Abstract
While the phenomenon of polyadenylation has been well-studied, the dynamics of poly(A) tail size and its impact on transcript function and cell biology are less well-appreciated. The goal of this review is to encourage readers to view the poly(A) tail as a dynamic, changeable aspect of a transcript rather than a simple static entity that marks the 3′ end of an mRNA. This could open up new angles of regulation in the post-transcriptional control of gene expression throughout development, differentiation and cancer.
Keywords: polyadenylation, poly(A) tail, deadenylation, hyperadenylation, mRNA 3′ end processing
Introduction
Poly(A) tails are added to the majority of mRNAs during 3′ end processing stages in a co-transcriptional fashion [1]. Despite their simple sequence composition, 3′ terminal polyadenosine tracts have critical roles in multiple aspects of a transcript’s life cycle. For one, the poly(A) tail is important for mediating the translocation of a completely processed mRNA to the cytoplasm [2]. In addition, poly(A) tails can play key regulatory roles in enhancing translation efficiency [3–8], particularly in certain developmental stages [9]. Finally, poly(A) tails are vital for regulating the efficiency of mRNA quality control and degradation [10,11]. When an mRNA is no longer being used for translation, poly(A) tail shortening is one of the key steps initiating decay of the body of the message [12]. Thus a full appreciation of the polyadenylation process and the poly(A) tail as a regulatory entity is crucial for understanding numerous aspects of gene expression.
While it is generally believed that most mRNAs have 150-250 nt poly(A) tails [13], several recent global analyses provide evidence that a significant population of mRNAs lack or have shortened poly(A) tails [14–17]. This phenomenon has been well-described in oocytes and embryonic cells as a means of regulating translation, but the functions of and mechanisms to generate short poly(A) tails in adult cells remain elusive. The poly(A) tail is not simply a static entity at the end of mRNAs waiting to be removed as its final act. Clearly, the hypothesis that poly(A) tail length is a potentially important aspect of post-transcriptional control of gene expression is worth evaluating [18]. In this article, we will review the data assessing the short poly(A) tail-containing transcript population, the sequences and proteins critical for regulating poly(A) tail length, and the potential functions of shortened poly(A) tails.
Does size matter?
Does it truly matter to a transcript whether its poly(A) tail is 15 or 150 or 250 nucleotides in length? There are indeed observations arguing that poly(A) length might have a dramatic impact on cell biology and thus may be an understudied aspect of gene expression regulation. The poly(A) tail lengths of hundreds of mouse liver mRNAs change in a rhythmic fashion along with the circadian rhythm of a cell [19]. Furthermore, a correlation was observed between longer poly(A) tails and increased protein expression at specific times in the circadian cycle. In a recent extensive global analysis of poly(A) lengths in multiple eukaryotic organisms, housekeeping genes tended to have shorter poly(A) tails [9]. Still, this study could not identify any significant correlation between poly(A) tail length and translational efficiency beyond the embryonic stages. This recent work from the Bartel laboratory provides further support for the well-documented evidence that changes in poly(A) tail length dictate translational activity on an mRNA in early development [20]. However, it is somewhat at odds with the circadian rhythm study as well as the association between poly(A) tail length and translation that have been observed in neurons [21]. Thus additional work is undoubtedly needed to sort out the tantalizing connections between the variations in poly(A) tail length and mRNA translational efficiency.
As noted above, recent global analyses have demonstrated that the poly(A) tail of many mRNAs may actually be much shorter (50-100 nts) than originally believed (150-250 nts) [14–17]. Therefore small variations of 10-20 nucleotides in poly(A) tail size could have a substantial impact on the poly(A) tail size (e.g. increase it by 40%) and its subsequent function. A rich diversity in poly(A) binding protein types and dynamics has recently been appreciated, which can perhaps exploit these seemingly minor tail differences for biologically relevant purposes [22]. Furthermore, the addition of short tracts of A-residues to RNAs by the TRAMP complex [23] serves as a likely landing pad for exonucleases [24]. Could increasing the poly(A) tail size by a few residues provide a similar landing pad for the exonucleases at the end of mRNAs devoid of poly(A) binding proteins and daunting secondary structures? There is evidence that terminal uridylation of certain transcripts can serve as such a mark for Dis3L2-mediated degradation [25]. To fully uncover such possibilities, changes in poly(A) tail size need to be carefully assessed through cellular development, differentiation and stages of the cell cycle.
