Abstract
IMPORTANCE
Targeting oncogenic drivers (genomic alterations critical to cancer development and maintenance) has transformed the care of patients with lung adenocarcinomas. The Lung Cancer Mutation Consortium was formed to perform multiplexed assays testing adenocarcinomas of the lung for drivers in 10 genes to enable clinicians to select targeted treatments and enroll patients into clinical trials.
OBJECTIVES
To determine the frequency of oncogenic drivers in patients with lung adenocarcinomas and to use the data to select treatments targeting the identified driver(s) and measure survival.
DESIGN, SETTING, AND PARTICIPANTS
From 2009 through 2012, 14 sites in the United States enrolled patients with metastatic lung adenocarcinomas and a performance status of 0 through 2 and tested their tumors for 10 drivers. Information was collected on patients, therapies, and survival.
INTERVENTIONS
Tumors were tested for 10 oncogenic drivers, and results were used to select matched targeted therapies.
MAIN OUTCOMES AND MEASURES
Determination of the frequency of oncogenic drivers, the proportion of patients treated with genotype-directed therapy, and survival.
RESULTS
From 2009 through 2012, tumors from 1007 patients were tested for at least 1 gene and 733 for 10 genes (patients with full genotyping). An oncogenic driver was found in 466 of 733 patients (64%). Among these 733 tumors, 182 tumors (25%) had the KRAS driver; sensitizing EGFR, 122 (17%); ALK rearrangements, 57 (8%); other EGFR, 29 (4%); 2 or more genes, 24 (3%); ERBB2 (formerly HER2), 19 (3%); BRAF, 16 (2%); PIK3CA, 6 (<1%); MET amplification, 5 (<1%); NRAS, 5 (<1%); MEK1, 1 (<1%); AKT1, 0. Results were used to select a targeted therapy or trial in 275 of 1007 patients (28%). The median survival was 3.5 years (interquartile range [IQR], 1.96-7.70) for the 260 patients with an oncogenic driver and genotype-directed therapy compared with 2.4 years (IQR, 0.88-6.20) for the 318 patients with any oncogenic driver(s) who did not receive genotype-directed therapy (propensity score–adjusted hazard ratio, 0.69 [95% CI, 0.53-0.9], P = .006).
CONCLUSIONS AND RELEVANCE
Actionable drivers were detected in 64% of lung adenocarcinomas. Multiplexed testing aided physicians in selecting therapies. Although individuals with drivers receiving a matched targeted agent lived longer, randomized trials are required to determine if targeting therapy based on oncogenic drivers improves survival.
The introduction of targeted therapy has transformed the care of patients with lung cancers by incorporating tumor genotyping into therapeutic decision making. Adenocarcinoma, the most common type of lung cancer, is diagnosed in 130 000 patients in the United States and 1 million persons worldwide each year.1 It is also the type of lung cancer with a higher than 50% estimated frequency of actionable oncogenic drivers.2,3 The Lung Cancer Mutation Consortium (LCMC) collectively termed these molecular abnormalities oncogenic drivers to include multiple types of genomic changes and emphasize that unlike many biomarkers and “passenger” mutations, these alterations are critical to cancer development and maintenance. The LCMC further defined these drivers as actionable based on the demonstration that the downstream effects of these abnormalities that initiate or maintain the neoplastic process can be negated by agents directed against each genomic alteration. Testing for somatic mutations in the epidermal growth factor receptor (EGFR) gene4-6 and rearrangements of the anaplastic lymphoma kinase (ALK) gene7 is now routine.8
With additional oncogenic drivers being detected, testing for targets sequentially lessens the efficiency of the process.9 The need to genotype lung adenocarcinomas for EGFR and ALK, the emergence of new targets, and the ability to perform multiplex genotyping, have led institutions to systematically characterize genetic aberrations.10-15 The LCMC selected oncogenic drivers based on the ability to detect the change within Clinical Laboratory Improvement Amendments (CLIA)–certified laboratories, a reported frequency of at least 1% in lung adenocarcinomas, and availability of a drivertargeted agent(s), either as an approved agent or as part of a trial when this study was designed in 2009. The LCMC proposed to determine the frequency of oncogenic drivers, demonstrate the practicality of routine genetic analyses, and use the information to guide treatment and facilitate studies of targeted therapies.
Methods
Patients
Institutional review board approval was obtained at all 14 study sites. Patients with stage IV16 or recurrent adenocarcinomas of the lung and SWOG (Southwest Oncology Group) performance status of 0 (asymptomatic), 1 (symptomatic, fully ambulatory), or 2 (symptomatic, in bed <50% of a day) were enrolled. All patients provided written informed consent for this study and the analysis reported in this paper. The LCMC analyzed 1 specimen per patient. Those with adequate tumor tissue for genomic characterization remained eligible. Patients who had been previously tested for oncogenic drivers that were clinically indicated were allowed to enroll. Prospectively defined testing for this study was carried out after enrollment. Adenocarcinoma was centrally confirmed. No immunohistochemistry tests were routinely used. Adenosquamous carcinomas were ineligible. Age, sex, smoking history, and previous treatment data were collected.
