Abstract
Background
Landsteiner-Wiener (LW) is the human blood group system no. 16, which comprises 2 antithetical antigens, LWa and LWb and the high prevalence antigen LWab. LW is encoded by the Intracellular Adhesion Molecule 4 (ICAM4) gene. The ICAM4 protein is part of the Rhesus complex in the red cell membrane and is involved in cell-cell adhesion.
Methods
We developed a method to sequence the whole 1.9 kb ICAM4 gene from genomic DNA in 1 amplicon. We determined the nucleotide sequence of exons 1 to 3, the 2 introns and 402 bp 5′-UTR and 347 bp 3′-UTR in 97 Caucasian and 91 African American individuals.
Results
Seven variant ICAM4 alleles were found, distinct from the wild type ICAM4 allele (GenBank KF712272), known as LW*05 and encoding LWa. An effect of the LWa/LWb amino acid substitution on the protein structure was predicted by 2 of the 3 computational modeling programs used.
Conclusions
We describe a practical approach for sequencing and determining the ICAM4 alleles using genomic DNA. LW*05 is the ancestral allele, which had also been observed in a Neandertal sample. All 7 variant alleles are immediate derivatives of the prevalent LW*05 and caused by 1 single nucleotide polymorphism (SNP) in each allele. Our data were consistent with the NHLBI GO Exome Sequencing Project (ESP) and the dbSNP databases, as all SNPs had been observed before. Our study has the advantage over the other databases in that it adds haplotype (allele) information for the ICAM4 gene, clinically relevant in the field of transfusion medicine.
Introduction
Blood group systems are determined by the presence or absence of specific antigens (proteins, carbohydrates, glycoproteins, or glycolipids) on the surface of red blood cells (RBC). Transfusions incompatible for blood group antigens can lead to life-threatening clinical complications. To date, 33 blood group systems are identified in humans 1. Landsteiner-Wiener (LW), discovered in 1940, is the 16th blood group system and consists of two antithetical antigens, LWa and LWb, and the high prevalence antigen LWab 2,3. The molecular basis of LWa/LWb antigen polymorphism is an A>G change at nucleotide 299 resulting in a Gln100Arg (Q100R) amino acid substitution 4. The LW antigens are more strongly expressed on D antigen positive than D antigen negative RBCs 5 and are absent on Rhnull cells 6. Alloanti-LWa and -LWb have been associated with mild hemolytic transfusion reactions (HTR) 7–10, while alloanti-LWa, -LWb and -LWab are associated with mild hemolytic disease of the fetus and newborn (HDFN) 8,11. Beside the 3 alloantibodies, autoantibodies against the 3 LW antigens are common in individuals with warm type autoimmune hemolytic anemia (AIHA) 2,12 and autoanti-LWa has been reported in a case of HDFN 13.
The LW antigens reside on a 42 kilodalton (kDa) RBC membrane glycoprotein known as LW or intercellular adhesion molecule 4 (ICAM4) 14. The ICAM4 glycoprotein is expressed on RBCs, erythroid precursor cells and on other blood cells including T and B cells 15. It belongs to the immunoglobulin superfamily (IgSF) consisting of five members designated ICAM1 to ICAM5 16. The ICAM4 protein is part of the Rh macromolecular complex consisting of Rh polypeptides (RhD and RhCE) and the Rh-associated glycoprotein (RhAG). In addition CD47, glycoprotein B (GPB) and Duffy glycoproteins (FY) interact with the complex by non-covalent bonds 17. The ICAM4 protein is known to maintain close contact between the RBC surface and vascular endothelium and also plays a role in various normal and pathological conditions, including erythropoiesis and microvascular occlusions during painful crises of sickle cell disease (SCD), respectively 12,14,18–23.
An allele, broadly defined as a haplotype, is one of a number of alternative forms of the same gene or genetic locus. A comprehensive population based collation of ICAM4 alleles and their protein products was missing, because the online databases such as dbSNP 24 and NHLBI GO Exome Sequencing Project (ESP) 25 lack haplotype (allele) and population data. In this study, we sequenced the whole ICAM4 gene in 97 Caucasians and 91 African American individuals to identify alleles defined by nucleotide variations in the full length of the gene.
Materials and Methods
Blood samples
EDTA blood samples were drawn from blood donors at the NIH Blood Bank and genomic DNA was isolated from the buffy coat (Qiagen EZ1 DNA blood kit on the BioRobot EZ1; Qiagen, Valencia, CA).
Primers
The primers were designed using the online version of Primer3 26. Primers LWF (5′-GCAACATTGCCCAGACTTCC-3′) and LWR (5′-TCCTCCGAAGAAGGGCAGTA-3′) were used for the amplification and primers LWF, LWR, LW1R (5′-CCAGGCTTTCGGAATAGATG-3′) and LW2R (5′-CCACCACACCAGGCTAATTT-3′) were used for sequencing.