Poly(A) tail size appears to play a major role in mRNA quality control as dramatic changes to the status quo of the poly(A) tail can serve as a signal to degrade mRNAs. Small oligo-adenylate tails provide binding sites for the LSm complex [26] and target RNAs for degradation [27]. Likewise, hyperadenylation of mRNAs is also associated with rapid decay of mRNAs in the nucleus [10,11,28,29]. Interestingly, not all small poly(A) tails signal decay as several ncRNAs contain terminal triple helix structures that use these short oligo(A) tails to provide stability [30–32]. Could triple helices also play a role with longer poly(A) tails? Curiously, A-A-U triple helices in solution depend on the size of the poly(A) strand. If the A strand is too short or too long, they do not form efficiently [33], and the size of the poly(A) strand that forms these duplexes (28-150 nts) is approximately the physiological length of the mRNA tail. Thus poly(A) length dependent triple helices/RNA structures involving the 3′ UTR or other sequences could conceivably play a broader role in RNA biology.
How does the poly(A) tail measure up?
As discussed above, there is accumulating evidence that the length of a poly(A) tail may be much more than a nuance in mRNA characterization. Instead, it may play major roles in cell biology that are yet to be defined. In order to determine if fluctuations in poly(A) tail length matter in the life of a transcript, there needs to be methods to accurately measure poly(A) tail size. In theory, this task should be relatively straightforward; however, in practice it is far from trivial. All of the available methods, whether they target one or multiple genes, share two main components. First, they can specifically identify the transcripts of interesting transcript-specific hybridization probes PCR primers, or target-specific sequence tags. Second, they all employ some method to determine the number of nucleotides in the poly(A) tail. This is where the real challenge of accurately assessing poly(A) tail dynamics begins. Thus, numerous approaches have been developed in the attempt of measuring poly(A) tail length. These include, but are not limited to, RNaseH/Oligo(dT) northern blot analysis, various iterations of the poly(A) test or PAT assay, oligo(dT) selection, and next-generation sequencing protocols. Depending on the experimental goals and required resolution, each of these approaches is capable of measuring poly(A) tail length to some degree. However, each method has its own set of limitations and drawbacks as well. In this section we will discuss several of these methods, including their advantages and disadvantages as appropriate.
The RNase H/Oligo(dT) northern blot is a gel-based method for direct assessment of poly(A) tail length by comparing mRNA treated with RNase H in the presence and absence of oligo(dT) [34–36]. Oligo(dT) hybridizes with the poly(A) tail, forming an RNA/DNA duplex. RNase H then specifically degrades RNA/DNA hybrid strands. The resolution of this assay can be improved by adding a second, transcript-specific oligo, which will allow RNase H to cleave the transcript in two locations, leaving a shorter RNA product. Both reactions (plus and minus oligo(dT)) are visualized by electrophoresis and northern blot using a probe specific for the gene of interest. The difference in product size gives a measure of poly(A) tail length for that transcript. The disadvantages of this technique are that it is labor intensive, requires large amounts of RNA, and works best on highly abundant transcripts. In addition, the results can be confounded by poly(A) tracks within the transcripts [35]. Because it does not rely on amplification of the RNA and provides a “direct” measure of polyadenylation, RNase H/Oligo(dT) analysis has the advantage of avoiding the biases inherent to PCR and can often be used when the PCR-based approaches for measuring polyadenylation (described below) fail or generate ambiguous results [36].