Interventions
Sites performed multiplex genotyping for mutation detection using any of 3 methods: (1) matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Sequenom, Arizona Research Laboratories), (2) multiplexed single-nucleotide extension sequencing (SNaPshot, Applied Biosystem), or (3) Sanger sequencing with peptide nucleic acid probes.11-14 Additionally, all sites performed sizing electrophoresis to detect avian erythroblastic leukemia (ERBB2 [formerly HER2]) insertions and EGFR deletions.17 Along with EGFR (NG_007726.3) and ALK (NG_009445.1), the LCMC identified mutations in KRAS (NG_007524.1), NRAS (NG_0075 72.1) , BRAF (NG_007873.3) , ERBB2 (NG_007503.1), PIK3CA (NG_012113.2), MEK (NG_008305.1), and AKT1 (NG_012188.1 ), and amplific ation o f MET (NG_008996.1).2,11,12,14,17-23 The LCMC prioritized genotyping as follows: (1) EGFR, (2) KRAS, ERBB2, AKT1, BRAF, MEK1, NRAS, PIK3CA, (3) ALK, (4) MET. When the LCMC designed the study, trials demonstrating the superiority of EGFR tyrosine kinase inhibitors (EGFR TKIs) over chemotherapy were reported, making EGFR testing the first priority. Once sufficient DNA was extracted for EGFR mutation testing, we were able to assess other oncogenic drivers with modest additional resources and DNA. The LCMC tested for ALK rearrangements and MET amplification by fluorescence in situ hybridization (FISH). ALK was prioritized over MET because of the availability of crizotinib for ALK rearranged tumors.24,25 Specimens obtained by surgery, core, and fine needle biopsy or pleural fluid were acceptable. Submitted slides and blocks were assessed for diagnosis and for adequacy of tumor sample at the site where testing was performed. Criteria for specimen adequacy were not prespecified. Generally, 100 cells per slide were required for FISH and mutational testing. Approximately 200 ng DNA were needed for testing by multiplex mutational profiling and 120 ng DNA for time-of-flight mass spectrometry. The LCMC pathologists shared blinded samples, with a positive sample included for all 10 drivers. Interlaboratory and interassay variability in proficiency testing will be reported in a future publication. Mutation information was recorded in a GeneInsight database.26 The LCMC clinical committee chairs (MGK and BEJ) solicited industry-sponsored trials for 9 drivers (eTable 1 in the Supplement). These trials were subsequently reviewed and approved by representatives from all 14 LCMC sites. The numbers of patients enrolled as well as those treated on any other genotype-driven study or receiving an agent targeted to the driver are displayed in eTable 2 in the Supplement. The decision to recommend a targeted therapy to a patient with a tumor harboring an oncogenic driver was left to the treating physician. The LCMC clinical committee chairs reviewed the records of each patient with an oncogenic driver and the targeted agent administered to verify that the treatment was directed against that driver(s) detected in the patient’s tumor. Sites reported whether or not each patient received therapy directed against a detected driver and the duration of survival. In all survival analyses, the calculation was performed from the date of metastatic disease diagnosis. We collected vital status and treatment data at least once a year and in 2012 when the study data were analyzed. Survival figures show 5 years of data, to facilitate viewing within the timeframe where most events occurred.
Primary and Secondary Outcomes
The primary objective was to determine the frequency of 10 oncogenic drivers in patients with lung adenocarcinomas. The secondary objectives were to study as many tumors as possible for all 10 genes to define the co-occurrence of drivers in a single tumor, document our ability to use the data to select treatment or a clinical trial targeting the driver(s) identified, and measure survival.
Statistical Methods
Sample size estimation was based on the 95% CI method using the binomial probability density function. With the proposed sample size of 1000, the half-width of the 95% CI is less than 5% if the oncogenic driver rate is less than 40%. Descriptive statistics, including median and range for continuous variables, as well as percentages and frequencies for categorical variables, were tabulated and presented here. Overall survival time was defined as the time from date of diagnosis of metastatic cancer to date of death or last follow-up. Survival curves were calculated by the Kaplan-Meier method for groups of interest and were compared using the log-rank test. The association of targeted therapy with overall survival was estimated by multivariable Cox proportional hazard modeling. Because of limited sample size, propensity score and stage were included in the Cox model to control for selection bias, and the proportional hazards assumption was assessed. The knowledge-based confounding variables adjusted in the propensity model were sex, age at enrollment, performance status, smoking history, stage at diagnosis, prior therapy (surgery, chest radiotherapy, or chemotherapy), and elapsed time from metastatic disease diagnosis to study enrollment (years). Adjusted hazard ratio (HR) and adjusted 95% CI were reported. All statistical tests were 2-sided, with a P-value less than .05 considered to indicate statistical significance. For analysis, R (R-project, Institute for Statistics and Mathematics), version 3.0.2, was used.
Results
From 2009 to 2012, 1537 patients were enrolled and 1102 were eligible. The primary reason for ineligibility was the lack of tumor tissue for genomic testing. In 85 cases, the slide received contained insufficient material to permit a diagnosis of adenocarcinoma. Among the 1017 patients with confirmed adenocarcinoma, 1007 had tumors with at least 1 gene studied for genomic changes and 733 individuals had tumors fully-genotyped with all 10 oncogenic drivers assessed (eFigure 1 in the Supplement). Enrollment and genotyping frequencies by site are reported in eTable 3 in the Supplement. Sixty percent of patients were women and 89% had performance status of 0 or 1 (Table 1). Most patients were former smokers and 34% were never smokers. Sixty-four percent had stage IV disease at diagnosis. Characteristics of patients with oncogenic drivers are similar whether or not they received a targeted therapy as presented in Table 1.
Table 1.
