Abstract
Hepatitis C virus (HCV) establishes frequently persistent infections. Chronic carriers can develop severe liver disease. HCV has been intensely studied in a variety of cell culture systems. However, commonly used cell lines and primary hepatocyte cultures do not or only in part recapitulate the intricate host environment HCV faces in the liver. HCV infects readily only humans and chimpanzees, which poses challenges in studying HCV infection in vivo. Consequently, tractable small animal models are needed that are not only suitable for analyzing HCV infection but also for testing novel therapeutics. Here, we will focus our discussion on humanized mice, i.e. mice engrafted with human tissues or expressing human genes, which support HCV infection. We will further highlight novel methods that can be used to unambiguously detect HCV infected cells in situ, thereby facilitating a spatio-temporal dissection of HCV infection in the three dimensional context of the liver.
Keywords: Hepatitis C virus, viral hepatitis, animal model, humanized mice, imaging
1. Hepatitis C and hepatitis C virus
At least 150 million people are chronically infected with hepatitis C virus (HCV). Only 20-30% of exposed individuals clear an HCV infection spontaneously while the majority becomes persistently infected. If untreated chronic carriers are at significant risk of developing severe liver disease including liver fibrosis and liver cirrhosis and development of hepatocellular carcinoma.
HCV was first identified as the etiologic agent for non-A non-B hepatitis, later classified as hepatitis C in 1989 (Choo et al., 1989). HCV is an enveloped, positive-strand RNA virus in the family Flaviviridae, containing a 9.6-kb RNA genome (Moradpour et al., 2007). Translation of the genome initiates at an internal ribosome entry site (IRES) in the 5’ UTR and produces a major 3,000-aa polyprotein, which is further cleaved into three structural proteins (core, E1 and E2) and seven nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) by cellular and viral proteases. HCV core protein can bind HCV genomic RNA to form the viral nucleocapsid, enveloped by glycoproteins E1 and E2 (Moradpour et al., 2007). The envelope proteins (E1 and E2) are responsible for mediating viral entry, followed by uncoating in the cytoplasm (reviewed in (Lindenbach and Rice, 2013)). p7 functions as the viral ion channel in planar lipid bilayers which plays an important role for HCV infection and therefore it is an attractive target for antiviral drug development (Griffin et al., 2003). The nonstructural protein 2 (NS2) is an autoprotease that catalyzes the cleavage at the NS2-3 junction, and NS3-4A serine protease processes the remainder of the nonstructural proteins. NS3/4A is not only vital for viral replication (Grakoui et al., 1993; Moradpour et al., 2007) but also cleaves multiple cellular targets (reviewed in (Morikawa et al., 2011)), that block the downstream interferon activation (Li et al., 2005; Meylan et al., 2005). NS4B is a highly hydrophobic protein that induces rearrangement of intracellular membranes to form a membranous web, which is the site of viral RNA replication (Tellinghuisen et al., 2007). NS5A is an RNA binding protein that plays an essential role in the RNA replication and virus assembly. Phosphorylation of NS5A appears to be the switch between viral replication and infectious virus production (Reed et al., 1997). NS5B is the viral RNA-dependent RNA polymerase (RdRp), which catalyzes the replication of the viral genome (Tellinghuisen et al., 2007; Lindenbach and Rice, 2013). Several HCV encoded proteins, that serve essential functions in the HCV lifecycle, including NS3/4A, NS5A and NS5B, are targets of choice for the development of anti-HCV drugs.
Since the first treatment attempts of chronic HCV with interferon alpha in the late 1980s (Schvarcz et al., 1989) significant progress has been made. Subsequently, a combination of pegylated interferon (IFN) alpha 2a or 2b and ribavirin (RBV) was the standard therapeutic regimen, until the approval of the first directly acting antivirals (DAAs) in 2011, which interfere specifically with the function of the viral NS3/4A protease (boceprevir and telaprevir). Combining peg-IFN and RBV with one of the first generation protease inhibitors increased significantly sustained virological response (SVR) rates. However, under this treatment regimen HCV frequently develops resistance leading to viral breakthrough. Furthermore, addition of the first generation DAA to the HCV therapy exacerbated previously observed substantial side effects including anemia and rash. Some of these issues have been addressed with the development and approval of second generation protease inhibitors (e.g. simeprevir) and a very potent, first in class inhibitor of the RdRP (sofosbuvir). Current clinical trials (reviewed in (Schinazi et al., 2014)) are focusing on the combination of different classes of DAA's to exploit synergetic effects and minimize the risk of emerging resistances. One of the largest interferon-free trials for treatment-naïve genotype 1 patients so far was the SOUND-C2 trial, that combined the NS3/4A inhibitor faldaprevir, the NS5B inhibitor deleobuvir and ribavirin. After 28 weeks of treatment 69% of the patients achieved a SVR (Zeuzem et al., 2013). Even more encouraging were the results of a recent once daily all-oral trial of the NS5A replication complex inhibitor daclatasvir in combination with sofosbuvir for 24 weeks. It showed SVR's of 100% for genotype 1 patients, even if they had a previous virologic failure under the treatment with boceprevir or telaprevir and interferon alpha plus ribavirin (Sulkowski et al., 2014).
The encouraging results of these trials suggest that in the near future, HCV treatment regimens will not only be interferon-free, but will also allow shorter duration of treatment and will have activity against a broad spectrum of genotypes. Finally, it seems that traditional predictors of treatment outcome like fibrosis score, prior treatments, IL-28B genotype become less and less important for treatment decisions.
While the advances in anti-HCV therapy are astounding their impact on containing the global HCV epidemic remains to be seen. The considerable costs of currently approved drug cocktails, along with inadequate infrastructure for medical supervision and distribution may diminish the impact of future therapies in resource poor environments. Consequently, development of vaccines, which may offer cost-effective therapeutic and prophylactic intervention options, remains critical to contain HCV infection globally.
