aggregate of all of the classes with no threshold).
Cis-BP: a catalogue of direct and inferred sequence binding preferences
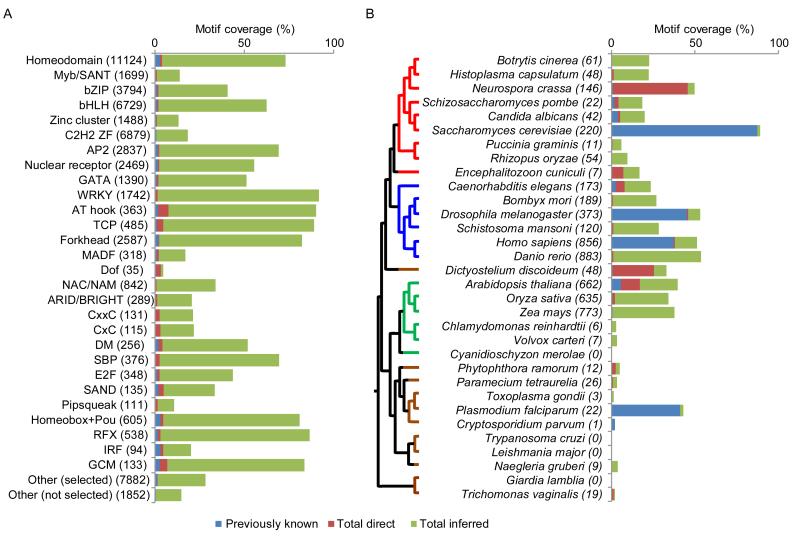
Using the DBD identity thresholds generated above, we globally assigned probable motifs (and other sequence preference data, if available) to TFs from 290 eukaryotic genomes. To do this, we supplemented the PBM data collected in this study with 4,234 additional published motifs (674 from PBMs, and 3,560 from other sources – see Table S1) derived from 1,850 different proteins, mainly in human (623 proteins), mouse (409), fly (316), or yeast (216). Altogether, there are experimentally determined motifs for only 1.7% (2,750) of the 166,851 unique TFs within these genomes. Using the thresholds determined here, it is possible to infer a motif for roughly one third of all TFs encoded by sequenced eukaryotes, bringing the total (known + inferred) to 57,165. Figures 4A and 4B summarize the coverage by DBD class and by species (see Table S3 for all DBD classes and species). Lineages that benefit most from the inference scheme include vertebrates, plants, fungi, and insects, which contain many orthologs conserved in the model species analyzed most heavily. For example, the motif collection for zebrafish (Danio rerio), which largely consists of inferred motifs, is as complete as that of mouse and human (Figure 4B).
Figure 4. TF motif coverage.
TFs with multiple protein isoforms are counted as a single gene. (A) Motif coverage by DBD class. DBD classes sorted top to bottom by number of TFs characterized in this study. Those with fewer than eight proteins characterized in this study are grouped into “Other”. “Other (selected)” indicates DBD classes selected for characterization in this study. “Other (not selected)” indicates DBD classes not characterized here. “Direct” includes those experimentally characterized in this study, but not previously known. “Total inferred” excludes those experimentally characterized in this or previous studies. (B) Motif coverage by species. Tree at left, phylogenetic relationships between organisms (Baldauf et al., 2000). See also Table S3.
To facilitate use of the known and inferred motifs by the scientific community, we created a database called Cis-BP (catalogue of inferred sequence preferences of DNA-binding proteins) (http://cisbp.ccbr.utoronto.ca). In addition to the new experimental data reported here, Cis-BP contains comprehensive 8-mer binding scores, position weight matrices, and IUPAC consensus motifs from publicly available sources (JASPAR (Portales-Casamar et al., 2010), Transfac (public data only) (Matys et al., 2006), FlyFactorSurvey (Zhu et al., 2011), FactorBook (Wang et al., 2013), and data from 674 PBM experiments taken from other studies (compiled in UniPROBE (Newburger and Bulyk, 2009)).