ICAM4 gene amplification
One amplification reaction (total volume 25 μl) covering the complete ICAM4 gene (Chr.19:10,397,239-10,399,246 on NCBI Build GRCh37/hg19; 2008 bp) contained 25 ng genomic DNA, 2x Master Mix (OneTaq Hot Start; New England Biolabs, Beverly, MA), 10 μM each forward and reverse primers, and nuclease free water. Thermocycling conditions were 94 °C for 30 sec; 30 cycles of 94 °C for 30 sec, 61 °C for 1 min, 68 °C for 3 min; and a final extension at 68 °C for 5 min (Bio-Rad C1000; Bio-Rad, Hercules, CA). The 2008 bp PCR amplicon was cleaned and eluted in a 20 μl volume (QIAquick PCR purification kit; Qiagen).
ICAM4 nucleotide sequencing
Four sequencing reactions (total volume 20 μl each; (Chr.19:10,397,278-10,399,197 on NCBI Build GRCh37/hg19; 1920 bp) contained 2.5 μl PCR product, 1.8 μl Master Mix (BigDye Terminator v3.1; Applied Biosystems, Carlsbad, CA), 1.25 μl of 10 μM sequencing primer (LWF, LWR, LW1R or LW2R), and nuclease free water. Thermocycling conditions were: 25 cycles of 96 °C for 15 sec, 58 °C for 10 sec, and 60 °C for 4 min. Unincorporated dye was removed (DyeEx 96 well plates; Qiagen), sequencing reaction products were dehydrated (Savant SPD 2010 SpeedVac Concentrator; ThermoScientific, Wilmington, DE), suspended in 10 μl formamide (Hi-Di; Applied Biosystems) and analyzed (3500xL Genetic Analyzer; Applied Biosystems). Nucleotide sequences were aligned (CodonCode Aligner; CodonCode, Dedham, MA) to NCBI RefSeq NG_007728.1 and nucleotide positions defined using the first nucleotide of the coding sequence (CDS) of NM_001039132.2 (ICAM4 isoform 3).
Phylogenetic tree
The topologic associations between various ICAM4 alleles were analyzed using Neighbor-Joining method (CodonCode Aligner).27 Each variation was counted as 1 event. The ICAM4 sequence from chimpanzee (Pan troglodytes, NC_006486.3) was used for external rooting as previously described 28.
Neandertal genome
The published DNA sequence of the Neandertal genome (www.eva.mpg.de/neandertal/; Altai, Southern Siberia)29 was analyzed using Integrative genomics viewer version 2.3.20 30 and aligned to the human genome (NCBI Build GRCh37/hg19). The other 5 Neandertal fossil genomes lacked nucleotides sequences for ICAM4 and were excluded from analysis 31–33.
Protein structure
Representative models of the 3 ICAM4 protein isoforms were predicted with TMHMM2.0,34 Phobius,35 InterProScan36 and SOSUI37 (using default settings) and were based on Isoform Long (1) as described in UniProt38 and AceView39 databases.
Computational modeling of amino acid substitutions
Polymorphism Phenotyping algorithm (PolyPhen-2) 40, Sorting Tolerant From Intolerant (SIFT) 41 and Protein Variation Effect Analyzer (PROVEAN) 42 were used to predict the functional impact of amino acid substitutions.
Statistical description
95% confidence intervals (CI) for allele frequencies were calculated using the Poisson distribution (Statistics Calculators, online). The Fisher’s exact test was performed to compare the allele frequency distributions between Caucasians and African Americans; because of the multiple testing, we applied the Bonferroni multiple comparisons correction.
Results
A method was developed to amplify one stretch of 2,008 nucleotides encompassing the whole ICAM4 gene. We applied the method to determine the full length ICAM4 nucleotide sequence in a survey of 97 Caucasian and 91 African American blood samples.
ICAM4 alleles
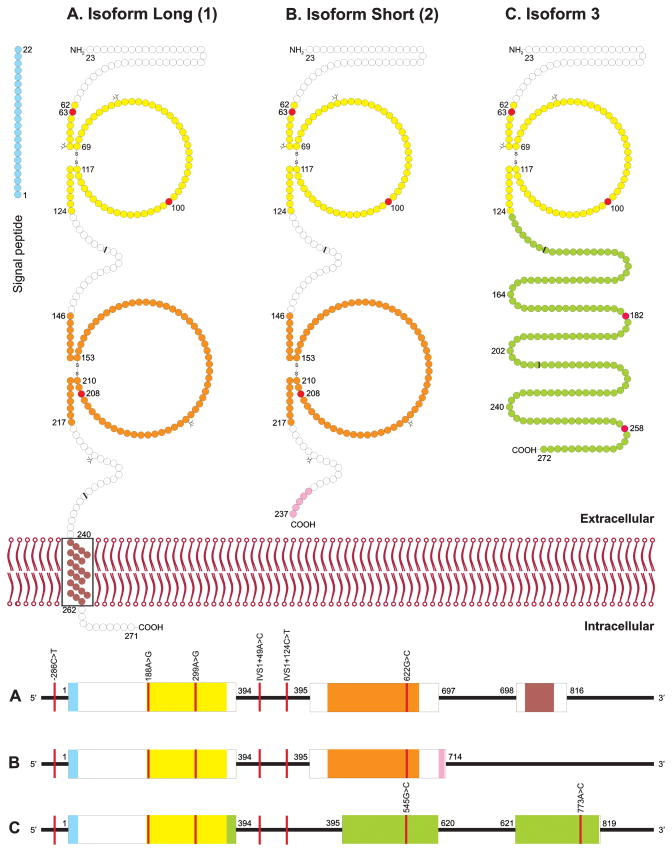
We observed a total of 8 ICAM4 alleles including the wild type allele KF712272 (Table 1). Among the 6 previously identified alleles, 1 allele carried the single nucleotide polymorphism (SNP) in the promoter region (rs3093030), 2 in intron 1 (rs34385135, rs35165411) and 3 in the exons (rs150654072, rs77493670, rs36023325) (Table S1). Another allele with the non-synonymous variation c.773A>C (p.Lys258Thr) was not documented in the dbSNP database at the time of detection but added since (rs201399464). The amino acid positions differ in the 3 isoforms for two of the 4 exonic variants (Fig. 1 and Table S2).