The study of HCV and the development of novel therapeutics have progressed tremendously in the last years. Development of robust cell culture systems has been instrumental for these advances, which have been recently reviewed elsewhere (Taylor, 2013). However, study of HCV in vitro heavily relies on the use of human hepatoma cells, which do not adequately mimic hepatocytes because of their transformed nature leading to aberrant host responses (reviewed in (Sheahan et al., 2010)). In attempts to analyze HCV infection in a more physiologically relevant context cultures of primary human hepatocytes, including adult (Ploss et al., 2010; Podevin et al., 2010), fetal (Andrus et al., 2011; Marukian et al., 2011; Sheahan et al., 2014) and stem-cell derived hepatocytes (Roelandt et al., 2012; Schwartz et al., 2012; Wu et al., 2012), have been developed, that support HCV infection transiently at low levels. Although primary hepatocyte cultures are arguably a better approximation of HCV's host environment than hepatoma cell lines they dedifferentiate within a few days in tissue culture and moreover are largely uncoupled from cues provided within the 3 dimensional context of the liver. Those cues include interactions with non-parenchymal cells, nutrient and oxygen gradients and circadian rhythms influencing host gene expression and thus contributing to hepatocyte heterogeneity in vivo. Likewise, studies of host responses at the organismal level, such as cellular and humoral immune responses and virus-induced pathogenesis mandate the study of HCV in vivo. Clinical specimens, i.e. liver biopsies and/or peripheral blood samples could be used, but studies using patient material are hampered by the limited control over important experimental parameters, i.e. time of infection, dose, inoculum, and the inherent heterogeneity within a given study cohort. Consequently, the use of experimental animal models could play an important role in providing insights into host responses to HCV infections in a “real life” setting.
2. Animal models for HCV infection
Historically, it has been challenging to study HCV infection in vivo. The only species beside humans that is readily susceptible to HCV are chimpanzees. Chimpanzees have played an important role in the early characterization of transfusion-mediated hepatitis and the subsequent identification of HCV as the etiologic agent for non-A non-B hepatitis (Bukh, 2004). The natural course of HCV infection observed in patients is mirrored well in chimpanzees, although more severe manifestations of liver disease are rare presumably due to the lack of co-morbidities, such as alcohol-abuse, obesity or HIV co-infections, all of which accelerate progression and exacerbate severity of liver disease in humans. Despite their utility studies in large apes are challenging. Experimental cohorts are usually heterogeneous in terms of age, sex, weight and as an outbred species also genetically, thereby broadening the observed response patterns. Studies are further confounded by usually small cohort sizes, which is driven by the scarcity of the animals and their high costs. Terminal experiments are prohibited in chimps limiting the analysis to sampling of peripheral blood and occasional liver biopsies. Public opposition has led to a ban of biomedical research using chimpanzees in many countries and growing ethical concerns in the US have led to an NIH memorandum that severely restricts all federally-funded (HCV) research involving chimpanzees (NIH 2011, posting date. http://grants.nih.gov/grants/guide/notice-files/NOT-OD-12-025.html).
A multitude of approaches has been undertaken in efforts to model hepatitis C in vivo (reviewed in (Billerbeck et al., 2013)). Those include mice expressing transgenically individual or combinations of HCV proteins, surrogate models relying on infection of small non-human primates with viruses related to HCV and humanized mice, i.e. mice transplanted with human hepatocytes or expressing essential HCV host factors. Here, our discussions will focus on the latter. The rationale is the following: First, humanized mice are susceptible to HCV, the virus causing diseases in humans as opposed to viral surrogates such as GB virus B (Karayiannis et al., 1989; Schaluder et al., 1995; Bukh et al., 2001; Lanford et al., 2003) or HCV-like hepaciviruses (Kapoor et al., 2011; Burbelo et al., 2012) that can differ in genome organization and vary in their sequence. Second, in contrast to e.g. HCV transgenic mouse lines, which stably express HCV gene products often under the control of cellular promoters (reviewed in (Kremsdorf and Brezillon, 2007; Billerbeck et al., 2013)), humanized mice can be infected and thus better approximate the inflammatory milieu in the liver. Third, although some reports suggest that tree shrews (Tupaia belangeri) are susceptible to HCV infection (Xu et al., 2007; Amako et al., 2010), as an outbred species, they are genetically heterogeneous, and they cannot easily be genetically manipulated and tupaia-specific reagents are scarce.
2.1. Xenotransplantation models for HCV
To overcome species barriers in mice, a number of host and viral adaptation approaches have been undertaken (reviewed in (Ploss and Rice, 2009)). To render the murine host more susceptible to HCV infection, the liver has been humanized in specially conditioned xenorecipients. To facilitate efficient engraftment, mice must be immunocompromised to prevent rejection of the transplanted human graft. Further, recipients must suffer from an endogenous liver injury, which allows to selectively ablate endogenous murine hepatocytes and provides the proliferative stimulus for transplanted hepatocytes to expand in the murine parenchyma.
Liver injury can be inflicted chemically, surgically or genetically. The latter is most commonly used, and a number of murine liver injury models have been developed. One of the best-characterized systems is the urokinase plasminogen activator (uPA) transgenic (tg) mouse in which uPA expression is driven by a liver-specific albumin promoter. uPA transgene expression is acutely hepatotoxic and leads to blood coagulopathies resulting in high neonatal mortality (Heckel et al., 1990). However, AlbuPA mice are frail and hypofertile (Brezillon et al., 2008), decreasing the throughput at which animals available for transplantation can be produced. In efforts to create more robust human liver-chimeric models, a variety of alternatives have been tested. Under the control of the major urinary protein (MUP) promoter uPA expression and consequently the onset of liver injury occurs later in post-natal development. This allows a delay of the hepatocyte injection to a time point when the animals are older and more likely to recover from the surgical procedure (Heo et al., 2006). As an alternative to expressing uPA, which is directly hepatotoxic, two transgenic strains have been engineered allowing to selectively ablate mouse liver cells by administration of a prodrug. Liver specific expression of HSV-1 thymidine kinase (HSV-TK) phosphorylates acyclovir or ganciclovir, converting them into toxic drugs thereby killing murine hepatocytes (Hasegawa et al., 2011). Likewise, liver specific expression of caspase 8 fused to an FK506 binding protein in AFC8 transgenic mice allows for selective ablation of mouse liver cells following administration of FK506 (Washburn et al., 2011).
Another liver injury model relies on a disruption in the fumaryl acetoacetate hydrolase (FAH-/-) gene (Grompe et al., 1993), which leads to hepatic toxicity and renal insufficiencies resulting in neonatal lethality. However, FAH-/- animals can be pharmacologically rescued by administration of the 4-hydroxyphenyl pyruvate dioxygenase inhibitor (2-(2-nitro-4-fluoromethylbenzoyl)-1,3-cyclohexanedione) (NTBC) and FAH-/- mice breed normally while on this liver protective drug. In contrast to the uPA models, in FAH mice liver injury can be induced at will by NTBC withdrawal.