In vitro-derived motifs predict ChIP-seq peaks
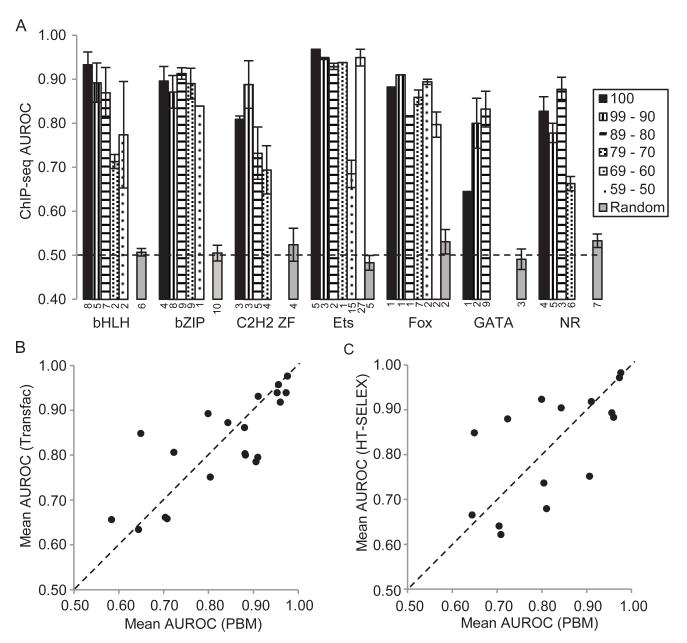
To examine the relationship between in vitro-defined motifs (both measured and inferred) and in vivo binding sites, we asked how well the motifs in Cis-BP predict in vivo binding, based on ENCODE human ChIP-seq data (see Experimental Procedures). To gauge the ability of a motif to discriminate between real ChIP peaks (positives) and peak sequences permuted using an algorithm that maintains all dinucleotide frequencies (negatives), we used the Area Under the Receiver Operating Characteristic (AUROC) summary statistic, in which perfect discrimination between positives and negatives scores 1.00, and random guessing scores 0.50. Nearly all (111) of the 114 PBM-derived motifs achieved AUROC scores exceeding the 0.50 random expectation level in at least one ChIP dataset (Figure 5A, Table S4). Strikingly, over one-third of the motifs (43 of 114), in a variety of DBD classes and cell types, achieve AUROCs exceeding 0.90 in at least one ChIP dataset, including GABPA in H1-hESC cells (Ets family, AUROC=0.99), USF2 in GM12878 cells (bHLH, 0.97), and FOS in K562 cells (bZIP, 0.97). Motifs inferred from in vitro data from related proteins achieve AUROC values similar to those obtained using in vitro motifs from exactly the same protein that was ChIPped (Figure 5A). Overall, PBM-derived motifs are equally accurate as those derived from other sources; the mean AUROCs across the 19 TFs with at least one PBM-derived motif and one Transfac motif are 0.834 and 0.827, respectively (Figure 5B and Table S4), and 0.826 vs. 0.831 for the 14 TFs with both PBM and HT-SELEX-derived motifs (Figure 5C and Table S4). The small number of cases in which motifs derived from these other sources performed better than PBM motifs appear to correspond to multimeric binding that was not detected in the PBM assays. From this analysis, we conclude that there is generally no fundamental discrepancy between motifs obtained from different assays, and between the in vivo and in vitro sequence preferences of TFs.
Figure 5. PBM-derived motifs identify in vivo TF binding locations.
(A) AUROC analysis, showing ability of directly determined and inferred motifs to distinguish ChIP-seq peak sequences from scrambled sequences. We identified TFs with available ENCODE ChIP-seq data that also have PBM data available either for that TF, or for related TFs (based on the inference threshold for the DBD class). We then gauged the ability of the PBM-derived motifs to distinguish real ChIP peaks from scrambled sequences (maintaining all dinucleotide frequencies) using the AUROC (see Experimental Procedures). For each DBD class, results are binned by DBD %AA ID (key indicated at upper right). Numbers below each bar indicate the count in each bin. Error bars indicate standard error. ‘Random’ indicates results obtained with a randomly assigned, unrelated TF motif. Abbreviation: Fox, Forkhead box. Figure S7 shows results obtained using an alternative null model. (B) Comparison of AUROC for PBM-derived motifs and literature-derived motifs. We identified TFs with ENCODE ChIP-seq experimental data that also have both Transfac and PBM-derived motifs available. For each TF, we calculated the best AUROC obtained by any PBM or any Transfac motif on any of the ENCODE cell line ChIP experiments for that TF. For TFs with multiple motifs from the same source, the plot shows the mean AUROC across the motifs. (C) PBM-derived motifs vs. HT-SELEX-derived motifs. Same as for (B), but including only TFs with motifs available both from PBMs and a recent HT-SELEX study (Jolma et al., 2013). See also Table S4.