Table 1.
ICAM4 alleles identified in the present study
| Allele † | ISBT name | Nucleotide substitution (position) *
|
||||||
|---|---|---|---|---|---|---|---|---|
| 5′UTR
|
Exon 1
|
Intron 1 (IVS1)
|
Exon 2
|
Exon 3
|
||||
| -286C>T | c.188A>G (p.Lys63Arg) | c.299A>G (p.Gln100Arg) | IVS1+49A>C | IVS1+124C>T | c.545G>C (p.Arg182Pro) | c.773A>C (p.Lys258Thr) | ||
| KF712272 | LW*05 | C | A | A | A | C | G | A |
| KF725837 | NA | T | A | A | A | C | G | A |
| KF725836 | NA | C | G | A | A | C | G | A |
| KF725831 | LW*07 | C | A | G | A | C | G | A |
| KF725834 | NA | C | A | A | C | C | G | A |
| KF725835 | NA | C | A | A | A | T | G | A |
| KF725832 | NA | C | A | A | A | C | C | A |
| KF725833 | NA | C | A | A | A | C | G | C |
relative to NCBI Reference Sequence NG_007728.1 within the 1920 nucleotides of the ICAM4 gene. Numbering is according to ICAM4 Isoform 3 (NM_001039132.2, NP_001034221.1). Variant nucleotides are in bold
GenBank nucleotide database accession number
NA – Not applicable, because LW serology was not confirmed
Figure 1. ICAM4 protein and ICAM4 pre-mRNA.
Schematic models of ICAM4 protein are depicted for the 3 known isoforms (upper panel). Variations were found at 3 amino acid positions (red circles). The first 22 amino acid positions are predicted to be a signal peptide (blue circles). Additional predicted structural features are the 2 immunoglobulin (Ig) domains (yellow and orange circles) and a transmembrane segment (brown circles). Isoform Long (1) is a single-pass transmembrane protein (A), while Isoform Short (2) and Isoform 3 are secreted (B and C). Some protein segments in isoforms 2 and 3 differ from isoform 1 (purple, pink and green circles). The projections on circle surfaces denote the positions of the 4 N-glycosylation sites. The 7 variant nucleotide positions are depicted in the 3 pre-mRNA ICAM4 isoforms (bottom panel). Three variants were found in the exons (boxes) and 4 in the introns (lines). The nucleotide stretches of the exons are colored according to the encoded protein segments. The exon boundaries in the ICAM4 cDNA, as reflected in the amino acid sequence, are indicated (black bars).
Two African American individuals were found to harbor 2 different SNPs each (Table S1, patterns 9 and 10): -286C>T/c.299A>G and -286C>T/c.545G>C. Using allele specific PCRs, we determined that the 2 SNPs in each individual were caused by the heterozygous occurrence of 2 known alleles (KF725837/KF725831 and KF725837/KF725832). Hence, no new allele was found in our study. A search returned no ICAM4 alleles that differed from the currently described alleles or the LW*05N.01 null allele43 in the GenBank database (updates inclusive of 2013-11-06).
Population frequencies
Based on the number of analyzed samples, we calculated the variant frequency and allele frequency and its 95% confidence interval (CI) in our cohort and compared it with the data from NHLBI GO Exome Sequencing Project (ESP) 25 (Table 2 and Table S3).
Table 2.