When crossed to highly immunodeficient backgrounds, Alb-uPA (Mercer et al., 2001; Meuleman et al., 2005), MUP-uPA (Tesfaye et al., 2013), HSV-tk (Hasegawa et al., 2011) and FAH-/- (Azuma et al., 2007; Bissig et al., 2007; Su et al., 2011) can be robustly engrafted with human adult hepatocytes. In immunodeficient AFC8 mice, hepatic chimerism was so far only reported with fetal hepatoblasts (Washburn et al., 2011). Importantly, human hepatocytes integrate into and proliferate within the murine parenchyma forming functional bile canaliculi that connect to mouse canaliculi. Since only human hepatocyte cell suspensions are usually being injected other nonparenchymal cell subsets, including oval cells, stellate cells, cholangiocytes, fibroblast and Kupffer cells, are not replaced. Resulting human liver chimeric Alb-uPA (Mercer et al., 2001; Meuleman et al., 2005), MUP-uPA (Tesfaye et al., 2013), HSV-tk (Kosaka et al., 2013) and FAH-/- (Bissig et al., 2010) mice are used for infection with cell culture derived HCV and patient isolates. Susceptibility to HCV infection correlates with a high human hepatic chimerism (Bissig et al., 2010; Kawahara et al., 2010; Vanwolleghem et al., 2010).
Although currently less well characterized, human hepatocytes can also be engrafted into xenorecipients in ectopic sites. In early studies it was shown that small pieces of human liver tissue can be maintained for a few weeks following transplantation under the renal capsule in so called “trimera mice” (Ilan et al., 2002). When HCV infected tissue was used, animals were reported to remain viremic for short periods of time (Ilan et al., 2002). Considerable progress has also been made in engineering increasingly complex, artificial liver organoids (reviewed in (Palakkan et al., 2013)) which can be sustained for a few weeks in the intraperitoneal cavity (Chen et al., 2011). However, it has yet to be shown whether mice harboring “human ectopic artificial livers” are indeed susceptible to HCV infection.
While human liver chimeric mice have great utility as challenge models for HCV and other human hepatotropic pathogens, histopathology such as fibrosis, cirrhosis or hepatocellular carcinoma observed in patients chronically infected with HCV has not been reported in chronically infected animals. As an ongoing inflammatory response is thought to contribute to disease progression, the presence of a functional human immune system would be necessary. Proof-of-concept for dually engrafting human liver and components of a human hematopoietic system has recently been shown in two studies. Alb-uPA transgenic mice (Suemizu et al., 2008) were crossed to non-obese diabetic (NOD), recombinase activating gene 2 (Rag2-/-) and the interleukin 2 receptor gamma-chain (IL2RγNULL) background, consequently lacking functional murine B, T and NK cells. The long-term dual reconstitution was achieved in Alb-uPA NOG injected with human hematopoietic stem cells (HSCs) and allogeneic adult hepatocytes but not fetal hepatoblasts. Of note, even major histocompatibility complex mismatched transplantation was sustained without any evidence of hepatocyte rejection by the human immune system (Gutti et al., 2014), suggesting some level of immune tolerance. Similarly, AFC8 Rag2-/- IL2RγNULL mice were repopulated with a mixture of donor matched human hematopoietic progenitor cells and a mixture of human fetal hepatoblasts and other non-parenchymal cells. Dually reconstituted AFC8 Rag2-/-IL2RγNULL mice supported HCV infection at low levels and mounted virus specific immune responses, which even led to an early onset of liver fibrosis (Washburn et al., 2011). Future work will have to focus on improving the function of a transplanted human immune system and increasing throughput while minimizing variability within and between cohorts of engrafted mice.
2.2. Genetically humanized mice
Xenotransplantation is time-consuming, requires survival surgery and is subject to significant donor-to-donor variations, which has spurned efforts to render mice genetically susceptible to HCV infection. Mice are generally resistant to HCV because multiple steps of the viral life-cycle are not efficiently supported in mouse cells (reviewed in (Sandmann and Ploss, 2013)). An increasingly large number of host molecules have been implicated in the uptake of HCV into hepatocytes. HCV forms complexes with host-encoded apo-lipoproteins (reviewed in (Bartenschlager et al., 2011)). Initial attachment to hepatocytes of such lipo-viro-particles is mediated by glycosaminoglycans (Barth et al., 2003; Koutsoudakis et al., 2006) before virions engage the low-density lipoprotein receptor (LDLR) (Agnello et al., 1999; Monazahian et al., 1999; Molina et al., 2007; Owen et al., 2009), the scavenger receptor class B type I (SCARB1; (Scarselli et al., 2002)) and the tetraspanin CD81 (Pileri et al., 1998). Virally triggered signaling through the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2; (Lupberger et al., 2011)) has been proposed to lead to the formation of complexes between CD81 and claudin 1 (CLDN1) (Evans et al., 2007; Diao et al., 2012). This may eventually shuttle the virus towards tight junctions facilitating the interaction with occludin (OCLN; (Liu et al., 2009; Ploss et al., 2009)) and ultimately resulting in an uptake via receptor-mediated endocytosis. The cholesterol uptake receptor Niemann Pick C1 like 1 (NPC1L1, (Sainz et al., 2012)) and transferrin receptor (TfR, (Martin and Uprichard, 2013)) have been implicated in HCV uptake, but their exact roles have yet to be determined.
Of this large set of host factors, CD81, SCARB1, CLDN1 and OCLN are all required, but only CD81 and OCLN have to be of human origin to facilitate viral uptake into murine cells lines (Ploss et al., 2009). It was subsequently shown that transient adenovirally mediated (Dorner et al., 2011) or stable transgenic expression (Dorner et al., 2013) of human CD81 and OCLN in mice enables HCV uptake into murine hepatocytes in vivo. To visualize HCV infection in these genetically humanized mice, sensitive reporter systems were developed, which will be discussed in subsequent sections.