Motifs display positional bias in promoters across eukaryotes
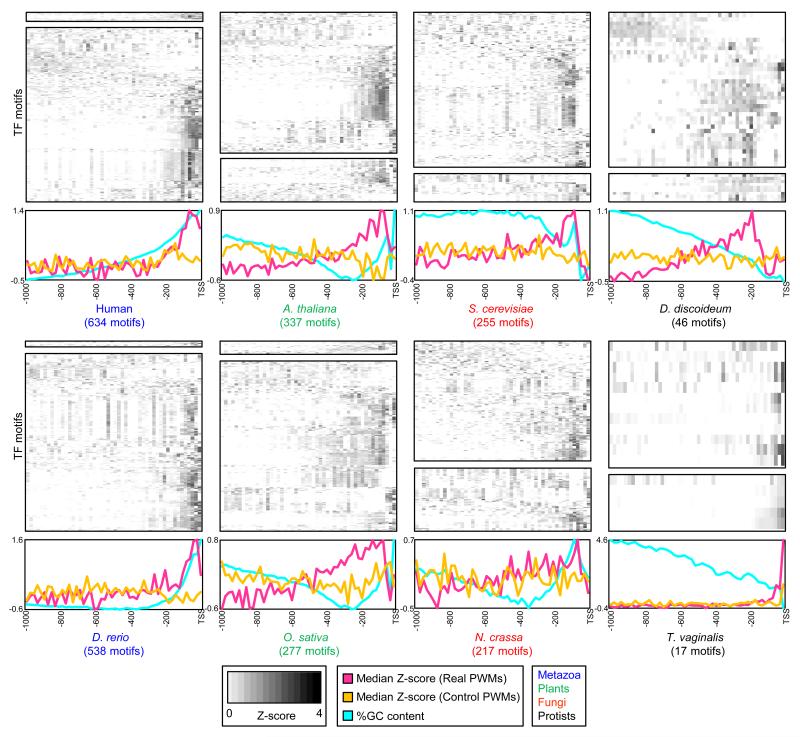
Enrichment and positional bias of TF motifs in promoters has been reported in several model species and human (FitzGerald et al., 2004; Lee et al., 2007; Marino-Ramirez et al., 2004; Ohler et al., 2002), and in some cases TFs appear to be involved in determining promoter identity (de Boer et al., 2014; Megraw et al., 2009). However, it is unknown whether this is a general property of eukaryotic promoters. We found that genomes from vertebrates, plants, fungi, and protists all displayed enrichment of binding sites for their TFs just upstream of the TSS, relative to permuted sequences, and also relative to unrelated (control) motifs taken from other lineages (Figure 6). The enrichment involved both directly-determined and inferred motifs (Figure 6). Strikingly, the TF classes with motifs enriched in promoters differed between lineages of organisms (Figure S3), indicating that these trends either arose independently in different lineages, or have evolved considerably in most of them. Furthermore, we found little to no correspondence between overall motif enrichment and motif sequence composition (i.e., overall GC-content) (Table S5), and the enrichment was observed whether the promoters were characteristically AT-rich or GC-rich (i.e., predicted to be nucleosome favoring or disfavoring, respectively). We observed similar trends using a reduced set of non-redundant motifs for each organism (Figure S4), showing that the phenomenon is not due to expansion of a small number of TFs families that bind promoters. These observations indicate that, across eukaryotes, the region just upstream of the TSS typically is enriched for binding sites that may function in either promoter definition or gene regulation.
Figure 6. Positional bias of motif matches in eukaryotic promoters.
PBM-derived PWMs (direct, top; inferred, bottom) scored in 20-bp bins, normalized to dinucleotide-permuted controls, averaged across all promoters, and displayed as Z-scores (see Experimental Procedures). Each row in the heatmap corresponds to one PWM. Rows were clustered using hierarchical clustering (Pearson correlation, average linkage). Summary plots at the bottom indicate the median Z-score, taken across all PWMs from the indicated species (‘Real PWMs’), or across a set of PWMs from unrelated lineages (‘Control PWMs’) (see Experimental Procedures). See also Table S5 and Figures S3 and S4.
Arabidopsis eQTL SNPs are enriched for TF binding sites
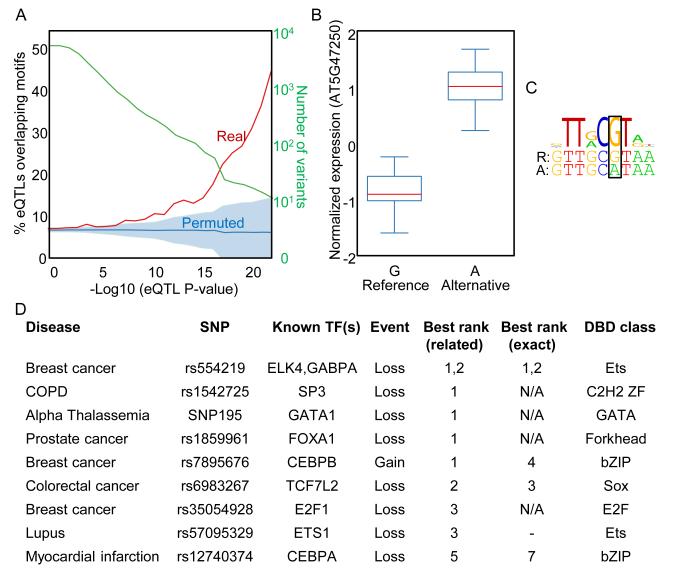
Identifying causal genetic variants and their mechanisms of action is a fundamental challenge in association studies. As our data greatly expand the motif collection in the model plant Arabidopsis, we examined a dataset of expression Quantitative Trait Loci (eQTLs) defined from Arabidopsis genomic sequences and matched seedling RNA-seq taken data from 19 strains (Gan et al., 2011). Among highly significant eQTLs found within 1 kb upstream of a TSS, there was a striking enrichment (>3-fold for the SNPs with strongest association) for overlap with TF motif matches (Figure 7A), strongly suggesting that these SNPs impact transcription factor binding. As an example, Figures 7B and C show a SNP in the promoter of the AT5G47250 gene, in which loss of a potential binding site for the VNI2 TF in the “A” allele correlates almost perfectly with a dramatic increase in AT5G47250’s expression level, consistent with the well-characterized role of VNI2 as a transcriptional repressor (Yamaguchi et al., 2010; Yang et al., 2011). Both VNI2 and AT5G47250 were associated with the plant defense response to the pathogenic oomycete H. arabidopsidis in a recent genome-wide association study examining 107 different phenotypes (Atwell et al., 2010). Taken together, these results suggest that VNI2 represses the expression of AT5G47250 in a pathogen response context, a mechanism dependent on the specific SNP present in the AT5G47250 promoter region. We expect that the Cis-BP motif collection will be useful for similar analyses in other organisms: eQTL analysis can be performed in virtually any species for which there are multiple strains (or individuals), and does not require well-developed genetic systems. In fact, in this analysis, a similar level of enrichment was observed using only a set of 65 motifs inferred from organisms other than Arabidopsis thaliana (Figure S5), suggesting that the motif data need not be derived from the organism in question (see also below).