A number of PCR-based methods have been developed for assessing poly(A) tail length. Advantageously, these techniques are straightforward (in concept) and fast, while also allowing the investigation of transcripts with low abundances. Without optimization of the PCR reaction conditions, these techniques are susceptible to bias introduced by amplification artifacts and reaction efficiencies weighted towards shorter products. The PCR-based ‘PAT’ assays include RACE-PAT (also call the poly(A) length assay), ligation-mediated or LM-PAT, and extension or ePAT [35–38]. The basic principle of these methods is the same, but they differ in how poly(A) tail length is captured and measured. RACE-PAT uses an oligo(dT) primer with a G/C-rich 5′ anchor sequence for cDNA synthesis in combination with a target specific primer for PCR [35,36]. The oligo(dT) primer ideally binds anywhere along the length of the poly(A) tail, though the presence of the anchor sequence should encourage primer annealing at the 3′ end of the transcript, and prevent shortening of the tail during PCR amplification [35]. The PCR products are then visualized by standard gel electrophoresis. In the LM-PAT assay, high concentrations of the oligo(dT) primer containing a 5′ phosphate are used to saturate the entire poly(A) tail [35,36]. Then an anchored oligo(dT) primer-adapter is added to specifically ligate to the 5′ phosphorylated oligo(dT) primer at the 3′-most end of the poly(A) tail. The ligated oligos serve as a primer for the cDNA reaction. Then PCR is used withthe anchor sequence and a transcript-specific primer to determine tail length. The ePAT assay relies on an anchored oligo(dT) primer to target the 3′ end of the poly(A) tail and then Klenow polymerase extends the transcript in a template-dependent manner [37]. The reverse transcription reaction is performed at an elevated temperature to ensure that only transcripts primed at the 3′ end then extended are converted to cDNA. As with the other PAT assays, the anchor sequence is combined with a transcript-specific primer and PCR is used to assess tail length. A recent variation of this protocol uses yeast poly(A) polymerase to add guanosine and inosine (G and I) to the transcript’s 3′ end to provide a specific template for a PCR reaction [38]. The addition of fluorescent dyes to the 5′ ends of the primers targeting the anchor sequences enables analysis of the PCR products by capillary electrophoresis, which can yield more accurate measurements of poly(A) length [38]. As mentioned earlier, the main limitations of these techniques can be a bias towards shorter PCR products and difficulty in accurately quantifying the sizes of longer poly(A) tails.
A full assessment of poly(A) tail dynamics on a cellular level requires global analysis of poly(A) tail length, but such measurements are fraught with technical challenges. Rather than attempting to specifically measure the lengths of many poly(A) tails at once, conventional genome-wide analyses have focused on differences in polyadenylation status. This categorization is generally accomplished by transcript fractionation using oligo(dT) magnetic beads to first bind poly(A) tails. Then, different elution conditions are used to collect transcripts with long or short tails [15,16,39]. These transcripts can also be compared with the population of mRNA which does not bind to oligo(dT) during the fractionation and presumably have no, orvery short, poly(A) tails [15,16] As with the PAT assays, multiple variations of this general approach have been developed. Polyadenylation state array (PASTA) analysis [40] enhances the ability to detect differences in poly(A) tail length by increasing the number of different fractions collected and analyzed. An alternative approach for determining polyadenylation state is to isolate the mRNA using an antibody against the 5′ cap binding protein eIF4E and comparing this population of transcripts with the population isolated by the more traditional methods selecting for the poly(A) tail [14]. These genome-wide techniques are able to give insight into the general trends of polyadenylation state; however, they are limited in their ability to resolve small differences or changes in tail length and have a general bias towards longer poly(A) sequences.