Heterokaryons of human and mouse cell lines support HCV RNA replication, suggesting that dominant restriction factors do not exist in mouse cells (Frentzen et al., 2011). These observations are corroborated by studies showing HCV replication can be established at low efficiency in murine cell lines with drug-selectable HCV genomes so-called HCV replicons (Zhu et al., 2003; Uprichard et al., 2006). Genetic incompatibilities between murine orthologues of host factors essential for HCV RNA replication and virally encoded components of the HCV replicase complex may explain in part suboptimal levels of viral replication. In addition, the innate immune defense of murine cells further limits HCV RNA replication. The observation that HCV can propagate its genome more efficiently in mouse cell lines derived from mouse mutant lines, harboring targeted disruptions of genes critical for type I and III interferon signaling, supports this hypothesis (Chang et al., 2006; Nandakumar et al., 2013; Vogt et al., 2013; Frentzen et al., 2014). Although HCV has an intricate arsenal of tools to evade antiviral defenses in human cells (Keller et al., 2007), these do not seem to suffice to readily establish infection in mouse cells. This may be explained by the combination of low levels of e.g. the HCV NS3/4A, which is critical for blunting antiviral immunity, due to low RNA copy numbers in infected cells and conceivably more rapid kinetics and a greater magnitude of antiviral signaling. While there are several blocks at the level of entry and viral replication in mouse cells, later stages of the HCV life-cycle including virion assembly and release are supported in mouse cell lines. Evidence was provided that mouse cells can support the late stages for the HCV life cycle if critical components of the VLDL pathway are present (Long et al., 2011; Vogt et al., 2013; Frentzen et al., 2014). These in vitro observations have paved the path for the development of an inbred mouse model for HCV infection. Indeed it was shown that mice transgenically expressing human CD81 and human OCLN crossed to innate immune deficiency background can be infected with cell culture derived HCV (Dorner et al., 2013). Infection resulted in measurable HCV RNA levels in both serum and liver over several weeks (Dorner et al., 2013). RNA replication in these genetically humanized mice was dependent on the presence of cyclophilin A, a host factor which is essential for HCV RNA replication and could also be pharmacologically suppressed (Dorner et al., 2013). At times when animals were viremic, infectious HCV particles could be detected in the serum demonstrating that the entire HCV life-cycle can be recapitulated in adequately conditioned mice. Mice with inbred susceptibility to HCV infection allow to apply mouse genetic tools to dissect the viral life-cycle. However, in order to elevate the utility of the model additional improvements are necessary. Compared to infection in patients, chimpanzees and human liver chimeric mice HCV RNA levels are still low in these mice with inbred susceptibility to HCV infection. It will also be important to show that diverse HCV genotypes can replicate in mice. For studying HCV immunity and pathogenesis it will also be important to establish HCV's replicative cycle in fully immunocompetent mice. Despite remaining shortcomings of both human liver chimeric and genetically humanized mice, both systems offer unprecedented opportunities to analyze interactions of HCV with its host cells in the three-dimensional context of the liver. The liver has a complex architecture, which can be anatomically and functionally divided into lobules that refer to liver cells surrounding hepatic blood vessels and bile ducts. Hepatocytes along the porto-central axis of the liver are heterogeneous dictated by environmental factors such as oxygen and nutrients levels, which form zonal gradients around the arteries and veins (reviewed in (Torre et al., 2010)). This translates on the transcriptional level to specific pericentral versus periportal transcriptomic profiles. Liver-specific transcription is further governed by organ–specific circadian rhythms (reviewed in (Tong and Yin, 2013)). The influence of these spatio-temporal expression patterns on physiological processes such as liver metabolism is well appreciated but the impact on hepatic inflammation, specifically HCV infection, is not understood. Since both liver zonation and circadian rhythmicity is optimally studied in the intact organ, humanized mice offer a unique approach to dissect the impact of organ-specific spatio-temporal gene regulation exemplary for HCV infection. For such analyses it is critical to unambiguously detect HCV infected cells, which may be facilitated by some of the methods discussed in the following sections.
3. Visualizing HCV infected cells in situ
Detection of HCV RNA and proteins in situ is notoriously difficult because of their low abundance and unfavorable imaging properties of liver tissue. Thus, despite considerable efforts, information on the frequency, type and distribution of HCV infected cells in the liver are limited and conflicting (Lau et al., 1996). The technical difficulties of detecting HCV antigens in situ reliably has spurned a considerable controversy about the existence of extrahepatic reservoirs of HCV, including for example intestinal epithelial cells (Deforges et al., 2004), brain (Seifert et al., 2008; Wilkinson et al., 2009) and hematopoietically derived cells (Navas et al., 1998; Laskus et al., 2000; Ducoulombier et al., 2004; Pham et al., 2008).
In human biopsies, HCV antigen detection is in part hampered by the quantity and quality of tissue specimens. In contrast, sampling of tissue from HCV infected humanized mice allows for optimal preservation of tissues under controlled experimental settings.
Viral RNA is routinely quantified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). However, standard RT-qPCRs on RNA extracted from whole tissues only provides an average across the entire cell population and includes non-infected bystander cells, but it does not provide any spatial information on where the infected cells may have been located within the tissues. These caveats have been in part addressed by employing laser capture microdissection (LCM) to isolate small clusters of infected cells (Stiffler et al., 2009; Kandathil et al., 2013) thus yielding a more refined representation of the frequency of HCV of infected and non-infected cells. Detection of negative strand RNA generated during asymmetric genome replication is desirable and would give the distinction between cells in which infection has ceased and residual RNA is still remaining and cells actively replicating HCV RNA. However, this method is technically challenging and error prone. PCR based methods are hampered by false-positive detection influenced by contaminations in an environment where HCV is frequently handled, and the inability to determine infection frequency or to allow for the spatial visualization of infected cells.
3.1. HCV reporter genomes
To simplify the analysis it would be desirable to simply utilize HCV genomes expressing reporter proteins. The advent of an infectious cell culture system for HCV has allowed to test different strategies to express fluorescent and luminescent genes in the context of the HCV genome (reviewed in (Murray and Rice, 2011)). Identification of flexible regions within non-structural proteins, and mutations that did not impair or only minimally impacted HCV RNA replication, allowed the insertion of reporter genes. This provided tools for studying multiple aspects of viral replication, for example the formation and turnover of HCV replication complexes in living cells (Moradpour et al., 2004; McCormick et al., 2006; Jones et al., 2007). Inclusion of luciferase with a dominant selectable marker in bi-cistronic HCV replicons was also shown to be functional (Vrolijk et al., 2003; Ikeda et al., 2005). This configuration yielded a useful tool for the study of HCV replication in high throughput. It was further shown that reporter proteins can be inserted between NS5A and NS5B with minimal impact on viral fitness (Horwitz et al., 2013). However, while recombinant HCV reporter viruses have great utility in vitro, they have a number of drawbacks that complicate their use in vivo. Recombinant viruses often require cell culture adaptive mutations to compensate for reduced viral titers that do not always improve viral fitness in vivo. Further, recombinant HCV often relies on an artificial genomic organization that uncouples viral protein expression from recombinant gene expression and reporter gene expression is not stable.