Figure 7. Overlap of predicted TF binding sites with cis-eQTLs.
(A) Number and percentage of Arabidopsis cis-eQTLs overlapping motifs, as a function of eQTL significance. Shaded region indicates one standard deviation in the expected distribution (see Experimental Procedures). (B) A cis-eQTL affecting the expression of the AT5G47250 gene. Boxplots indicate the median normalized gene expression level for each allele of the cis-eQTL. ‘Reference’ indicates the allele present in the Arabidopsis reference genome assembly. (C) The same cis-eQTL “breaks” a putative binding site for the VNI2 transcriptional repressor. Sequence logo depicts the DNA-binding motif we obtained for VNI2. Sequences below indicate the reference (top) and alternative (bottom) alleles of the cis-eQTL SNP (boxed), and its flanking bases. (D) Prediction of human TF binding events altered by disease risk alleles. We created a method for using PBM data to predict TFs whose binding is affected by disease associated genetic variants, and applied it to 16 known examples. Shown here are the ten cases in which we ranked the correct TF (column labeled ‘exact’) or a highly related TF from the same DBD class (column labeled ‘related’) within the top five TFs. The ‘Event’ column indicates whether the risk allele results in a ‘Loss’ or ‘Gain’ of binding of the TF. ‘N/A’ indicates that PBM data is not available for the corresponding TF. ‘-’ indicates that the TF did not receive a rank because both alleles had E-score > 0.45. See also Figure S5.
Identification of TFs affected by disease-associated genetic variants
Recent analyses indicate that between 85 and 93% of disease and trait associated variants are located in non-coding regions (Hindorff et al., 2009; Maurano et al., 2012), suggesting that many might alter TF binding events. However, the identification of specific TFs whose binding might be affected by a given variant remains a challenging and laborious task. We devised a system that utilizes all available PBM data to produce a ranked list of human TFs whose binding might be affected by any given genetic variant (see Experimental Procedures). To examine the utility of this system for analyzing human disease-associated variants, we collected a set of 15 SNPs whose alleles have been experimentally demonstrated to affect the binding of a specific TF. One of these SNPs affects two TFs, bringing the total of analyzed TF/SNP pairs to 16. Strikingly, in ten of these 16 cases this procedure ranked the correct TF (or a highly related TF from the same DBD class) in the top five, and often #1 (Figure 7D and Supplementary Data 1). We note that most of the novel high-ranking TFs we identify have likely never been experimentally examined, and thus might also represent bona fide cases.
In one example, a recent study implicated rs554219 in estrogen-receptor-positive breast cancer tumors, and used a series of experiments to predict, and eventually establish that this SNP causes the differential binding of two Ets family TFs, ELK4 and GABPA (French et al., 2013). Since rs554219 does not overlap ChIP-seq binding peaks from ENCODE or other sources, ELK4 and GABPA were identified using a series of EMSA competition experiments involving known TF binding sites, a laborious process involving substantial guesswork. Our automated computational procedure correctly ranked ELK4 #1, and GABPA #2 out of all human TFs (their EMSA data demonstrate strong and moderate differential binding of ELK4 and GABPA, respectively). The ELK4 prediction is based on an inference from the pufferfish Tetraodon nigroviridis ELK4 protein (81% AA ID to human ELK4), further illustrating the utility of cross-species inferences. Moreover, differential binding of ELK4 to the alleles of rs554219 can also be predicted using data inferred from Drosophila melanogaster Ets21C (57% AA ID to human ELK4) and Caenorhabditis elegans F19F10.1 (only 33% AA ID to ELK4). Thus, our system (available at http://cisbp.ccbr.utoronto.ca/TFTools.php) can accurately predict the specific TFs whose binding is affected by risk alleles of disease associated SNPs, even when using PBM data inferred from distantly related organisms.