More recently, novel next-generation sequencing-based methods, including TAIL-seq and PAL-seq, have been developed which improve our capacity to accurately measure poly(A) tail length on a global scale. Sample preparation in both techniques is similar and both share a particular emphasis on avoiding the use of oligo(dT) for transcript selection. The differences between them relate to how the poly(A) tail is characterized. The TAIL-seq protocol [17] uses an unbalanced paired-end sequencing approach. The first read of the pair specifically identifies individual transcripts, while the second read of the pair is used to directly sequence the poly(A) tail. The TAIL-seq approach relies on a novel solution to the challenge of sequencing through a long homopolymer by looking for an inflection in the fluorescent signal intensities to help identify the true start of the poly(A) sequence at the end of the 3′UTR. The primary limitation of this approach is that it cannot accurately measure poly(A) tails longer than 231 nucleotides or shorter than 8 nucleotides. This technique does offer the advantage of being able to identify uridine and guanosine stretches, which sometimes tail poly(A) sequences on transcripts and may have important roles in regulation. In contrast to TAIL-seq, PAL-seq [9] is based on a single-end read approach which is analyzed in two directions. PAL-seq does not rely on directly sequencing the poly(A) tail. After transcript sequences are captured on the flow cell and sequence clusters are generated, transcript sequences are primed at the 3′ end immediately adjacent to the poly(A) tail. The first half of the PAL-seq reaction uses the poly(A) tail as a template to extended the primer sequence from the 5′ end meanwhile incorporating biotin-labeled dUTP into the reaction product. The second step uses the same primer for standard Illumina sequencing of 36 nucleotides for transcript identification. In the final step, the incorporated biotin is conjugated with a fluorescent dye. Fluorescent signal intensity is directly related to tail length through the incorporation of the biotin-labeled nucleotides. In theory, the PAL-seq approach could be used to measure poly(A) tails of any length, but this method could also be limited by the efficiency of biotin-labeled nucleotide incorporation during the primer extension step.
Sequences that control poly(A) tail length
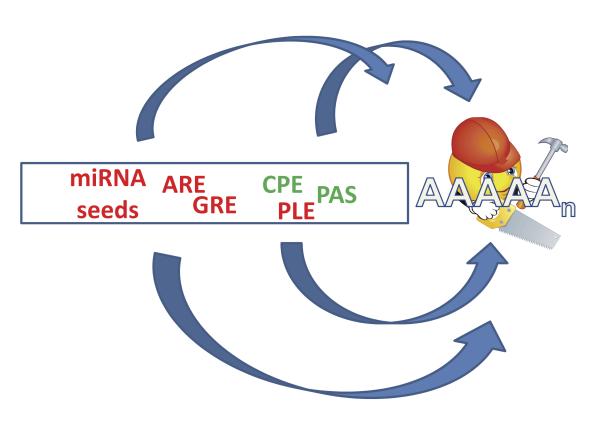
Rather than a simple static entity that marks the 3′ end of an mRNA, some evidence suggests that the poly(A) tail is a dynamic entity whose length can have a major impact on the biological fate of a transcript. The length of poly(A) tail added to an mRNA can be determined at the time of its initial synthesis or by remodeling post synthesis through the concerted action of deadenylases and poly(A) polymerases [42]. Individual mRNAs contain a series of signals that collectively influence the length of the poly(A) tail at a given time in cellular growth, differentiation and/or development. These sequence elements are outlined in Figure 1 and described below.
Figure 1. Numerous sequence elements in the 3’UTR of mRNAs influence poly(A) tail length.
Sequence elements that result in increased poly(A) tail length are indicated in green. Those that generally result in shorter or shortening of the poly(A) tail are indicated in red. ARE, Au-rich element; GRE, GU-rich element; CPE, cytoplasmic polyadenylation element; PLE, poly(A) limiting element; PAS, poly(A) signal (AAUAAA).
The process of cleavage/polyadenylation is regulated by specific sequences in the 3′ end of unprocessed transcripts that direct the actions of various proteins and protein complexes This process has been extensively reviewed previously [43]. Briefly, the canonical poly(A) signal (PAS) is an AAUAAA hexamer located 10-30 nt upstream of the poly(A) cleavage site [44]. An approximately 5 base U-rich or GU-rich element (DSE) located 15-30 bases downstream of the cleavage site works in concert with the PAS to determine the precise site of cleavage [45] . Upstream or downstream auxiliary elements can further enhance polyadenylation and 3′ end processing [46,47]. While the majority of poly(A) sites contain a canonical PAS hexamer (AAUAAA, AU/GUAAA or UAUAAA) multiple large-scale sequencing analyses have revealed that an estimated 8 to 18% do not, indicating the possibility of uncharacterized signal sequences [48–50]. For some genes, the presence of an A-rich sequence upstream of a robust DSE is sufficient for inducing polyadenylation [51]. Identifying variations of polyadenylation signal sequences that influence 3′ end processing efficiency has become increasingly important in recent years as the large extent and dramatic regulation of poly(A) site choice in most mRNAs has been realized [52,53].