3.2. In situ staining methods
Antibody based staining and microscopic detection methods are widely used to detect viral antigens in situ. However, these conventional methods had limited utility for detecting reliably HCV in situ because of the usually low protein antigen load in infected cells and the high autofluorescence of liver tissue. To overcome some of these challenges, two-photon excitation microscopy was combined with quantum dot technology yielding very high sensitivity of HCV antigen detection in the liver of chronically infected patients (Liang et al., 2009). Sporadic clusters of infected cells were visualized only in livers of patients with chronic hepatitis C but not of HCV-negative controls, thereby highlighting the specificity of the immunostaining.
A highly sensitive fluorescence in situ hybridization (FISH) system was developed suitable not only for detecting HCV RNA but simultaneously mRNAs of interferon stimulated genes in human liver biopsies (Wieland et al., 2013). Similar to previously published protocols primarily focusing on detecting host RNA species (Zenklusen and Singer, 2010; Lyubimova et al., 2013), this method relies on hybridizing sets of short oligonucleotides to a target mRNA in tissue sections. Signals stemming from fluorophores binding to the probes can be quantified at the single cell resolution in a series of random high-power fields.
Although 2-photon microscopy and FISH have so far only been used to detect HCV antigens in human samples, conceivably these methods can be applied to HCV infected tissues from humanized mice. However, the use of in vivo systems, which are tractable for genetic manipulations, opens up opportunities to use of host host and virally encoded reporters to track HCV infection in vivo.
3.3. Cre based reporter systems
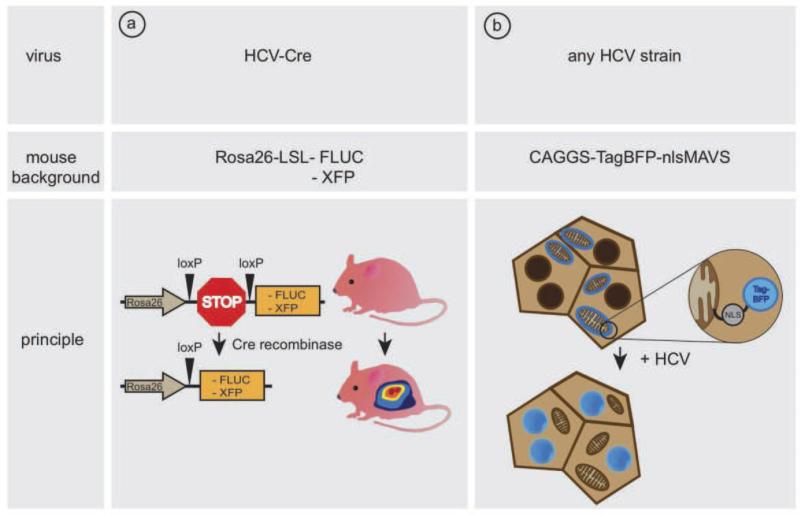
The advent of mice with inheritable susceptibility to HCV infection has made it possible to design a cellularly encoded reporter for imaging HCV infection in vivo. The inability of HCV to replicate efficiently in mouse cells (Ploss et al., 2009) has motivated the development of a Cre-loxp based system to detect HCV entry events in vivo (Safran et al., 2003) (Figure 1a). Here, detection of HCV uptake is uncoupled from the need for continuous viral replication. Mice expressing the essential human HCV entry factors following adenoviral delivery (Dorner et al., 2011) or expressing transgenically the entry factors (Dorner et al., 2013) can take up intravenously injected recombinant HCV expressing Cre recombinase. Upon initial translation of the HCV polyprotein, Cre recombinase relocalizes from the cytoplasm to the nucleus and excises a transcriptional stop cassette which blocks expression of a reporter gene, such as firefly luciferase (Safran et al., 2003), lacZ (Soriano, 1999), or fluorescent proteins (Srinivas et al., 2001; Stoller et al., 2008). Low levels of Cre recombinase are sufficient to permanently mark cells which have taken up HCV.
Fig. 1.
Methods for the visualization of HCV-infection in vivo. (a) A recombinant bicistronic HCV genome, expressing Cre recombinase excises a loxP site-flanked transcriptional stop cassette, leading to expression of a reporter gene (FLUC/XFP). (b) Upon HCV cell entry the virus protease NS3/4A cleaves mitochondrial bound TagBFP-nlsMAVS, resulting in a nuclear translocation.
3.4. HCV-dependent cellular reporter systems
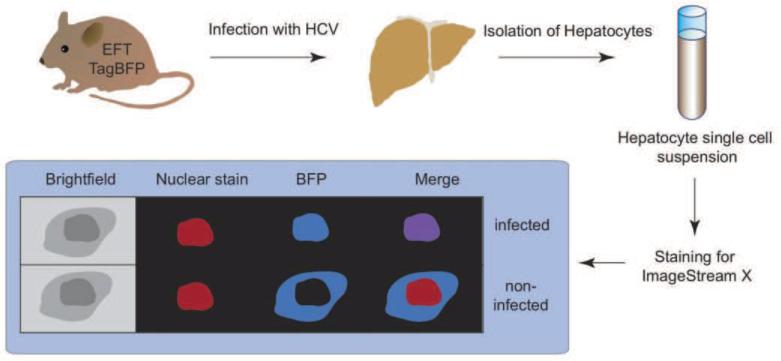
The Cre-based reporter allows to sensitively mark individual HCV infected cells solely based on productive entry and HCV RNA translation. However, activation of the reporter does not require continuous viral replication, and consequently the signal intensity does not correlate with levels of HCV RNA replication. Alternative detection systems were developed based on activation of a cellularly encoded reporter directly by HCV encoded NS3-4A protein leading to secretion of secreted alkaline phosphatase (Pan et al., 2009), gene induction (Tanaka et al.) or subcellular protein relocalization (Jones et al.). The HCV NS3/4A protease cleaves the innate immune signaling protein, MAVS (also known as IPS1, VISA, Cardif), from the mitochondrial and peroxisomal outer membrane, thereby interrupting an antiviral cascade allowing the viral infection to remain unnoticed by the host cell (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Based on this natural phenomenon, a fluorescent reporter was engineered consisting of a fusion of MAVS and a fluorescent protein (Jones et al.). Upon infection, the viral protease cleaves the fluorescent reporter from the mitochondria, exposing a nuclear localization signal that results in trafficking of the fluorescent protein to the nucleus. This system was shown to be very effective to monitor HCV infection in fetal (Sheahan et al., 2014), adult (Jones et al., 2010; Ploss et al., 2010) and stem cell-derived (Schwartz et al., 2012) primary hepatocytes. Because of the conserved nature of this immune evasion response, reporter systems relying on cleavage of MAVS derived sequences have the ability to detect multiple HCV genotypes, potentially allowing the assay of infection by unadulterated clinical isolates (Ploss et al.). Transgenic expression of a TagBFPnlsMAVS fusion protein allowed for sensitive detection of HCV infection in genetically humanized mice (Dorner et al., 2011) (Figure 1b). HCV protease-mediated nuclear relocalization can be visualized by fluorescent microscopy or in higher throughput in hepatocyte single cell suspensions using an imaging flowcytometer that combines the speed, sensitivity and phenotyping abilities of flow cytometry with the detailed imagery and functional insights of microscopy. In TagBFPnlsMAVS transgenic mice expressing also human CD81 and OCLN but not mice lacking expression of these human entry factors (EFT), HCV infection causes overlap of TagBFP fluorescence with a nuclear counterstain (Figure 2).