Patient Characteristics
| Genotyping |
Driver Present |
||||
|---|---|---|---|---|---|
| No. (%) | Any (n = 1007)a | Full (n = 733)a | No Driver (n = 382)b,c |
No Treatment (n = 342)b,c |
Treatment (n = 275)b,c |
| Sex | |||||
|
| |||||
| Men | 407 (40) | 293 (40) | 177 (46) | 132 (39) | 95 (35) |
|
| |||||
| Women | 600 (60) | 440 (60) | 205 (54) | 210 (61) | 180 (65) |
|
| |||||
| Age at enrollment, median (IQR), y | 63 (55-70) | 64 (54-72) | 63 (54-70) | 65 (57-72) | 62 (53-67) |
|
| |||||
| Performance statusd | |||||
|
| |||||
| 0 | 345 (34) | 245 (33) | 122 (32) | 112 (33) | 110 (40) |
|
| |||||
| 1 | 549 (55) | 411 (56) | 217 (57) | 185 (54) | 142 (52) |
|
| |||||
| 2 | 97 (10) | 68 (9) | 38 (10) | 40 (12) | 17 (6) |
|
| |||||
| Missing | 16 (2) | 9 (1) | 5 (1) | 5 (1) | 6 (2) |
|
| |||||
| Cigarette smoking history | |||||
|
| |||||
| Never | 341 (34) | 240 (33) | 102 (27) | 74 (22) | 161 (59) |
|
| |||||
| Former | 589 (58) | 440 (60) | 248 (65) | 231 (68) | 106 (39) |
|
| |||||
| Current | 73 (7) | 49 (7) | 28 (7) | 37(11) | 8 (3) |
|
| |||||
| Missing | 4 (0) | 4 (1) | 4 (1) | 0 (0) | 0 (0) |
|
| |||||
| Stage at diagnosis | |||||
|
| |||||
| I | 103 (10) | 84 (11) | 39 (10) | 41 (12) | 22 (8) |
|
| |||||
| II | 67 (7) | 58 (8) | 28 (7) | 24 (7) | 15 (5) |
|
| |||||
| III | 169 (17) | 120 (16) | 71 (19) | 61 (18) | 37 (13) |
|
| |||||
| IV | 643 (64) | 452 (62) | 231 (60) | 208 (61) | 198 (72) |
|
| |||||
| Missing | 25 (2) | 19 (3) | 13 (3) | 8 (2) | 3 (1) |
|
| |||||
| Prior therapy | |||||
|
| |||||
| Surgery | 435 (43) | 337 (46) | 174 (46) | 149 (44) | 110 (40) |
|
| |||||
| Chest radiotherapy | 202 (20) | 145 (20) | 83 (22) | 76 (22) | 40 (15) |
|
| |||||
| Chemotherapy | 594 (59) | 435 (59) | 226 (59) | 191 (56) | 173 (63) |
|
| |||||
| Time from metastatic disease diagnosis to enrollment, median (range), y |
0.36 (0.07-1.48) | 0.33 (0.07-1.46) | 0.37 (0.07-1.34) | 0.25 (0.06-1.35) | 0.53 (0.07-1.80) |
Abbreviation: IQR, interquartile range.
Any genotyping indicates 1 to 10 total genes assayed; full genotyping, 10 genes assayed.
Driver categorization from the any genotyping group; because not all genes were assayed in all cases in this group, the no driver group represents combination of negative findings and no findings.
Sum of the patients in these 3 groups is 999; the remaining 8 patients had a driver, but data on targeted treatment was unknown or not reported.
The SWOG (Southwest Oncology Group) performance status of 0 (asymptomatic), 1 (symptomatic, fully ambulatory), or 2 (symptomatic, in bed <50% of a day).
Genomic data for each of the 10 genes were generated from a minimum of 833 patients’ tumors (MET) to as many as 987 patients’ tumors (EGFR) (eTable 2 in the Supplement). The primary reason for the inability to test for all 10 genes was insufficient tissue. Among the 733 specimens tested for all 10 genes, 466 (64% [95% CI, 60%-67%) had an oncogenic driver (442 with 1 driver and 24 with 2 drivers; eFigure 2 in the Supplement). As illustrated in Table 2, KRAS mutations were the most frequent, found in 182 of 733 specimens (25%), followed by sensitizing EGFR (EGFR[s]) mutations (exon 19 deletions, L858R, L861Q, and G719X) in 122 of 733 specimens (17%). The frequency of EGFR(s) was 17% in the full genotyping group and 17% in the any genotyping group. ALK rearrangements occurred in 57 of 733 specimens (8%). Other EGFR (EGFR[o]) mutations not associated with sensitivity to kinase inhibitors (18 of 733 exon 20 insertions, 7 of 733 de novo T790M, 4 other) were identified in 29 of 733 specimens (4%).27,28 The frequencies of ERBB2 (all exon 20 insertions) were 3% (19 of 733 specimens), BRAF mutations were 2% (16 of 733 specimens), and PIK3CA (6 of 733 specimens), NRAS (5 of 733 specimens), and MEK1 (1 of 733 specimens) were identified in less than 1%, no AKT1 (0 of 733 specimens) mutations. Amplification of MET (MET/ CEP7 >2.2) was found in 5 of 733 patients’ tumors (<1%). A listing of the specific point mutations is presented in eTable 4 in the Supplement. The frequencies of drivers in the 1007 specimens in which at least 1 gene was assessed are presented in Table 2 and mirror the results with the patients’ tumors with full genotyping. In the full genotyping group, we found drivers in 2 genes in 24 tumors (3%). Of these 24 specimens, 20 (83%) included either a mutation in PIK3CA (n = 12) or MET amplification (n = 8). Gene pairings and specific mutations for these “doubletons” are presented in eTable 5 in the Supplement. Mutation frequencies in current, former, and never smokers are presented in eTable 6 in the Supplement.