Interestingly, there is in fact evidence for sequence elements that specify poly(A) tail length to favor the formation of short poly(A) tails on nascent mRNAs. The first evidence of a cis-acting element that limits poly(A) tail length was described in albumin mRNA from Xenopus. In contrast to most eukaryotic mRNAs, Xenopus albumin has a mere 17 nt poly(A) tail. Despite its minimal poly(A) tail length, the mRNA stability and translation efficiency of the albumin transcript are not significantly altered compared to typical mRNAs [54]. Additionally, the albumin mRNA poly(A) tail is not shortened simply as a result of cytoplasmic deadenylation; the albumin pre-mRNA receives a short poly(A) tail during transcriptional 3′ end processing [55].
Two poly(A)-limiting elements (PLE A and PLE B) regulate the albumin mRNA short poly(A) tail. The PLE is composed of a pyrimidine-rich region followed by an AG dinucleotide located in the last exon. To determine if PLEs are specific to albumin, the Schoenberg laboratory analyzed transferrin mRNA, another highly abundant liver transcript with a short poly(A) tail. Transferrin mRNA contains a sequence similar to the albumin PLE B located in the terminal exon that specifies a short poly(A) tail. Using PLE B as a query sequence, analysis of ESTs from multiple species uncovered putative PLEs in numerous other transcripts, including those encoding zinc finger transcription factor genes. Further analysis of the HIV-EB/Schnurri-2 zinc finger mRNA uncovered a functional PLE that confers a short poly(A) tail during nuclear processing in Jurkat cells [56]. In addition, the PLE was found to interact with the U2 snRNP auxiliary factor (U2AF), a nuclear protein involved in splicing [57]. To our knowledge, the PLE is thus far the only sequence attributed to specifically regulating short poly(A) tail length during nuclear 3′ end processing. There are likely numerous other transcripts with short poly(A) tails that have not specifically been shown to contain a pyrimidine-rich PLE-type sequence element. Notably, terminal uridylation may help stabilize these short poly(A) tails on mRNAs [58]. Further investigation to uncover specific cis-acting elements that produce transcripts with short poly(A) tails could provide key information on how such sequences affect polyadenylation dynamics.
As mentioned previously, the poly(A) tail is not static, but can undergo remodeling in the cytoplasm as is the case for oocytes, embryonic cells, and neurons [20]. The cytoplasmic polyadenylation element (CPE) is the cis-acting element that directs cytoplasmic polyadenylation in conjunction with the poly(A) signal. The CPE has been best characterized in Xenopus oocytes and can be a UA-rich sequence (UUUUA1-3U) that directs polyadenylation during the maturation of oocytes or a U-rich sequence of up to 18 U-residues that leads post-fertilization poly(A) tail elongation [59,60]. Analogous to the multiple sequences governing nuclear polyadenylation, an additional C-rich element has also been identified in Xenopus oocytes that acts in conjunction with the UA-rich CPE to regulate cytoplasmic polyadenylation [61]. Other cis-elements that direct cytoplasmic polyadenylation include the MSI-binding element (MBE), which interacts with Musashi (MSI1) [62], and the translation control sequence (TCS) [63]. As with the CPE, the MBE and the TCS also require the PAS to induce polyadenylation in the cytoplasm. Both the MSE and the TCS interact with proteins to prevent translation until oocyte maturation [20]. Cytoplasmic polyadenylation is a critical mechanism for regulating translation in cells that are no longer transcriptionally active, such as oocytes, or to induce the localized translation seen at neuronal synapses [64]. As of yet there is no concrete evidence suggesting that cytoplasmic polyadenylation is widely used in somatic cells as a means of post-transcriptional regulation of gene expression; however, improvements in the technology to detect and measure changes in poly(A) tail status on a global scale may reveal otherwise.