ICAM4 allele frequencies in 97 Caucasian and 91 African American blood donors
| Allele* | Allele frequencies in the population
|
|||||
|---|---|---|---|---|---|---|
| Caucasian
|
African American
|
|||||
| Observed (n) | Mean † | 95% CI † | Observed (n) | Mean † | 95% CI ‡ | |
| KF712272 | 120¶ | 62% | 24.5% – 41.0% | 147¶ | 81% | 35.3% – 55.3% |
| KF725837 | 70¶ | 36% | 13% – 25.5% | 26¶ | 14% | 4.2% – 12.9% |
| KF725836 | 0 | 0% | 0% – 1.9% | 1 | 0.55% | 0.01% – 3.1% |
| KF725831 | 0 | 0% | 0% – 1.9% | 1 | 0.55% | 0.01% – 3.1% |
| KF725834 | 1 | 0.52% | 0.01% – 2.9% | 0 | 0% | 0% – 2% |
| KF725835 | 2 | 1% | 0.01% – 2.9% | 0 | 0% | 0% – 2% |
| KF725832 | 0¶ | 0% | 0% – 1.9% | 7¶ | 4% | 0.6% – 5.6% |
| KF725833 | 1 | 0.52% | 0% – 1.9% | 0 | 0% | 0% – 2% |
| Total | 194 | 182 | ||||
GenBank nucleotide database accession number
Number of observed alleles (n)/Total number of alleles
95% confidence interval (CI), Poisson distribution, two sided
Statistically significant difference by the Fisher’s exact test, two sided (p < 0.016); Bonferroni multiple comparison correction, n = 3, 0.05/3 = 0.016.
Phylogenetic tree of ICAM4 alleles
Using the ICAM4 sequence of chimpanzee (NC_006486.3) 44 for external rooting, we found the wild type ICAM4 allele (KF712272) to be the ancestral allele from which all the other alleles (KF725831 to KF725837) derived (Figure 2). The only other LW allele listed by the International Society of Blood Transfusion (ISBT) Working Party on Red Cell Immunogenetics and Blood Group Terminology 45 is LW*05N.01 (not shown, because the whole ICAM4 allele sequence was not determined).
Figure 2. Phylogeny of ICAM4 alleles in humans.

A phylogenetic tree of ICAM4 is shown for the 8 alleles found in this study using the ICAM4 sequence from Pan troglodytes (NC_006486.3) for external rooting. Clustering of the described ICAM4 alleles is based on Neighbor-Joining method. For each evolutionary step, the event is indicated; depicted distances of the alleles are arbitrary.
ICAM4 alleles in a Neandertal sample
We analyzed the 99.9% complete Neanderthal genome (Altai, Southern Siberia; 50-fold coverage) for the ICAM4 sequence (Table 3). The wild type ICAM4 allele (KF712272) identified in our study was also present in the Neandertal genome without any variation in the 1920 bp nucleotides sequenced.
Table 3.
Comparison of the ICAM4 genes in human, Neandertal and chimpanzee genomes
| Species | Nucleotide position*
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ UTR | Exon 1 | Intron 1 (IVS1) | Exon 2 | Intron 2 (IVS2) | Exon 3 | |||||||||||||||||
|
| ||||||||||||||||||||||
| −286 | −170 | 97 | 129 | 188 | 261 | 299 | +49 | +98 | +124 | 545 | +40 | +41 | +52 | 629 | 654 | 773 | *55 | *111 | *116 | *121 | *363 | |
| H. sapiens* | C | T | A | G | A | A | A | A | G | C | G | G | G | T | C | G | A | G | C | T | T | G |
| Neandertal† | C | T | A | G | A | A | A | A | G | C | G | G | G | T | C | G | A | G | C | T | T | G |
| P. troglodyte† | NA | C | G | A | A | G | A | A | A | C | G | C | del | del | T | T | A | A | A | G | G | C |
nucleotide positions relative to NCBI Reference Sequence NM_001039132.2 and NP_001034221.1 in the human nucleotide sequence
nucleotide sequences from http://cdna.eva.mpg.de/neandertal/altai/AltaiNeandertal/bam/ (Neandertal) and NCBI Reference Sequence NC_006486.3 (chimpanzee)
NA – data not available
del – nucleotide deleted
Effect on protein structure
The Lys63Arg (rs150654072) and Gln100Arg (rs77493670) amino acid substitutions were located in the first Ig domain of all 3 isoforms (Fig. 1). The SNP rs36023325 encoded a Val208Leu in the second Ig domain of Isoform Long (1) and Isoform Short (2), while this SNP encoded an Arg182Pro in Isoform 3. The SNP rs201399464 encoded Lys258Thr in Isoform 3 only, as it did not occur in the cDNA of Isoform Long (1) and Isoform Short (2) (Fig. 1).
Computational modeling using PolyPhen-2 and SIFT predicted structural changes induced by the Gln100Arg substitution (Table 4), which PROVEAN did not predict. However, the only 3 missense variations occurring between chimpanzee and humans (Table 3: c.97A>G, Ser33Gly; c.706C>T, Pro236Ser; and c.731G>T, Gly244Val) were consistently predicted to be “benign”, “tolerated” and “neutral” by PolyPhen, SIFT and PROVEAN, respectively (data not shown).
Table 4.