Table 2.
Oncogenic Drivers Identified and Targeted Treatments Received for Patients With Any and Full Genotyping
| Gene With Mutational or Structural Change |
Genotyping, No. (%) [95% CI] |
Patients Receiving Targeted Therapy No. (%)c |
|
|---|---|---|---|
| Any (n = 1007)a,b | Full (n = 733)b | ||
| Any gene(s) | 623 (62) [59-65] | 466 (64) [60-67] | 275 (44) |
|
| |||
| Singletonsd | |||
|
| |||
| KRAS | 245 (24) [22-27] | 182 (25) [22-28] | 22 (9) |
|
| |||
| EGFR (sensitizing)e | 175 (17) [15-20] | 122 (17) [14-20] | 146 (83) |
|
| |||
| exon19 del | 103 (10) [9-12] | 68 (9) [7-12] | |
|
| |||
| L858R | 64 (6) [5-8] | 47 (6) [5-9] | |
|
| |||
| G719X | 5 (0.5) [0.2-1] | 5 (0.7) [0.3-2] | |
|
| |||
| L861Q | 5 (0.5) [0.2-1] | 4 (0.5) [0.2-2] | |
|
| |||
| ALK (rearrangement) | 80 (8) [6-10] | 57 (8) [6-10] | 52 (65) |
|
| |||
| EGFR (other)f | 35 (4) [3-5] | 29 (4) [3-6] | 23 (66) |
|
| |||
| ERBB2 (formerly HER2) | 23 (2) [2-4] | 19 (3) [2-4] | 11 (48) |
|
| |||
| BRAF | 18 (2) [1-3] | 16 (2) [1.3-3.6] | 3 (17) |
|
| |||
| V600E | 14 (1) [0.8-2] | 12 (2) [0.9-3] | 2 (14) |
|
| |||
| Non-V600E | 4 (0.4) [0.1-1] | 4 (0.5) [0.2-2] | 1 (25) |
|
| |||
| PIK3CA | 7 (0.7) [0.3-2] | 6 (0.8) [0.3-2] | 0 |
|
| |||
| MET (amplification) | 6 (0.6) [0.2-1] | 5 (0.7) [0.3-2] | 3 (50) |
|
| |||
| NRAS | 5 (0.5) [0.2-1] | 5 (0.7) [0.3-2] | 0 |
|
| |||
| MEK1 | 2 (0.2) [0.03-1] | 1 (0.1) [0-1] | 0 |
|
| |||
| AKT1 | 0 [0-1] | 0 [0-1] | 0 |
|
| |||
| Doubletons | |||
|
| |||
| >1 gene | 27 (3) [2-4] | 24 (3) [2-5] | 15 (56) |
For percentages in this column, 1007 is used as the denominator. Because not all cases in this group were tested for all genes, percentages reflect rate of detection of mutation where nondetection is a combination of negative findings and no findings.
Any genotyping indicates 1 to 10 total genes assayed; full genotyping, 10 genes assayed.
Percent among cases with detected driver in any genotyping group.
Per-gene count (percent) of patients with mutations occurring in a single gene (singletons). Patients with oncogenic drivers in more than 1 gene (doubletons) are included as their own category. For detailed information on mutations occurring within doubletons, see eTable 5 in the Supplement.
The sum of counts for the 4 categories of EGFR (sensitizing) mutations differs from the patient-level count of total number of patients with EGFR (sensitizing) mutations due to 2 patients with the co-occurrence of 2 different sensitizing mutations in the same specimen.
Patients are counted in the EGFR (other) group based on a finding of any one or more mutations in EGFR other than exon 19 deletions, L858R, G719X, or L861Q, with or without co-occurrence in the same tumor specimen of 1 of these sensitizing mutations. See eTable 5 in the Supplement for a detailed count of co-occurring EGFR mutations.
For the 27 patients with tumors with 2 oncogenic drivers, individuals could participate in a trial for either driver at the physician’s discretion. Fifteen of the 27 patients received a targeted agent, including 10 with erlotinib for their EGFR mutation and 3 with crizotinib for ALK rearrangements. Overall, 28% (95% CI, 24%-30%) of eligible patients and 44% of individuals with drivers detected received a targeted therapy. Of the 175 patients (83%) with EGFR(s) mutations, 146 were treated with targeted therapy: 130 with erlotinib alone, and 16 with another EGFR inhibitor alone or in combination. Of the 35 patients (66%) with EGFR(o) mutations, 23 were treated with an EGFR inhibitor alone, another targeted agent, or a combination. We treated 52 of the 80 patients (65%) with ALK rearrangements with crizotinib. Of the 23 patients (48%) with ERBB2-mutant lung cancers, 11 received an ERBB2-targeted agent. Of the 245 (9%) with KRAS mutations, 22 were treated with investigational targeted agents.