Fig. 2.
Analysis of HCV-infection in vivo using ImageStream X. Entry factor transgenic (EFT) mice are infected with HCV, followed by hepatocyte isolation, fixation, permeabilization and nuclear counterstaining. Nuclear dyes and TagBFP colocalize in nuclei of infected cells.
The above described methods enable unambiguous detection of HCV infected cells. Multiplex analysis including co-staining for host transcripts and gene products will shed light on the frequency, spatial location and phenotype of infected cells at single cell resolution.
Highlights.
Chronic hepatitis C remains a major global health problem
HCV has a narrow host range limited to humans and chimpanzees
Humanized mice offer unique opportunities to dissect HCV infection in vivo
Low HCV antigen load and high autofluorescence in the liver pose challenges to visualize HCV infection
Novel reporters facilitate reliable detection of HCV infection in situ
Acknowledgements
The authors thank Brigitte Heller and Benjamin Winer for edits and critical discussion of the manuscript. Work in the laboratory is supported in part by grants from the National Institutes of Health (2 R01 AI079031-05A1, 1 R01 AI107301-01, 1 R56 AI106005-01, 1R21 AI 106000-01), the Walter Reed Army Institute of Research, the Bill and Melinda Gates Foundation and the Grand Challenges Program at Princeton University. M.v.S. is a recipient of a fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang Q-X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amako Y, Tsukiyama-Kohara K, Katsume A, Hirata Y, Sekiguchi S, Tobita Y, Hayashi Y, Hishima T, Funata N, Yonekawa H, Kohara M. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J Virol. 2010;84:303–11. doi: 10.1128/JVI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, Bhatia SN, Rice CM. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–12. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol. 2007;25:903–10. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends in microbiology. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–12. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- Billerbeck E, de Jong Y, Dorner M, de la Fuente C, Ploss A. Animal models for hepatitis C. Curr Top Microbiol Immunol. 2013;369:49–86. doi: 10.1007/978-3-642-27340-7_3. [DOI] [PubMed] [Google Scholar]
- Bissig KD, Le TT, Woods NB, Verma IM. Repopulation of adult and neonatal mice with human hepatocytes: a chimeric animal model. Proc Natl Acad Sci U S A. 2007;104:20507–11. doi: 10.1073/pnas.0710528105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–30. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezillon NM, DaSilva L, L'Hote D, Bernex F, Piquet J, Binart N, Morosan S, Kremsdorf D. Rescue of fertility in homozygous mice for the urokinase plasminogen activator transgene by the transplantation of mouse hepatocytes. Cell Transplant. 2008;17:803–12. doi: 10.3727/096368908786516800. [DOI] [PubMed] [Google Scholar]
- Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–75. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- Bukh J, Apgar CL, Govindarajan S, Purcell RH. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J Med Virol. 2001;65:694–7. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. Serology enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012 doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80:7364–74. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AA, Thomas DK, Ong LL, Schwartz RE, Golub TR, Bhatia SN. Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci U S A. 2011;108:11842–7. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Deforges S, Evlashev A, Perret M, Sodoyer M, Pouzol S, Scoazec JY, Bonnaud B, Diaz O, Paranhos-Baccala G, Lotteau V, Andre P. Expression of hepatitis C virus proteins in epithelial intestinal cells in vivo. J Gen Virol. 2004;85:2515–23. doi: 10.1099/vir.0.80071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, Kapadia SB. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86:10935–49. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T, Akira S, Law M, Rice CM, Ploss A. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–41. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–11. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoulombier D, Roque-Afonso AM, Di Liberto G, Penin F, Kara R, Richard Y, Dussaix E, Feray C. Frequent compartmentalization of hepatitis C virus variants in circulating B cells and monocytes. Hepatology. 2004;39:817–25. doi: 10.1002/hep.20087. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Frentzen A, Anggakusuma, Gurlevik E, Hueging K, Knocke S, Ginkel C, Brown RJ, Heim M, Dill MT, Kroger A, Kalinke U, Kaderali L, Kuehnel F, Pietschmann T. Cell entry, efficient RNA replication, and production of infectious hepatitis C virus progeny in mouse liver-derived cells. Hepatology. 2014;59:78–88. doi: 10.1002/hep.26626. [DOI] [PubMed] [Google Scholar]
- Frentzen A, Huging K, Bitzegeio J, Friesland M, Haid S, Gentzsch J, Hoffmann M, Lindemann D, Zimmer G, Zielecki F, Weber F, Steinmann E, Pietschmann T. Completion of Hepatitis C Virus Replication Cycle in Heterokaryons Excludes Dominant Restrictions in Human Non-liver and Mouse Liver Cell Lines. PLoS Pathog. 2011;7:e1002029. doi: 10.1371/journal.ppat.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–8. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, Soriano P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- Gutti TL, Knibbe JS, Makarov E, Zhang J, Yannam GR, Gorantla S, Sun Y, Mercer DF, Suemizu H, Wisecarver JL, Osna NA, Bronich TK, Poluektova LY. Human hepatocytes and hematolymphoid dual reconstitution in treosulfan-conditioned uPA-NOG mice. Am J Pathol. 2014;184:101–9. doi: 10.1016/j.ajpath.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kawai K, Mitsui T, Taniguchi K, Monnai M, Wakui M, Ito M, Suematsu M, Peltz G, Nakamura M, Suemizu H. The reconstituted 'humanized liver' in TK-NOG mice is mature and functional. Biochem Biophys Res Commun. 2011;405:405–10. doi: 10.1016/j.bbrc.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel JL, Sandgren EP, Degen JL, Palmiter RD, Brinster RL. Neonatal bleeding in transgenic mice expressing urokinase-type plasminogen activator. Cell. 1990;62:447–56. doi: 10.1016/0092-8674(90)90010-c. [DOI] [PubMed] [Google Scholar]
- Heo J, Factor VM, Uren T, Takahama Y, Lee JS, Major M, Feinstone SM, Thorgeirsson SS. Hepatic precursors derived from murine embryonic stem cells contribute to regeneration of injured liver. Hepatology. 2006;44:1478–86. doi: 10.1002/hep.21441. [DOI] [PubMed] [Google Scholar]