DISCUSSION
We anticipate that the new data collected here – as well as the inferred motifs across eukaryotes - will be an invaluable resource and knowledge base for functional genomics and analysis of gene regulation. In addition to the data itself, the Cis-BP database also contains web-based interfaces to tools for scanning DNA sequences for putative motifs, reporting the TFs with motifs similar to a given motif, predicting the motif recognized by a given TF based on its DBD AA sequence, and identifying TFs that will bind differentially to two different DNA sequences (e.g., disease risk and non-risk alleles). Cis-BP will enable dissection of regulatory mechanisms from expression data even in the vast majority of species for which there are currently no genetic tools.
The analyses here present a strategy to rapidly populate TF motif collections across the eukaryotes. As we previously proposed for RNA-binding proteins (Ray et al., 2013), targeting members of the largest groups of uncharacterized proteins for experimental analysis will allow the largest number of inferred motifs to be obtained. Motif inference based on DBD identity alone is only a first approximation, but it is remarkably cost effective: the analyses described here indicate that motifs can be inferred for 34% of all TFs, using data from only 1.7%. We extrapolate that 1,032 additional successful experiments (less than 0.5% of all TFs, and a number identical to that in this study) – one from each of the largest groups of orthologs and paralogs with no known motifs – would increase coverage across the eukaryotes from 34 to 48%.
The inference scheme described here relies on the high degree of conservation among DBDs. Indeed, our analyses confirm the “deep homology” that has been described for metazoan developmental processes and the TFs that regulate them (e.g. homeodomains) (Berger et al., 2008; Carroll, 2008; Noyes et al., 2008), and furthermore indicate that deep homology is a property of the sequence preferences of many TFs in all eukaryotic kingdoms. Our initial analyses (data not shown) suggest that many motifs likely date to the base of metazoans, land plants, angiosperms (flowering plants), or euteleostomi (bony vertebrates), consistent with well-established TF expansions in these lineages (de Mendoza et al., 2013; Weirauch and Hughes, 2011).
Despite widespread conservation, TF repertories do change over evolutionary time, and these changes likely shape eukaryotic evolution (de Mendoza et al., 2013). TFs that tend to diversify, such as the large metazoan C2H2 class (Stubbs et al., 2011), will present an ongoing challenge to a complete characterization of eukaryotic TF motifs. In other lineages, other classes of TFs have expanded and diversified, including the nuclear receptor class in C. elegans (Maglich et al., 2001), the zinc cluster/GAL4 class in fungi (Shelest, 2008), and several classes in plants (Lang et al., 2010). The data described here confirm that the sequence specificities of at least some of these factors have also diversified. Mapping recognition codes represents an alternative approach to more complete cataloguing of TF motifs, and the data presented here provide many new examples for the study of TF-DNA recognition. Exceptions to the simple AA similarity rules described here should also be informative regarding mechanisms of sequence recognition, since they will identify AA residues critical for DNA sequence specificity (De Masi et al., 2011; Noyes et al., 2008). Ongoing efforts to further the collective knowledge of TF binding specificities will greatly advance our understanding of TF-DNA interactions, as well as our ability to interpret the function of DNA sequences, including understanding the functional impact of natural genetic variants in human and other species.
EXPERIMENTAL PROCEDURES
Full details are provided in Extended Experimental Procedures.
Data availability
PBM data is available in the GEO database under accession GSE53348. PBM data, clone information, and other data from analyses carried out in this study are available on the project website: http://hugheslab.ccbr.utoronto.ca/supplementary-data/CisBP/. Additional data (including 8-mer scores, PWMs, sequence logos, and information on TFs) is found on the CisBP web server (http://cisbp.ccbr.utoronto.ca/).
Selection, cloning, and PBM analysis of TFs
We compiled the predicted proteomes of 290 eukaryotic organisms from a variety of sources, and supplemented them with an additional 49 known TFs from organisms without fully sequenced genomes. We scanned all protein sequences for putative DNA-binding domains (DBDs) using the 81 Pfam (Finn et al., 2010) models listed in (Weirauch and Hughes, 2011) and the HMMER tool (Eddy, 2009). Each protein was classified into a family based on its DBDs and their order in the protein sequence. We selected 2,913 individual TFs to analyze, using several different criteria, including a relatively even balance among DBD classes and species, a survey of different levels of sequence identity among proteins, and a deeper focus on several model organisms and abundant DBD classes. For most constructs, we designed primers to clone the region encompassing all DBDs plus the 50 flanking endogenous AAs on either side (or until the termini of the protein) by conventional PCR methods into one of a panel of T7-GST vectors for expression in E. coli (referred to hereafter as “plasmid constructs”). PBM laboratory methods were identical to those described in (Lam et al., 2011; Weirauch et al., 2013). Each plasmid was analyzed in duplicate on two different arrays with differing probe sequences (denoted ‘ME’ and ‘HK’). Calculation of 8-mer Z- and E-scores was performed as previously described (Berger et al., 2006). To obtain a single representative motif for each protein, we used a procedure similar to a recent study from our group in which we generated motifs for each array using four different algorithms, and chose the best-performing single motif based on cross-replicate array evaluations (Weirauch et al., 2013).