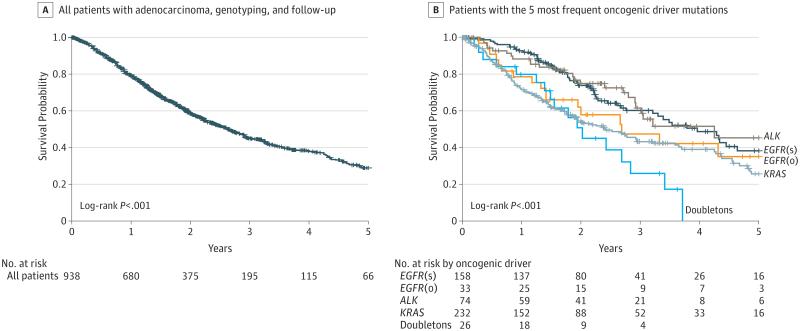
Among the 1007 patients tested for at least 1 driver, 93% had sufficient information to be included in the survival analysis (456 were alive and 482 had died); among this group, median follow-up was 1.67 years (IQR, 0.9-2.69); range, 0-18.56. Seven percent of patients (69) were not included because of the lack of treatment information or they did not have at least 1 follow-up visit. The median potential follow-up was 2.0 years. The median survival for all 938 patients with adequate data was 2.7 years (95% CI, 2.4-2.9) (Figure 1A). The median survival of individuals with each of the 5 most common oncogenic drivers ranged from 2.0 years (mutations in 2 genes) to 4.3 years (ALK) (Figure 1B; P ≤ .001). Survival by mutation type for the 7 drivers identified in at least 10 patients is shown in eFigure 3 in the Supplement (P = .001).
Figure 1. Survival of Patients.
ALK indicates anaplastic lymphoma kinase gene; EGFR(s), epidermal growth factor receptor gene (sensitizing); EGFR(o), epidermal growth factor receptor gene (other); KRAS, Kirsten rat sarcoma; NA, not applicable.
A, The median survival 2.65 years (95% CI, 2.35-2.93). B, Median survival (95% CI): EGFR(s), 3.97 years (3.21-4.64); EGFR(o), 2.70 years (1.42-NA); ALK, 4.25 years (2.92-NA); KRAS, 2.41 years (1.87-3.21); doubletons (oncogenic drivers in 2 genes), 2.03 years (1.39-2.84). Vertical tick marks are censoring events.
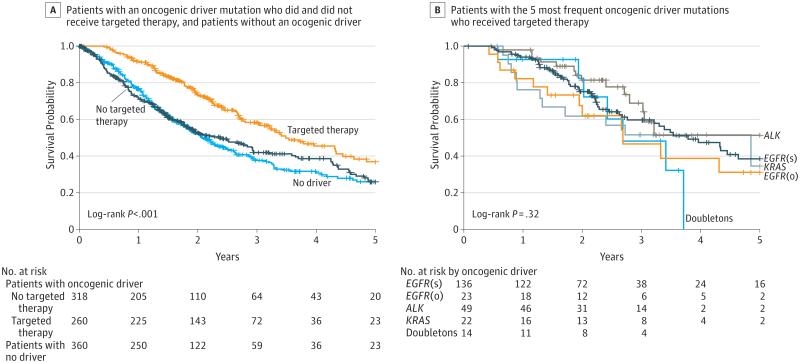
The association of targeted therapy and survival in patients with tumors with oncogenic drivers is shown in Figure 2A. The 260 patients with an oncogenic driver and treatment with a targeted agent had a median survival of 3.5 years; the 318 patients with a driver and no targeted therapy, 2.4 years; and the 360 patients with no driver identified, 2.1 years (P < .001; Figure 2A). We recomputed these analyses for the full genotype set for the patients with at least 1 follow-up date (n = 689). The 190 patients with an oncogenic driver and treatment with a targeted agent had a median survival of 3.5 years (IQR, 1.96-7.70); the 248 with a driver and no targeted therapy, 2.5 years (P < .001, eFigure 4 in the Supplement). In addition, we presented propensity score–adjusted Cox models to examine the group differences. Sex, age, performance status, smoking history, stage, prior therapy, and the time of diagnosis of metastatic disease to enrollment were included in the propensity score–matching. For the propensity modeling, we analyzed 275 patients with targeted therapy and 734 without; setting the acceptable distance for any matched propensity score as 0.0001, we found 11 matched case-control pairs. Compared with patients with any oncogenic driver(s) who did not receive genotype-directed therapy (n = 318, 169 deaths), patients with a driver and genotype-directed therapy (n = 260, 111 deaths) showed a decreased risk of death (HR, 0.69 [95% CI, 0.53-0.9], P = .006). We recomputed the survival data comparing patients with drivers who received and did not receive a targeted therapy, excluding all patients with drivers with a diagnosis of metastatic disease more than 6 months before entry. For the 442 patients with metastatic disease diagnosed at 6 months or less prior to the LCMC start date, the 189 with drivers who received targeted therapy had a median survival of 2.7 years, whereas the 253 with drivers who did not receive targeted treatment had a median survival of 1.5 years (P < .001; eFigure 5 in the Supplement). We also recomputed the survival data excluding individuals with EGFR- and ALK-positive cancers. The 49 patients with oncogenic drivers other than EGFR or ALK who received a targeted therapy had a median survival of 4.9 years compared with the 678 with (318 patients) or without (360 patients) a driver identified, but not receiving a targeted agent, whose median survivals were 2.4 years with a driver (IQR, 0.88-6.20) and 2.1 years without (P = .14; eFigure 6 in the Supplement). Patients with the 5 most frequent drivers treated with targeted therapies showed similar median survival times varying from 2.7 years (mutations in 2 genes) to 4.9 years (KRAS), P = .32 (Figure 2B).