Amino acid substitution and predicted effect on protein structure
| Variant | Protein | Bioinformatics program and computational analysis results
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PolyPhen-2 † | SIFT ‡ | PROVEAN ¶ | ||||||||
|
|
|
|
|
|
||||||
| Allele | dbSNP reference no. | Isoform | Amino acid substitution* | Classification | Score | Classification | Score | MIC | Classification | Score |
| KF725836 | rs150654072 | 1 | Lys63Arg | benign | 0.000 | tolerated | 0.11 | 1.97 | neutral | −0.147 |
| 2 | Lys63Arg | benign | 0.001 | tolerated | 0.2 | 2.74 | neutral | −0.147 | ||
| 3 | Lys63Arg | benign | 0.002 | tolerated | 0.09 | 3.20 | neutral | −0.339 | ||
| KF725831 | rs77493670 | 1 | Gln100Arg | possibly damaging | 0.95 | tolerated | 0.54 | 1.95 | neutral | −1.902 |
| 2 | Gln100Arg | probably damaging | 0.97 | damaging | 0.04 | 2.74 | neutral | −1.922 | ||
| 3 | Gln100Arg | probably damaging | 0.99 | tolerated | 0.21 | 3.20 | neutral | −1.869 | ||
| KF725832 | rs36023325 | 1 | Val208Leu | benign | 0.001 | tolerated | 0.77 | 1.97 | neutral | 0.333 |
| 2 | Val208Leu | benign | 0.002 | tolerated | 0.33 | 2.74 | neutral | 0.386 | ||
| 3 | Arg182Pro | benign | 0.000 | damaging | 0.01 | 3.46 | neutral | −1.261 | ||
| KF725833 | rs201399464 | 3 | Lys258Thr | benign | 0.053 | damaging | 0 | 4.32 | neutral | −0.412 |
relative to NCBI Reference Sequence NP_001034221.1
score 0.00–0.452 = benign, 0.453–0.956 = possibly damaging, 0.957–1.00 = probably damaging
score ≤0.05 = damaging, >0.05 = tolerated; MIC = median sequence information (range 0 to 4.32)
score >−2.5 = neutral, score ≤−2.5 = deleterious
Discussion
We presented a practical approach for sequencing the ICAM4 gene and determining the ICAM4 alleles using genomic DNA. In 188 Caucasian and African American blood donors, we identified a total of 8 ICAM4 alleles, including nucleotide substitutions in the promoter and intron 1 (Table 1). The allele frequencies differed significantly between the Caucasian and African American populations indicating that the overall profile of genetic differences in the ICAM4 gene may be distinguishable.
Based on population frequency (Table 2) and published data 46, the LW*05 and LW*07 alleles are known to encode the LWa and LWb antigen, respectively. It is likely that all other 6 alleles (Table 1) express the LWa antigen, which can eventually be documented. These alleles can also be analyzed for their role in the expression of the ICAM4 protein and its effect on RhD protein expression.
The ICAM4 glycoprotein is encoded by the ICAM4 gene located on chromosome 19p13.3 43,46. Three mRNA isoforms are transcribed from the ICAM4 gene, which are translated into 3 different proteins (Fig. 1, upper panel). Two of the 3 proteins are soluble (Table 5) 38. Isoform Short (2) has 2 exons, while Isoform Long (1) and Isoform 3 have 3 exons (Fig. 1, lower panel). In the common Isoform Long (1) coding for the membrane bound protein, exon 1 encodes the 5′ untranslated region (UTR) including the 22 amino acid signal peptide 47 and the first immunoglobulin superfamily (IgSF) domain (Fig. 1). Exons 1 and 2 are separated by intron 1, an intervening sequence (IVS), of 129 bp length and exon 2 encodes the second immunoglobulin superfamily (IgSF) domain. Exons 2 and 3 are separated by intron 2 of 147 bp length and exon 3 encodes the transmembrane domain, cytoplasmic tail and the 3′ UTR.
Table 5.
ICAM4 gene isoforms
| Isoform | Transcript
|
Protein
|
Supporting experimental evidence
|
|||||
|---|---|---|---|---|---|---|---|---|
| GenBank accession no. | mRNA (bp) | CDS* (bp) | Exons | Length (amino acids) | Predicted localization | Tissue | GenBank accession no.† | |
| Long (1) | NM_001544.4 | 1354 | 816 | 3 | 271 | membrane | lung lung, mucoepidermoid carcinoma embryonic stem cells colon tumor |
DA590748 and CA309669 BC029364 CN310683 AI916092 |
| Short (2) | NM_022377.3 | 1501 | 714 | 2 | 237 | secreted or extracellular | bone marrow | L27670 |
| 3 | NM_001039132.2 | 1277 | 819 | 3 | 272 | secreted or extracellular | blood choriocarcinoma | BU656201 BC000046 and BE278826 |
CDS – coding sequence
data according to http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ 39
We established the phylogeny of the observed 8 ICAM4 alleles (Fig. 2). LW*05, encoding the LWa antigen, is the primordial allele, from which all other infrequent alleles are derived by 1 distinct nucleotide substitution each (GenBank accession no. KF725831 to KF725837), including the LW*07 allele. This phylogenetic relationship among the 8 alleles may have been expected, because the LWb antigen (LW*07 allele) is rare (0.5% in Caucasians and 0.04% in African Americans) as compared to the LWa antigen (LW*05 allele).