- Horwitz JA, Dorner M, Friling T, Donovan BM, Vogt A, Loureiro J, Oh T, Rice CM, Ploss A. Expression of heterologous proteins flanked by NS3-4A cleavage sites within the hepatitis C virus polyprotein. Virology. 2013;439:23–33. doi: 10.1016/j.virol.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun. 2005;329:1350–9. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- Ilan E, Arazi J, Nussbaum O, Zauberman A, Eren R, Lubin I, Neville L, Ben-Moshe O, Kischitzky A, Litchi A, Margalit I, Gopher J, Mounir S, Cai W, Daudi N, Eid A, Jurim O, Czerniak A, Galun E, Dagan S. The hepatitis C virus (HCV)-Trimera mouse: a model for evaluation of agents against HCV. J Infect Dis. 2002;185:153–61. doi: 10.1086/338266. [DOI] [PubMed] [Google Scholar]
- Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 28:167–71. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–71. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J Gen Virol. 2007;88:470–5. doi: 10.1099/vir.0.82363-0. [DOI] [PubMed] [Google Scholar]
- Kandathil AJ, Graw F, Quinn J, Hwang HS, Torbenson M, Perelson AS, Ray SC, Thomas DL, Ribeiro RM, Balagopal A. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145:1404–13. e1–10. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:11608–13. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiannis P, Petrovic LM, Fry M, Moore D, Enticott M, McGarvey MJ, Scheuer PJ, Thomas HC. Studies of GB hepatitis agent in tamarins. Hepatology. 1989;9:186–92. doi: 10.1002/hep.1840090204. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Toso C, Douglas DN, Nourbakhsh M, Lewis JT, Tyrrell DL, Lund GA, Churchill TA, Kneteman NM. Factors affecting hepatocyte isolation, engraftment, and replication in an in vivo model. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2010;16:974–82. doi: 10.1002/lt.22099. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Keller BC, Johnson CL, Erickson AK, Gale M., Jr. Innate immune evasion by hepatitis C virus and West Nile virus. Cytokine Growth Factor Rev. 2007;18:535–44. doi: 10.1016/j.cytogfr.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka K, Hiraga N, Imamura M, Yoshimi S, Murakami E, Nakahara T, Honda Y, Ono A, Kawaoka T, Tsuge M, Abe H, Hayes CN, Miki D, Aikata H, Ochi H, Ishida Y, Tateno C, Yoshizato K, Sasaki T, Chayama K. A novel TK-NOG based humanized mouse model for the study of HBV and HCV infections. Biochem Biophys Res Commun. 2013;441:230–5. doi: 10.1016/j.bbrc.2013.10.040. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–20. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremsdorf D, Brezillon N. New animal models for hepatitis C viral infection and pathogenesis studies. World journal of gastroenterology : WJG. 2007;13:2427–35. doi: 10.3748/wjg.v13.i17.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Chavez D, Notvall L, Brasky KM. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology. 2003;311:72–80. doi: 10.1016/s0042-6822(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Piasek A, Nowicki M, Horban A, Cianciara J, Rakela J. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442–8. doi: 10.1086/315283. [DOI] [PubMed] [Google Scholar]
- Lau JY, Krawczynski K, Negro F, Gonzalez-Peralta RP. In situ detection of hepatitis C virus--a critical appraisal. J Hepatol. 1996;24:43–51. [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr., Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–58. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–4. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–66. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–95. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, van Oudenaarden A. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protoc. 2013;8:1743–58. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A. 2013;110:10777–82. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–23. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick CJ, Maucourant S, Griffin S, Rowlands DJ, Harris M. Tagging of NS5A expressed from a functional hepatitis C virus replicon. J Gen Virol. 2006;87:635–40. doi: 10.1099/vir.0.81553-0. [DOI] [PubMed] [Google Scholar]
- Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–33. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–56. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–9. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. Journal of Medical Virology. 1999;57:223–9. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional Hepatitis C virus replication complexes. J Virol. 2004;78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. Journal of viral hepatitis. 2011;18:305–15. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- Murray CL, Rice CM. Turning hepatitis C into a real virus. Annu Rev Microbiol. 2011;65:307–27. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- Nandakumar R, Finsterbusch K, Lipps C, Neumann B, Grashoff M, Nair S, Hochnadel I, Lienenklaus S, Wappler I, Steinmann E, Hauser H, Pietschmann T, Kroger A. Hepatitis C Virus Replication in Mouse Cells Is Restricted by IFN-Dependent and -Independent Mechanisms. Gastroenterology. 2013;145:1414–1423. e1. doi: 10.1053/j.gastro.2013.08.037. [DOI] [PubMed] [Google Scholar]
- Navas S, Martin J, Quiroga JA, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–6. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Huang H, Ye J, Gale M., Jr. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakkan AA, Hay DC, Anil Kumar PR, Kumary TV, Ross JA. Liver tissue engineering and cell sources: issues and challenges. Liver international : official journal of the International Association for the Study of the Liver. 2013;33:666–76. doi: 10.1111/liv.12134. [DOI] [PubMed] [Google Scholar]
- Pan KL, Lee JC, Sung HW, Chang TY, Hsu JT. Development of NS3/4A protease-based reporter assay suitable for efficiently assessing hepatitis C virus infection. Antimicrob Agents Chemother. 2009;53:4825–34. doi: 10.1128/AAC.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TN, King D, Macparland SA, McGrath JS, Reddy SB, Bursey FR, Michalak TI. Hepatitis C virus replicates in the same immune cell subsets in chronic hepatitis C and occult infection. Gastroenterology. 2008;134:812–22. doi: 10.1053/j.gastro.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–41. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–6. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 107:3141–5. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3141–5. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009 doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevin P, Carpentier A, Pene V, Aoudjehane L, Carriere M, Zaidi S, Hernandez C, Calle V, Meritet JF, Scatton O, Dreux M, Cosset FL, Wakita T, Bartenschlager R, Demignot S, Conti F, Rosenberg AR, Calmus Y. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139:1355–64. doi: 10.1053/j.gastro.2010.06.058. [DOI] [PubMed] [Google Scholar]