Inference scheme
We established a separate inference threshold for each DBD class. We first aligned the DBD sequences of all constructs within a DBD class using clustalOmega (Sievers et al., 2011). We then calculated the AA %ID for all construct pairs (i.e. the number of identical AAs in the alignments). Within each DBD class, we grouped all PBM construct pairs into bins, based on AA %ID. We used overlapping bins of size 10, ranging from 0 to 100, increasing by 5. We calculated the precision of each bin by comparing the DNA sequence preferences obtained from all characterized protein pairs contained in the bin. We quantified the similarity of the DNA sequence preferences of two proteins as the fraction of shared high-scoring 8-mers. We considered a prediction to be correct only if this fraction exceeded the value obtained at the 25th percentile of experimental replicates (i.e., the fraction of shared 8-mers between ME and HK arrays for the same protein). The proportion of predictions scored as correct (i.e. precision) for each bin of each DBD class is shown as magenta stars in Figure 2 and Supplementary Data 1.
We chose inference thresholds for each DBD class based on the precision scores of each AA %ID bin. Since we used the 25th percentile threshold to define precision, we would expect a precision of 0.75 or higher in each AA %ID bin. We therefore chose an inference threshold for each DBD class by identifying the final AA %ID bin before precision drops below 0.75 (vertical bars in Figure 2). Similar thresholds were obtained regardless of the E- and Z-score 8-mer thresholds used, and also regardless of the replicate overlap percentile considered (i.e., 25th percentile, requiring 0.75 precision or 20th percentile, requiring 0.80 precision) (Figure S6). The final threshold for a DBD class was chosen as the median threshold across the eight 8-mer similarity measures (see Figure S6 and Supplementary Data 1). We found this scheme to be appropriate for most DBD classes (all of which are depicted in Figure 2). For three DBD classes (IRF, CXXC zinc fingers, and Dof zinc fingers), we could not establish a threshold – these therefore received a threshold of 100%. We used a threshold of 40% for AT-hook TFs, which recognize AT-rich sequences, based on manual inspection of the data (see Supplementary Data 1). For the remaining classes, with suggestive but insufficient data, we chose a threshold of 70%, which is the mean, median, and mode threshold across all DBD classes.
We used the AA %ID of all pairs of proteins to infer motifs, 8-mer scores, and consensus sequences within each DBD class by simple transfer (i.e., aligning the DBD sequences of all proteins and all constructs in a given DBD class, as described above, and calculating the AA %ID of each protein with each construct). We also evaluated the effectiveness of our inference scheme in a leave-one-out cross validation framework, in which the PBM data for each characterized protein was held out, and compared to the PBM data of its nearest neighbor (i.e., the characterized protein with highest AA %ID), using a similar scoring scheme to that used to calculate the precisions.
Comparison to ChIP-seq data
We calculated AUROC scores on real and permuted (maintaining dinucleotide frequencies) ChIP-seq peak sequences, following (Weirauch et al., 2013). We obtained ENCODE consortium human ChIP-seq data from the UCSC Genome Browser (Rosenbloom et al., 2012). For each ChIP experiment, we extracted the top 500 scoring peak region sequences, and scored them (and the permuted sequences) using all direct and inferred PWM models for the given TF. For each PWM/experiment pair, we then calculated the AUROC using these sets of 500 positives and 500 negatives. Similar results were obtained using a negative set consisting of ChIP-bound peaks from unrelated TFs with matched GC-content (Figure S7).
Positional bias of motifs in eukaryotic promoters
We obtained the 1000 bases upstream of transcription start sites (or, if unavailable, translation start sites), and scored the PWMs of each organism at each position. We then placed the resulting scores into 20 bp bins, summed the scores for each bin, and took the average across all promoters for the given species for each bin. To correct for mono- and dinucleotide biases, we also scored shuffled promoter sequences, which were created by shuffling the sequences within each 20 bp bin (while maintaining dinucleotide frequencies). For each PWM, we then calculated the ratio of each bin’s real score relative to the score of the shuffled sequence. The resulting ratios were then normalized across all bins for the given PWM using a standard Z-score transformation. We also calculated Z-scores for a negative control set of TF PWMs for each organism, consisting of a collection of random motifs from species in other clades that were unrelated to any PWM from the given species.