The incomplete nucleotide sequence of the ICAM4 gene sequence in chimpanzee (Pan troglodyte) differed, where data were available, from the primordial human LW*05 allele by 15 nucleotide positions (Table 3). In contrast, these 15 nucleotide positions and the 7 variable positions identified in this study did not differ between humans and Neandertals (Table 3). The ICAM4 gene locus in the Neandertal individual and the human ICAM4 wild type allele (KF712272) were identical. LW*05 must have occurred in our common ancestors and may have been maintained by potential interbreeding. Because the human LW*05 allele sequence was homozygous in this individual (Table 3) who lived approximately 29,200 to 48,650 years ago 29 and has now been genotyped, we can conclude that this Neandertal carried the LW(a+b−) phenotype.
The prediction of an amino acid substitution to affect protein structure or clinical impact can be sensitive to the specific algorithm setting of a bioinformatics analysis program and to the specific alignment used 48, thus making it difficult to draw conclusions from any one tool alone 49. Hence, 3 common bioinformatics tools were applied to explore the impact of the 4 observed missense substitutions (Table 4). The PolyPhen-2 program classified the Gln100Arg variation, responsible for LWa/LWb antigen polymorphism, as “possibly” to “probably damaging” for all 3 isoforms; SIFT as “damaging” for isoform 2 only; and PROVEAN as “neutral” in all isoforms. Our results, in agreement with previous studies 50, suggest that the use of in silico platforms are not consistent enough to reliably predict the impact of amino acid substitution. Similarly, in vitro binding assays excluded protein misfolding caused by the Gln100Arg variation 22, despite its unequivocal clinical relevance effected by antibody binding. However, the only 3 missense variations occurring between chimpanzee and humans were predicted to have no structural effect, which may indicate, that the overall ICAM4 protein structure is quite conserved in the primate lineage.
We compared our results to the NHLBI GO Exome Sequencing Project (ESP) 25 and the dbSNP 24 and HapMap database 51. The ESP study identified 44 SNPs in the ICAM4 gene, of which 29 were in coding and 15 in noncoding gene segments. It documented 28 distinct SNPs in 4300 Caucasian and 24 in 2203 African American individuals. Five of these 44 SNPs (3 missense and 2 intronic) were also identified in the current study. With the exception of the LW*05 allele, all SNPs occurred with a frequencies of less than 2% (Table S3) and were explained by a combined effect of explosive, recent accelerated population growth and weak purifying selection 25.
The dbSNP database listed more than 170 SNPs, including those of the ESP study, in Isoform Long (1) of ICAM4 gene (NM_001544.4), most of them without any population frequency data 24. Because the genotypes deposited in the dbSNP database are unphased, no haplotype (allele) information was available. All 8 SNPs identified in the current study were listed in dbSNP. When analyzing the HapMap database 52, we did not find a significant linkage disequilibrium (LD) among our 8 SNPs. This lack of LD was compatible with our result that no more than 1 SNP occurred in any of the 376 alleles among the 188 individuals. Our study overcame shortfalls of the online databases, such as lack of haplotype and population data, by taking a systematic approach to sequencing the ICAM4 gene in a random sample of Caucasians and African Americans.
The ESP and dbSNP databases exemplify how allele identification is progressing rapidly by various initiatives independent of blood group research, yet still covering the genes of the 33 blood group systems 1. Hence, novel ICAM4 alleles are recognized almost monthly. For instance, the SNP underlying c.773A>C (p.Lys258Thr) was “novel”, when we identified the KF725833 structure in our study (Table 1), and has been independently submitted to the dbSNP database, while this manuscript was under preparation. Molecular immunohematology needs to be prepared and develop tools for utilizing the large datasets that are established without regard to blood group related information. We can contribute by establishing the alleles (haplotypes) and adding protein expression and antigen data, which are of clinical relevance.
Supplementary Material
Acknowledgments
We thank Harvey G Klein for critical review; Sherry L. Sheldon and Sharon D. Adams for sample coordination; Elizabeth J Furlong for English edits; and the staff of HLA laboratory in the Laboratory Services Section for DNA extraction.
Part of the work was done by Noorah Salman Almarry in fulfillment of the laboratory rotation (internship) in her master program of biochemistry and molecular biology at Georgetown University This research was supported by the Intramural Research Program of the NIH Clinical Center.
Footnotes
Conflict of interest: None.