While there are clearly multiple cis-acting elements that control polyadenylation, numerous other sequences are important for directing the removal of the poly(A) tail, generally to induce mRNA degradation. The 3′ UTRs of numerous mRNAs contain adenine-uridine rich elements (AREs) (composed of a multiple AUUUA pentamers within a U-rich region or overlapping UUAUUUA(U/A)(U/A) nonamers [65,66] and guanosine-uridine rich elements (GREs) [67]. AREs and GREs have been demonstrated to induce rapid deadenylation and subsequent decay of mRNAs, which may be mediated by recruitment of the exosome [68] or 5′-3′ decay machinery [27]. The two classes of 3′ UTR elements serve as binding sites for a multitude of AU-binding proteins (AUBPs) or GU-binding proteins, which can enhance or prevent mRNA decay, in some cases specifically via interaction with the poly(A) tail [65,69,70].
MicroRNA binding sites are also 3′UTR sequences involved in post-transcriptional regulation and have been extensively reviewed elsewhere [71]. The translational silencing that occurs when miRNAs bind to their target transcripts is generally attributed to induction of deadenylation-mediated decay [72]. It is interesting to note that miRNAs can also interact with AUBPs to have complementary or antagonistic effects on mRNA repression [73,74].
Finally, C-rich elements (CREs) are additional well-characterized sequences in the 3′UTRs of mRNAs that recruit poly-C binding proteins (PCBPs) and have myriad of functions including inducing mRNA stability and activating or silencing translation [75,76]. Members of the PCBP family can also interact with poly(A) tail-associated factors to exert their effects on mRNAs with CREs [76,77].
In summary, the multitude of sequences involved in determining how, when, where and to what extent polyadenylation occurs suggest that poly(A) tail is more than just a string of A-residues added to nearly every transcript during 3′ end processing. Considering the obvious homogeneity of the sequence of the tail itself, altering length would seem to be the only way to utilize the poly(A) tail as a regulatory element. Therefore, the presence of specific sequence elements (and associated trans-acting factors) in an mRNA could be critical for carefully governing perhaps the most dynamic and remarkable aspect of the poly(A) tail—its size.
Nuclear factors that regulate the poly(A) tail
Proteins are integral to all aspects of polyadenylation, beginning with the addition of the poly(A) tail in the nucleus during transcription. The PAS directs binding and assembly of the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulation factor (CstF), and other cleavage factors, which clip the 3′ end of the unprocessed transcript at the polyadenylation site [42,46]. The polyadenylate polymerases (PAPs) are also part of the cleavage machinery and slowly add the A-residues to form the initial poly(A) tail. The nuclear poly(A)-binding protein (PABPN1) binds to the newly formed poly(A) tail and to allow rapid addition of A-residues until the tail is about 200-250 nt in length [42]. Importantly, PABPN1 and CPSF are involved in controlling the number of A-residues added as demonstrated by siRNA-induced knockdown of PABPN1, which leads to shortened poly(A) tails [78,79]. PABPN1 and PAP can also stimulate hyperadenylation to signal nuclear mRNA decay by the exosome [10].
Once the poly(A) tail reaches its critical length (about 250 nt), the poly(A) protein complex is disrupted, eliminating the interaction between CPSF and the PAPs, and thus ending processive polyadenylation [80]. Two additional factors may be involved in determining the initial length of the poly(A) tail. First, during the termination steps of polyadenylation, nucleophosmin (NPM1 or B23) is deposited on the 3′UTR of the transcript near the PAS [81]. NPM1 interacts with the core CPSF complex in an RNA-independent fashion [82]. Interestingly, NPM1 deposition appears to play a significant role in regulating proper poly(A) tail length, as knocking down NPM1 leads to hyperadenylation and nuclear accumulation of mRNAs [82]. Thus NPM1 has been postulated to serve as a mark for transcripts that have been properly polyadenylated. Second, the Nab2 yeast protein and its human counterpart ZC3H14 are Cys3His-type zinc finger proteins that interact with multiple factors of the polyadenylation machinery to strictly regulate the poly(A) tail length. One model suggests that Nab2/ZC3H14 restricts PAP activity and that it may also recruit ribonucleases to trim excess A-residues [83]. Nab2/ZC3H14 , NPM1 and PABPN1 remain bound at or near the poly(A) tail and assist in the translocation of mRNAs from the nucleus to cytoplasm where transcripts interact with multiple other factors to become primed for translation [79,82,84].