Arabidopsis eQTL analysis
We used a publicly available dataset (Gan et al., 2011) containing genome-wide RNA-seq variance-stabilized expression levels (Huber et al., 2002) taken from 19 strains of seedling Arabidopsis thaliana, and matching genome sequences. We identified matches to each A. thaliana PWM within the 1,000 bases upstream of each TSS. We calculated the percentage of genetic variants that affect these putative binding sites, as a function of the cis-eQTL p-value of the variant (red line, Figure 7). We also created a null distribution (blue line and blue shaded region, Figure 7) to exclude the possibility that the observed percentages might solely be due to the higher density of TF binding sites in promoter regions.
Human disease SNP/TF analysis
We devised a system for utilizing our collection of PBM data to identify candidate human TFs whose binding might be affected by the allelic sequences of genetic variants. In this system, we score each variant (along with its flanking genomic bases) using 8-mer E-scores taken from the 3,132 PBM experiments contained in our database. For each PBM experiment, we identify the highest scoring 8-mer E-score attained by any of the risk allele sequences (Erisk), and the highest attained by any non-risk allele (Enon-risk). We then identify all PBM experiments where only one of Erisk and Enon-risk has an E-score value exceeding 0.45 (values above this threshold will likely be strongly bound by the given TF (Berger et al., 2008)), and map these experiments to human using the inference scheme. This procedure thus produces a ranked list of human TFs whose binding is likely to be affected by the alleles of a given SNP (e.g., strongly binding to one allele, but not binding to the other).
Supplementary Material
HIGHLIGHTS.
DNA binding motifs are known for <2% of eukaryotic transcription factors (TFs)
We assayed >1000 diverse TFs, to learn rules for transferring motifs between TFs
The resulting “motif library” covers ~34% of eukaryotic TFs across all major clades
We show how our library can be used to understand human and plant SNP mechanisms
ACKNOWLEDGEMENTS
We thank the following people for experimental and computational support on this project: Shaheynoor Talukder, Andrew Vorobyov, Anton van der Ven, Wilfred de Vega, Nicole Park, C. Alexander Valencia, Geanany Rasanathan, Yogesh Hooda, Sanie Mnaimneh, Kenneth Chu, Oliver Boright, Jerry Li, Agnieszka Janska, Esther Chan, Trevis Alleyne, Emily Stoakes, Oliver Stegle, Todd Riley, and Harmen Bussemaker. We are grateful to Martha Bulyk, Jussi Taipale, Raphael Kopan, Artem Barski, Ian Lewkowich, Brenda Andrews, and Charlie Boone for helpful conversations and advice, and the many investigators who generously contributed template DNA or RNA used in PCR cloning of DBDs (listed in Table S6). This work was supported by grants from the Canadian Institutes of Health Research to TRH (MOP-490425 and MOP-459114). MTW was supported by fellowships from CIHR and the Canadian Institute for Advanced Research (CIFAR) Junior Fellows Genetic Networks Program. TRH is a CIFAR scholar. EH and GS were supported by a grant from the NIH/NICHD P01 HD39691. JSR-H and AJMW were funded by NIH GM082971. MGL was supported by an EU Marie Curie International Outgoing Fellowship (project 252475). Research in JRE’s laboratory is supported by the NSF (grant MCB-1024999), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation (GBMF). JRE is a HHMI-GBMF Investigator. AM, AG and LFL were supported by Millennium Nucleus for Fungal Integrative and Synthetic Biology (NC120043) and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1131030), both to LFL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
MTW and TRH conceived of the study, oversaw analyses, and wrote the manuscript. MTW performed most of the computational analyses. AY performed and oversaw most of the experimental analyses. MTW and MA developed and implemented the database and web server. AC, AMM, IM, HZ, AG, JCL, MG, ML, EH, and JRH performed additional experiments. HSN, MA, HvB, and XC provided additional computational support. PD performed the Arabidopsis eQTL analysis of Figure 7. SL and MA performed the motif location enrichment analysis in Figure 6. KC performed motif analyses related to Figure 3. TM performed analysis for the ChIP-seq comparisons in Figure 5. SG, GS, AJMW, FYB, GR, LFL, and JRE developed and oversaw experimental and computational analyses. All authors provided critical feedback on the manuscript, with extensive contributions from AMM, HvB, JRH, AJMW, LFL, MTW, and TRH.