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Web resources
Primer3 software, version 4.0.0 (http://bioinfo.ut.ee/primer3-0.4.0/)
International Society Blood Transfusion (ISBT) (http://www.isbtweb.org/)
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)
SIFT (http://sift.jcvi.org/www/SIFT_enst_submit.html)
PROVEAN (http://provean.jcvi.org/seq_submit.php)
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP; 09, 2013) http://evs.gs.washington.edu/EVS/)
dbSNP database, Build ID: 138 Phase I (http://www.ncbi.nlm.nih.gov/SNP/)
Statistics Calculators, version 3.0 beta (http://www.danielsoper.com/statcalc3/calc.aspx?id=86)
AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=ICAM4)
Max Planck Institute for Evolutionary Anthropology (http://www.eva.mpg.de/neandertal/)
International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/)
References
- 1.Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, de Haas M, Hyland C, Lomas-Francis C, Moulds JM, Nogues N, Olsson ML, Poole J, Reid ME, Rouger P, van der Schoot E, Scott M, Tani Y, Yu LC, Wendel S, Westhoff C, Yahalom V, Zelinski T. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Cancun Report (2012) (in press) Vox Sang. 2013 doi: 10.1111/vox.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandstaff Moulds MK. The LW blood group system: a review. Immunohematology. 2011;27:136–42. [PubMed] [Google Scholar]
- 3.Byrne KM, Byrne PC. Review: other blood group systems--Diego, Yt, Xg, Scianna, Dombrock, Colton, Landsteiner-Wiener, and Indian. Immunohematology. 2004;20:50–8. [PubMed] [Google Scholar]
- 4.Hermand P, Gane P, Mattei MG, Sistonen P, Cartron JP, Bailly P. Molecular basis and expression of the LWa/LWb blood group polymorphism. Blood. 1995;86:1590–4. [PubMed] [Google Scholar]
- 5.Gibbs MB. The Quantitative Relationship of the Rh-like (LW) and D Antigens of Human Erythrocytes. Nature. 1966;210:642–3. doi: 10.1038/210642a0. [DOI] [PubMed] [Google Scholar]
- 6.Goossens D, Trinh-Trang-Tan MM, Debbia M, Ripoche P, Vilela-Lamego C, Louache F, Vainchenker W, Colin Y, Cartron JP. Generation and characterisation of Rhd and Rhag null mice. Br J Haematol. 2010;148:161–72. doi: 10.1111/j.1365-2141.2009.07928.x. [DOI] [PubMed] [Google Scholar]
- 7.Napier JAF, Rowe GP. Transfusion Significance of LWa Allo-Antibodies. Vox Sang. 1987;53:228–30. doi: 10.1111/j.1423-0410.1987.tb05071.x. [DOI] [PubMed] [Google Scholar]
- 8.Giles CM. The LW Blood Group: A Review. Immunol Commun. 1980;9:225–42. doi: 10.3109/08820138009065996. [DOI] [PubMed] [Google Scholar]
- 9.Sistonen P, Nevanlinna HR, Virtaranta-Knowles K, Pirkola A, Leikola J, Kekomäki R, Gavin J, Tippett P. Nea, a New Blood Group Antigen in Finland. Vox Sang. 1981;40:352–7. doi: 10.1111/j.1423-0410.1981.tb00720.x. [DOI] [PubMed] [Google Scholar]
- 10.Sistonen P. PhD thesis. University of Helsinki; 1984. The LW (Landsteiner-Weiner) blood group system: elucidation of the genetics of the LW blood groups based on the finding of a ‘new’ blood group antigen. [Google Scholar]
- 11.DeVeber LL, Clark GW, Hunking M, Stroup M. Maternal Anti-LW. Transfusion. 1971;11:33–5. doi: 10.1111/j.1537-2995.1971.tb04372.x. [DOI] [PubMed] [Google Scholar]
- 12.Vos GH, Petz LD, Garratty G, Fudenberg HH. Autoantibodies in Acquired Hemolytic Anemia With Special Reference to the LW System. Blood. 1973;42:445–53. [PubMed] [Google Scholar]
- 13.Davies J, Day S, Milne A, Roy A, Simpson S. Haemolytic disease of the foetus and newborn caused by auto anti-LW. Transfus Med. 2009;19:218–9. doi: 10.1111/j.1365-3148.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- 14.Delahunty M, Zennadi R, Telen MJ. LW protein: a promiscuous integrin receptor activated by adrenergic signaling. Transfus Clin Biol. 2006;13:44–9. doi: 10.1016/j.tracli.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira OL, Thomas DB, Lomas CG, Tippett P. Restricted expression of LW antigen on subsets of human B and T lymphocytes. J Immunogenet. 1984;11:297–303. doi: 10.1111/j.1744-313x.1984.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 16.Bailly P, Hermand P, Callebaut I, Sonneborn HH, Khamlichi S, Mornon JP, Cartron JP. The LW blood group glycoprotein is homologous to intercellular adhesion molecules. Proc Natl Acad Sci U S A. 1994;91:5306–10. doi: 10.1073/pnas.91.12.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flegel WA, Von Zabern I, Doescher A, Wagner FF, Vytisková J, Písačka M. DCS-1, DCS-2, and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion. 2008;48:25–33. doi: 10.1111/j.1537-2995.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 18.Celano MJ, Levine P. Anti-LW Specificity in Autoimmune Acquired Hemolytic Anemia. Transfusion. 1967;7:265–8. doi: 10.1111/j.1537-2995.1967.tb05515.x. [DOI] [PubMed] [Google Scholar]