Figure 2. Survival Comparisons.
ALK indicates anaplastic lymphoma kinase gene; EGFR(s), epidermal growth factor receptor gene (sensitizing); EGFR(o), epidermal growth factor receptor gene (other); KRAS, Kirsten rat sarcoma; NA, not applicable.
A, Median survival (95% CI): oncogenic driver + no targeted therapy, 2.38 (1.81-2.93); oncogenic driver + targeted therapy, 3.49 (3.02-4.33); no oncogenic driver, 2.08 (1.84-2.46). B, Survival by oncogenic driver detected for patients with the 5 most frequent oncogenic drivers and targeted treatment. Median survival (95% CI): EGFR(s), 3.78 (2.77-NA); EGFR(o), 2.70 (1.42-NA); ALK, NA (2.80-NA); KRAS, 4.85 (1.30-NA); doubletons (oncogenic drivers in 2 genes), 2.69 (1.94-NA). Vertical tick marks are censoring events.
Discussion
The LCMC demonstrated that it is possible to routinely generate multiplex genotyping information from patients with lung adenocarcinomas. The LCMC characterized at least 1 gene for 91% (1007 of 1102 patients) of eligible patients with genotyping of all 10 genes in 66% (733 of 1102 patients). This genotyping success rate compares favorably with other therapeutic studies for patients with lung cancers.29-31
The identification of oncogenic drivers has redefined how we describe these illnesses and care for persons with lung cancers. Since this trial began in 2009, up-front genotyping is now an essential step in choosing therapy. Chemotherapy is no longer a standard but a default if patients do not have an actionable driver. The use of targeted treatment in individuals with EGFR- and ALK-positive lung cancers (30 000 patients yearly in the United States) has been well documented. Our study is, to our knowledge, the first presentation of data from a prospective multi-institutional investigation supporting the concept that this can be accomplished with more drivers and drugs, with the potential to change the approach to lung cancer management. A listing of 11 oncological drugs approved for other indications that target 7 oncological drivers found in lung cancers is now included in the National Comprehensive Cancer Network guidelines.8 To our knowledge, this is the first study to show the lack of overlap between oncogenic drivers, only because we tested as many tumors as possible for all drivers. In other studies, once a driver was identified, testing for others was not pursued.
Our multi-institutional consortium identified patients with rare genomic changes and used the information to select treatments and facilitate trials. The ultimate goal of genomic testing is to use the information generated to select therapies and improve outcomes. Physicians do this by choosing therapies targeted to the specific oncogenic driver detected in a patient’s tumor specimen. Crizotinib was administered to the 8% of patients with ALK rearrangements and BRAF inhibitors to the 2% with BRAF mutations.7,25
Performing multiplex genomic characterization detected additional mutations with little need for additional resources. We identified 18 patients with BRAF mutations who were candidates for trials with BRAF inhibitors. The multiplexed genotyping platforms made effective use of patient tissue and identified actionable drivers sooner, giving patient’s earlier access to therapies or trials. Multiplexed testing also facilitates the rapid inclusion of new oncogenic drivers to existing panels. We have now added testing for RET32,33 and ROS134 fusions to the multiplex panels in the LCMC.
The LCMC effort differs from other genomic characterization efforts, like the Cancer Genome Atlas that studied patients with localized disease and obtained tissues exclusively from resection specimens.35-40 The frequencies of EGFR (21%) and KRAS mutations (25%) found here are similar to those seen in a series of 1118 patients with early-stage lung cancers (20% for EGFR and 25% for KRAS).39 Among 1017 cases, we characterized genetic aberrations across 10 genes in 72% of tumors and identified an oncogenic driver in 62%. The ability to carry out characterization of multiple genes in this 14-institution study compares well with 2 different single-institutional studies in which 6 or more genes could be characterized in 90% of the patients and driver mutations identified in more than half of the patients.11,14
The LCMC investigators tracked patients with oncogenic drivers and demonstrated that the majority of patients with EGFR mutations were treated with erlotinib or other EGFR TKIs and those with ALK rearrangements were treated with crizotinib.11,12 Patients with ERBB2 or MET were placed on targeted trials. The 38% of patients with drivers treated on genomically-driven trials was similar to the 29% to 41% observed in single-institution experiences.11,14 This percentage will grow as multiplexed genomic testing expands and more therapies for patients with tumors harboring KRAS mutations, the most common driver, become available.41
Our study findings should be interpreted with consideration of several limitations. As this was not a randomized study designed to compare survival in patients with a tumor with an oncogenic driver based on whether or not they received a targeted therapy, our observations should be considered a proof of concept rather than a definitive result. The study design is not appropriate to reach definitive conclusions about survival differences being attributable to the determination and use of oncogenic drivers. Because treatment was not randomly assigned and was at the discretion of the treating physician, there is the potential to introduce bias. Patients in the driver + treatment group had a 5% better performance status, a 3-year lower median age, and were more likely to be never smokers (Table 1); factors that could lead to an improved prognosis. The propensity score analysis (including sex, age at enrollment, performance status, smoking history, stage at diagnosis, prior therapy, and elapsed time from metastatic disease diagnosis to study enrollment) may not have adequately ruled out confounding, due to other confounding factors—known or unknown—not included in the model. Twenty-eight percent of patients enrolled were ineligible due to insufficient tissue because, when this project began, biopsies were performed solely to establish a diagnosis. This situation has changed substantially now that obtaining tissue to test for EGFR8,15 and ALK8 is part of treatment guidelines. Circumstances in which tumor genotyping is recommended for patients with squamous cell lung cancers is now part of guidelines as well.8 The median survival of persons on this trial is longer than is typically observed, allowing for treatment in a targeted-agent trial, often after chemotherapy. We calculated all survival data from the date of diagnosis of metastatic disease. Although the LCMC enrolled individuals with characteristics often associated with actionable drivers amenable to treatment with targeted drugs, KRAS mutations were the most common drivers detected, and observed survival differences remained significant in the propensity analysis. Although it is possible that improvements in survival are weighted by patients with EGFR-mutant and ALK-positive lung cancers treated with kinase inhibitors, survival analyses excluding these patients revealed that individuals with other drivers treated with targeted therapies still had an observed 2.5-year improvement in median survival over those who did not receive targeted therapy (eFigure 4 in the Supplement).