REFERENCES
- Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. The Plant cell. 2010;22:1174–1189. doi: 10.1105/tpc.109.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleyne TM, Pena-Castillo L, Badis G, Talukder S, Berger MF, Gehrke AR, Philippakis AA, Bulyk ML, Morris QD, Hughes TR. Predicting the binding preference of transcription factors to individual DNA k-mers. Bioinformatics. 2009;25:1012–1018. doi: 10.1093/bioinformatics/btn645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nature biotechnology. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard B, Thorsson V, Rovira H, Shmulevich I. Increasing coverage of transcription factor position weight matrices through domain-level homology. PLoS One. 2012;7:e42779. doi: 10.1371/journal.pone.0042779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Christensen RG, Enuameh MS, Noyes MB, Brodsky MH, Wolfe SA, Stormo GD. Recognition models to predict DNA-binding specificities of homeodomain proteins. Bioinformatics. 2012;28:i84–89. doi: 10.1093/bioinformatics/bts202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WJ, Mosley SP, Audino DC, Mullaney DL, Rovelli A, Stewart G, Denis CL. Mutations in the zinc-finger region of the yeast regulatory protein ADR1 affect both DNA binding and transcriptional activation. The Journal of biological chemistry. 1994;269:9374–9379. [PubMed] [Google Scholar]
- de Boer CG, Hughes TR. YeTFaSCo: a database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res. 2012;40:D169–179. doi: 10.1093/nar/gkr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer CG, van Bakel H, Tsui K, Li J, Morris QD, Nislow C, Greenblatt JF, Hughes TR. A unified model for yeast transcript definition. Genome Res. 2014;24:154–166. doi: 10.1101/gr.164327.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibes A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJ. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Sebe-Pedros A, Sestak MS, Matejcic M, Torruella G, Domazet-Loso T, Ruiz-Trillo I. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci U S A. 2013;110:E4858–4866. doi: 10.1073/pnas.1311818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci U S A. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al. Global analysis of Drosophila Cys(2)-His(2) zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013;23:928–940. doi: 10.1101/gr.151472.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, Khan S, Maranian MJ, O’Reilly M, Hillman KM, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. American journal of human genetics. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Jolma A, Taipale J. Methods for Analysis of Transcription Factor DNA-Binding Specificity In Vitro. Subcell Biochem. 2011;52:155–173. doi: 10.1007/978-90-481-9069-0_7. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Weiche B, Timmerhaus G, Richardt S, Riano-Pachon DM, Correa LG, Reski R, Mueller-Roeber B, Rensing SA. Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol Evol. 2010;2:488–503. doi: 10.1093/gbe/evq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome research. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, Carrick K, Kamdar K, Willson TM, Moore JT. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome biology. 2001;2:RESEARCH0029. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino-Ramirez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic acids research. 2004;32:949–958. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias JR, Zhong H, Jin Y, Vershon AK. Altering the DNA-binding specificity of the yeast Matalpha 2 homeodomain protein. J Biol Chem. 2001;276:32696–32703. doi: 10.1074/jbc.M103097200. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic acids research. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Pereira F, Jensen ST, Ohler U, Hatzigeorgiou AG. A transcription factor affinity-based code for mammalian transcription initiation. Genome Res. 2009;19:644–656. doi: 10.1101/gr.085449.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintseris J, Eisen MB. Design of a combinatorial DNA microarray for protein-DNA interaction studies. BMC Bioinformatics. 2006;7:429. doi: 10.1186/1471-2105-7-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT. Identification of Transcription Factor-DNA Interactions In Vivo. Subcell Biochem. 2011;52:175–191. doi: 10.1007/978-90-481-9069-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome biology. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic acids research. 2010;38:D105–110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic acids research. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelest E. Transcription factors in fungi. FEMS Microbiol Lett. 2008;286:145–151. doi: 10.1111/j.1574-6968.2008.01293.x. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Zhao Y. Determining the specificity of protein-DNA interactions. Nat Rev Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- Stubbs L, Sun Y, Caetano-Anolles D. Function and Evolution of C2H2 Zinc Finger Arrays. Subcell Biochem. 2011;52:75–94. doi: 10.1007/978-90-481-9069-0_4. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Bailey T, Grant CE, Noble WS, Keich U. Improved similarity scores for comparing motifs. Bioinformatics. 2011;27:1603–1609. doi: 10.1093/bioinformatics/btr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, Moore J, Pierce BG, Dong X, Virgil D, et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–176. doi: 10.1093/nar/gks1221. Factorbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Sub-cellular biochemistry. 2011;52:25–73. doi: 10.1007/978-90-481-9069-0_3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. The Plant cell. 2010;22:1249–1263. doi: 10.1105/tpc.108.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. The Plant cell. 2011;23:2155–2168. doi: 10.1105/tpc.111.084913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Stormo GD. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat Biotechnol. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, Brasefield JA, Zhu C, Asriyan Y, Lapointe DS, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011;39:D111–117. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PBM data is available in the GEO database under accession GSE53348. PBM data, clone information, and other data from analyses carried out in this study are available on the project website: http://hugheslab.ccbr.utoronto.ca/supplementary-data/CisBP/. Additional data (including 8-mer scores, PWMs, sequence logos, and information on TFs) is found on the CisBP web server (http://cisbp.ccbr.utoronto.ca/).