- 19.Zennadi R, Hines PC, De Castro LM, Cartron JP, Parise LV, Telen MJ. Epinephrine acts through erythroid signaling pathways to activate sickle cell adhesion to endothelium via LW-αvβ3 interactions. Blood. 2004;104:3774–81. doi: 10.1182/blood-2004-01-0042. [DOI] [PubMed] [Google Scholar]
- 20.Toivanen A, Ihanus E, Mattila M, Lutz HU, Gahmberg CG. Importance of molecular studies on major blood groups--Intercellular adhesion molecule-4, a blood group antigen involved in multiple cellular interactions. Biochim Biophys Acta. 2008;1780:456–66. doi: 10.1016/j.bbagen.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Ihanus E, Uotila LM, Toivanen A, Varis M, Gahmberg CG. Red-cell ICAM-4 is a ligand for the monocyte/macrophage integrin CD11c/CD18: characterization of the binding sites on ICAM-4. Blood. 2007;109:802–10. doi: 10.1182/blood-2006-04-014878. [DOI] [PubMed] [Google Scholar]
- 22.Hermand P, Huet M, Callebaut I, Gane P, Ihanus E, Gahmberg CG, Cartron JP, Bailly P. Binding Sites of Leukocyte β2 Integrins (LFA-1, Mac-1) on the Human ICAM-4/LW Blood Group Protein. J Biol Chem. 2000;275:26002–10. doi: 10.1074/jbc.M002823200. [DOI] [PubMed] [Google Scholar]
- 23.Mankelow TJ, Spring FA, Parsons SF, Brady RL, Mohandas N, Chasis JA, Anstee DJ. Identification of critical amino-acid residues on the erythroid intercellular adhesion molecule-4 (ICAM-4) mediating adhesion to αV integrins. Blood. 2004;103:1503–8. doi: 10.1182/blood-2003-08-2792. [DOI] [PubMed] [Google Scholar]
- 24.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, GOB, GOS Project obotNES. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Kumar S. Molecular Evolution and Phylogenetics. 1. Oxford University Press; USA: 2000. [Google Scholar]
- 28.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood. 2002;100:306–11. doi: 10.1182/blood-2002-01-0320. [DOI] [PubMed] [Google Scholar]
- 29.Mednikova MB. A proximal pedal phalanx of a Paleolithic hominin from denisova cave, Altai. Archaeology, Ethnology and Anthropology of Eurasia. 2011;39:129–38. [Google Scholar]
- 30.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotech. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, Hansen NF, Durand EY, Malaspinas A-S, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Höber B, Höffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Ž, Gušic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PLF, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Pääbo S. A Draft Sequence of the Neandertal Genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andrés AM, Eichler EE, Slatkin M, Reich D, Kelso J, Pääbo S. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinhasi R, Higham TF, Golovanova LV, Doronichev VB. Revised age of late Neanderthal occupation and the end of the Middle Paleolithic in the northern Caucasus. Proc Natl Acad Sci U S A. 2011;108:8611–6. doi: 10.1073/pnas.1018938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 35.Käll L, Krogh A, Sonnhammer ELL. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 36.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 38.The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D7. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Meth. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng PC, Henikoff S. Predicting Deleterious Amino Acid Substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermand P, Le Pennec PY, Rouger P, Cartron JP, Bailly P. Characterization of the gene encoding the human LW blood group protein in LW+ and LW− phenotypes. Blood. 1996;87:2962–7. [PubMed] [Google Scholar]
- 44.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 45.Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, Francis CL, Moulds JM, Moulds JJ, Olsson ML, Poole J, Reid ME, Rouger P, van der Schoot E, Scott M, Smart E, Tani Y, Yu LC, Wendel S, Westhoff C, Yahalom V, Zelinski T. International Society of Blood Transfusion Working Party on red cell immunogenetics and blood group terminology: Berlin report. Vox Sang. 2011;101:77–82. doi: 10.1111/j.1423-0410.2010.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermand P, Gane P, Mattei MG, Sistonen P, Cartron JP, Bailly P. Molecular basis and expression of the LWa/LWb blood group polymorphism. Blood. 1995;86:1590–4. [PubMed] [Google Scholar]
- 47.Molhoj M, Degan FD. Leader sequences are not signal peptides. Nat Biotech. 2004;22:1502. doi: 10.1038/nbt1204-1502. [DOI] [PubMed] [Google Scholar]
- 48.Williams S. Analysis of in silico tools for evaluating missense variants. National Genetics Reference Laboratory; Manchester: 2012. [Google Scholar]
- 49.Castellana S, Mazza T. Congruency in the prediction of pathogenic missense mutations: state-of-the-art web-based tools. Brief Bioinform. 2013;14:448–59. doi: 10.1093/bib/bbt013. [DOI] [PubMed] [Google Scholar]
- 50.Li M-X, Kwan JSH, Bao S-Y, Yang W, Ho S-L, Song Y-Q, Sham PC. Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies. PLoS Genet. 2013;9:e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.