Conclusions
Although the frequency of any individual oncogenic driver may be small, an actionable driver was detected in 64% of tumors from patients with lung adenocarcinomas. Multiplexed testing aided physicians in selecting therapies and patients for targeted trials. Although individuals with drivers receiving a matched targeted agent lived longer, randomized clinical trials are required to determine if selecting targeted therapy based on oncogenic drivers improves survival.
Supplementary Material
Acknowledgments
Funding/Support: This study was entirely supported by a grant from the National Institutes of Health, National Cancer Institute (HSS NIH NCI 1RC2CA148394-010).
Role of the Sponsor: The sponsor had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Dr Kris had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kris, Johnson, Kwiatkowski, Iafrate, Wistuba, Franklin, Aronson, Camidge, Khuri, Pao, Ladanyi, Kugler, Minna, Bunn.
Acquisition, analysis, or interpretation of data: Kris, Johnson, Berry, Kwiatkowski, Iafrate, Wistuba, Varella-Garcia, Franklin, Su, Shyr, Camidge, Sequist, Glisson, Khuri, Garon, Pao, Rudin, Schiller, Haura, Socinski, Shirai, Giaccone, Chen, Ladanyi, Kugler, Minna, Bunn.
Drafting of the manuscript: Kris, Johnson, Varella-Garcia, Shyr, Camidge, Pao, Socinski, Shirai, Giaccone, Chen, Kugler, Minna, Bunn.
Critical revision of the manuscript for important intellectual content: Kris, Johnson, Berry, Kwiatkowski, Iafrate, Wistuba, Varella-Garcia, Franklin, Aronson, Su, Camidge, Sequist, Glisson, Khuri, Garon, Pao, Rudin, Schiller, Haura, Giaccone, Ladanyi, Minna, Bunn.
Statistical analysis: Kris, Johnson, Berry, Su, Shyr, Chen.
Obtained funding: Kris, Kwiatkowski, Wistuba, Franklin, Khuri, Pao, Kugler, Bunn.
Administrative, technical, or material support: Kris, Berry, Kwiatkowski, Iafrate, Wistuba, Varella-Garcia, Franklin, Aronson, Camidge, Khuri, Garon, Pao, Rudin, Haura, Socinski, Giaccone, Ladanyi, Kugler, Bunn.
Study supervision: Kris, Kwiatkowski, Iafrate, Varella-Garcia, Franklin, Camidge, Khuri, Rudin, Socinski, Ladanyi, Minna, Bunn.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Kris reports consulting for Ariad, AstraZeneca, Bind Biosciences, Boehringer Ingelheim, Chugai Pharma, Clovis, Covidien, Daiichi Sankyo, Esanex, Exelixis, and Genentech; receiving grant funding from Boehringer Ingelheim, the National Lung Cancer Partnership (NLCP), Pfizer, PUMA, and Stand up to Cancer (Massachusetts General Hospital); and receiving payments for lectures from Boehringer Ingelheim, Novartis, Millenium, Pfizer, Roche, China, and Roche, Italy. Dr Johnson reports consulting for AstraZeneca and Genentech; holding patents for EGFR mutation testing as indication for EGFR TKI therapy; and receiving royalties from Dana-Farber Cancer Institute for EGFR mutation testing. Dr Berry reports receiving grant funding from NLCP. Dr Kwiatkowski reports consulting for Novartis. Dr Iafrate reports consulting for Bioreference Labs and Pfizer and receiving royalties from Bioreference Labs for licensed SNaPshot technology. Dr Wistuba reports consulting for Boehringer Ingelheim, GE, Genentech/Roche, GlaxoSmithKline, Novartis, Roche, Pfizer, and Ventana; receiving grant funding from AstraZeneca, Bayer, Genentech, Merck, Myriad, and Pfizer; receiving lecture payments from Boehringer Ingelheim and Medscape; and receiving payments for developing educational presentations from Pfizer. Dr Varella-Garcia reports receiving honoraria, research funding, and payments for developing educational presentations from Abbott Molecular. Mr Aronson reports receiving grant funding from the Lung Cancer Mutation Consortium to her institution; receiving lecture fees from Leerink Swann; his institution, Partners HealthCare, licensed the GeneInsight technology in November 2012 to a company in which it acquired 100% equity ownership. This transaction was reviewed by the Partners Committee on Conflicts of Interest in light of Partners acquisition of this financial interest, and the Committee, consistent with Partners policy, required that notice of Partners financial interest in the technology be provided to journals and in publications and presentations on the technology. Partners established a partnership with Illumina related to the GeneInsight software that was in effect during this work. Dr Sequist reports consulting for AstraZeneca, Boehringer Ingelheim, Clovis Oncology, GlaxoSmithKline, and Merrimack Pharmaceuticals. Dr Garon reports consulting for Boehringer Ingelheim and receiving grant funding from AstraZeneca, Eli Lilly, Genentech, Novartis, Pfizer, and Puma Biotechnology. Dr Pao reports consulting for AstraZeneca, Bristol-Myers Squibb, Champions, Clarient, Clovis Oncology, Evolution, Exelixis, MolecularMD, and Symphony; receiving research funding from AstraZeneca, Bristol-Myers Squibb, Clovis Oncology, Enzon, Symphogen, and Xcovery; receiving payment for the development of educational presentations for the International Association for the Study of Lung Cancer and WebMD; serving on panels and as a grant reviewer for the American Association for Cancer Research, Uniting Against Lung Cancer, the American Society of Clinical Oncology, and the National Comprehensive Cancer Network; and receiving remuneration and patent royalties from rights to EGFR T790M testing was licensed on behalf of himself and others