- Reed KE, Xu J, Rice CM. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J. Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P, Obeid S, Paeshuyse J, Vanhove J, Van Lommel A, Nahmias Y, Nevens F, Neyts J, Verfaillie CM. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. Journal of hepatology. 2012;57:246–51. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG., Jr. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- Sainz B, Jr., Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–5. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann L, Ploss A. Barriers of hepatitis C virus interspecies transmission. Virology. 2013;435:70–80. doi: 10.1016/j.virol.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21:5017–25. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaluder GG, Dawson GJ, Simons JN, Pilot-Matias TJ, Gutierrez RA, Heynen CA, Knigge MF, Kurpiewski GS, Buijk SL, Leary TP, et al. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- Schinazi R, Halfon P, Marcellin P, Asselah T. HCV direct-acting antiviral agents: the best interferon-free combinations. Liver international : official journal of the International Association for the Study of the Liver. 2014;34(Suppl 1):69–78. doi: 10.1111/liv.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarcz R, Weiland O, Wejstal R, Norkrans G, Fryden A, Foberg U. A randomized controlled open study of interferon alpha-2b treatment of chronic non-A, non-B posttransfusion hepatitis: no correlation of outcome to presence of hepatitis C virus antibodies. Scandinavian journal of infectious diseases. 1989;21:617–25. doi: 10.3109/00365548909021689. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, Bhatia SN. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:2544–8. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert F, Struffert T, Hildebrandt M, Blumcke I, Bruck W, Staykov D, Huttner HB, Hilz MJ, Schwab S, Bardutzky J. In vivo detection of hepatitis C virus (HCV) RNA in the brain in a case of encephalitis: evidence for HCV neuroinvasion. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2008;15:214–8. doi: 10.1111/j.1468-1331.2007.02044.x. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sheahan T, Imanaka N, Marukian S, Dorner M, Liu P, Ploss A, Rice CM. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe. 2014;15:190–202. doi: 10.1016/j.chom.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T, Jones CT, Ploss A. Advances and challenges in studying hepatitis C virus in its native environment. Expert Rev Gastroenterol Hepatol. 2010;4:541–50. doi: 10.1586/egh.10.53. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller JZ, Degenhardt KR, Huang L, Zhou DD, Lu MM, Epstein JA. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis. 2008;46:200–4. doi: 10.1002/dvg.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Liu C, Xiang D, Zhang H, Yuan S, Wang M, Chen F, Zhu H, He Z, Wang X, Hu Y. Xeno-repopulation of Fah −/− Nod/Scid mice livers by human hepatocytes. Sci China Life Sci. 2011;54:227–34. doi: 10.1007/s11427-011-4140-7. [DOI] [PubMed] [Google Scholar]
- Suemizu H, Hasegawa M, Kawai K, Taniguchi K, Monnai M, Wakui M, Suematsu M, Ito M, Peltz G, Nakamura M. Establishment of a humanized model of liver using NOD/Shi-scid IL2Rgnull mice. Biochem Biophys Res Commun. 2008;377:248–52. doi: 10.1016/j.bbrc.2008.09.124. [DOI] [PubMed] [Google Scholar]
- Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ, Schwartz H, Nelson DR, Everson GT, Eley T, Wind-Rotolo M, Huang SP, Gao M, Hernandez D, McPhee F, Sherman D, Hindes R, Symonds W, Pasquinelli C, Grasela DM, Group AIS. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. The New England journal of medicine. 2014;370:211–21. doi: 10.1056/NEJMoa1306218. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Mori Y, Tani H, Abe T, Moriishi K, Kojima H, Nagano T, Okabe T, Suzuki T, Tatsumi M, Matsuura Y. Establishment of an indicator cell system for hepatitis C virus. Microbiol Immunol. 54:206–20. doi: 10.1111/j.1348-0421.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- Taylor DR. Evolution of cell culture systems for HCV. Antiviral therapy. 2013;18:523–30. doi: 10.3851/IMP2593. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Evans MJ, von Hahn T, You S, Rice CM. Studying hepatitis C virus: making the best of a bad virus. J Virol. 2007;81:8853–67. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye A, Stift J, Maric D, Cui Q, Dienes HP, Feinstone SM. Chimeric mouse model for the infection of hepatitis B and C viruses. PLoS One. 2013;8:e77298. doi: 10.1371/journal.pone.0077298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Yin L. Circadian rhythms in liver physiology and liver diseases. Comprehensive Physiology. 2013;3:917–40. doi: 10.1002/cphy.c120017. [DOI] [PubMed] [Google Scholar]
- Torre C, Perret C, Colnot S. Molecular determinants of liver zonation. Progress in molecular biology and translational science. 2010;97:127–50. doi: 10.1016/B978-0-12-385233-5.00005-2. [DOI] [PubMed] [Google Scholar]
- Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwolleghem T, Libbrecht L, Hansen BE, Desombere I, Roskams T, Meuleman P, Leroux-Roels G. Factors determining successful engraftment of hepatocytes and susceptibility to hepatitis B and C virus infection in uPA-SCID mice. J Hepatol. 2010;53:468–76. doi: 10.1016/j.jhep.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Vogt A, Scull MA, Friling T, Horwitz JA, Donovan BM, Dorner M, Gerold G, Labitt RN, Rice CM, Ploss A. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology. 2013;444:1–11. doi: 10.1016/j.virol.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J Virol Methods. 2003;110:201–9. doi: 10.1016/s0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, Barry W, Ploss A, Rice CM, Su L. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–44. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, Chisari FV, Heim MH. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2013 doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J, Radkowski M, Laskus T. Hepatitis C virus neuroinvasion: identification of infected cells. J Virol. 2009;83:1312–9. doi: 10.1128/JVI.01890-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, Gilbert DM, Tang H. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS pathogens. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen H, Cao X, Ben K. Efficient infection of tree shrew (Tupaia belangeri) with hepatitis C virus grown in cell culture or from patient plasma. J Gen Virol. 2007;88:2504–12. doi: 10.1099/vir.0.82878-0. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Singer RH. Analyzing mRNA expression using single mRNA resolution fluorescent in situ hybridization. Methods in enzymology. 2010;470:641–59. doi: 10.1016/S0076-6879(10)70026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S, Soriano V, Asselah T, Bronowicki JP, Lohse AW, Mullhaupt B, Schuchmann M, Bourliere M, Buti M, Roberts SK, Gane EJ, Stern JO, Vinisko R, Kukolj G, Gallivan JP, Bocher WO, Mensa FJ. Faldaprevir and deleobuvir for HCV genotype 1 infection. The New England journal of medicine. 2013;369:630–9. doi: 10.1056/NEJMoa1213557. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–10. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]