by Memorial Sloan Kettering Cancer Center to MolecularMD. Dr Rudin reports consulting for Celgene and Oncothyreon. Dr Schiller reports consulting for AdventRX, Aggenix, Ariad, Arquile, AVEO, Biodesix, Boehringer Ingelheim, Clovis, Dekkun, EMD Serono, Genentech, GlaxoSmithKline, Merck, Novartis, Peregrine, Pfizer, Synta, and Threshold Pharmaceuticals; and receiving grant funding from Astex, Endocyte, Genentech, Geron, Merrimack, Novartis, and Synta. Dr Shirai reports receiving lecture fees from Bristol-Myers Squibb. Dr Ladanyi reports consulting for NanoString, Novartis, Puma Biotechnology and receiving lecture payments from Remedica Medical Education. Dr Minna reports consulting for Amgen; receiving royalties from the National Institutes of Health (NIH) and Genentech for the licensing of lung cancer cell lines; serving on the grant review board of the V Foundation; receiving travel and accommodations from the International Association for the Study of Lung Cancer; and holding NIH grants related to lung cancer. Dr Bunn reports consulting for Amgen, Astellas Pharma, AstraZeneca,Bayer,BoehringerIngelheim, Bristol-Myers Squibb, Celgene, Daiichi Sankyo, Eisai, Eli Lilly, GlaxoSmithKline, Merck, Merck Serono, Merrimack, Myriad Genetics, Pfizer, Roche/Genentech, sanofi-aventis, and Synta. No other disclosures are reported.
Previous Presentation: Presented in part at the 41st annual meeting of the American Society of Clinical Oncology; May 31, 2013, to June 4, 2013; Chicago, Illinois.
TRIAL REGISTRATION clinicaltrials.gov Identifier: NCT01014286
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Girard N. New driver mutations in non-small cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 3.Clinical Lung Cancer Genome Project, Network Genomic Medicine A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5(209):209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad SciUS A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Comprehensive Cancer Network NCCN clinical practice guidelines in oncology. 2014 http://www.nccn.org/professionals/physician_gls/f_guidelines.asp. Accessed April 21.
- 9.Atherly AJ, Camidge DR. The cost-effectiveness of screening lung cancer patients for targeted drug sensitivity markers. Br J Cancer. 2012;106(6):1100–1106. doi: 10.1038/bjc.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pao W, Kris MG, Iafrate AJ, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15(17):5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 11.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small cell lung cancer. J Mol Diagn. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardarella S, Ortiz TM, Joshi VA, et al. The introduction of systematic genomic testing for patients with non-small cell lung cancer. J Thorac Oncol. 2012;7(12):1767–1774. doi: 10.1097/JTO.0b013e3182745bcb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29(15):2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 16.Rami-Porta R, Ball D, Crowley J, et al. International Staging Committee; Cancer Research and Biostatistics; Observers to the Committee; Participating Institutions The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2007;2(7):593–602. doi: 10.1097/JTO.0b013e31807a2f81. [DOI] [PubMed] [Google Scholar]
- 17.Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18(18):4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012;11(2):485–491. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohashi K, Sequist LV, Arcila ME, et al. Characteristics of lung cancers harboring NRAS mutations. Clin Cancer Res. 2013;19(9):2584–2591. doi: 10.1158/1078-0432.CCR-12-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dziadziuszko R, Wynes MW, Singh S, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol. 2012;7(2):340–347. doi: 10.1097/JTO.0b013e318240ca0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 25.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13(10):1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32(5):532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcila MEKN, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12(2):220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8(2):179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366(9496):1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 30.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 31.Cappuzzo F, Ciuleanu T, Stelmakh L, et al. SATURN investigators Erlotinib as maintenance treatment in advanced non-small cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol. 2010;11(6):521–529. doi: 10.1016/S1470-2045(10)70112-1. [DOI] [PubMed] [Google Scholar]
- 32.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barletta JA, Perner S, Iafrate AJ, et al. Clinical significance of TTF-1 protein expression and TTF-1 gene amplification in lung adenocarcinoma. J Cell Mol Med. 2009;13(8B):1977–1986. doi: 10.1111/j.1582-4934.2008.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [published online September 27, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Angelo SP, Janjigian YY, Ahye N, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7(12):1815–1822. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 40.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small cell lung cancer. Nat Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant KRAS G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.