U2AF is the trans-acting factor shown to have a key role in regulating the addition of short poly(A) tails to nascent transcripts containing the poly(A)-limiting element [57]. U2AF is a nuclear protein composed of 65 and 35 kDa subunits that is involved in modulating 3′ end formation via its interaction with PAP [85]. Notably, U2AF was also found to bind mRNAs containing the poly(A)-limiting element [57]. Overexpression of U2AF65 leads to a dominant negative effect on the ability of the PLE to limit poly(A) tail length, but has no effect on the heterogeneous poly(A) tails of the control lacking the PLE. Mutations in the U2AF site that interacts with PAP did not affect poly(A) tail length in PLE-containing transcripts. In addition, when U2AF with a dysfunctional RNA-binding domain was overexpressed, it did not have an effect on PLE regulation of poly(A) tail length as compared to the wild-type, suggesting that U2AF competes with another PLE-binding protein. A C14G mutation in U2AF that enhances its binding to the 3′ splice site caused a mild increase in poly(A) tail length of PLE mRNAs, but monomeric U2AF65 and the PAP-interacting domain mutant version returned short poly(A). The C12G mutation therefore may increase U2AF’s binding affinity to the PLE, thereby preventing the unknown poly(A) regulatory protein from binding. The current hypothesis is that in the context of the PLE, U2AF may recruit another yet unknown protein responsible for limiting poly(A) tail length [57].
While ARE-binding proteins are involved in multiple aspects of cytoplasmic regulation of mRNAs and have been reviewed extensively [65,66,86,87], some AUBPs also contribute to restricting poly(A) tail length in the nucleus. Tristetraprolin (TTP) is notorious for promoting cytoplasmic decay of mRNAs containing AREs, but in the nucleus it can directly interact with PABN1 and PAP to inhibit polyadenylation, potentially contributing to a decrease in nuclear export of mRNAs [70,88]. The Hu proteins, which stabilize mRNAs in the cytoplasm, interact with CstF and CPSF during transcriptional 3′ end processing to prevent polyadenylation at poly(A) sites containing U-rich sequences [89]. There are undoubtedly additional RNA-protein interactions that influence polyadenylation that remain to be characterized.
Cytoplasmic factors that regulate the poly(A) tail
In the cytoplasm, another series of factors interacts with the poly(A) tail to regulate its length, which can have effects on translation and deadenylation-mediated mRNA decay. The cytoplasmic poly(A)-binding protein (PABPC1) binds to the poly(A) tail and forms a complex with translation initiation factor eIF4G and the cap-binding protein eIF4E. This mRNA-protein complex forms a circular structure, which facilitates recruitment of the 40S ribosomal subunit and initiates translation [80,90]. Translation terminates when eRF1 recognizes the stop codon and mediates the ribosomal release of the polypeptide chain while eRF3 utilizes GTP to catalyze the termination reaction [91]. Deadenylation can also be coupled to translation termination via competitive binding of eRF3 and the PAN3 deadenylase to the C-terminal domain of PABCP1. After eRF3 is released from PABPC1, the PAN2-PAN3 deadenylase is able to bind and recruit the CAF1-CCR4-NOT deadenylation complex leading to removal of the poly(A) tail, which stimulates 3′-to-5′ mRNA decay by the exosome [24,92] or decapping and 5′-to-3′ decay [27].
Multiple other trans-acting factors can augment or impede poly(A) tail shortening through interactions with specific sequence elements and/or the protein components of the decay machinery. Briefly, AUBPs regulate mRNAs that contain AREs within their 3′UTRs by either enhancing decay or increasing stability of targeted transcripts [65]. The RNA-binding protein HuR (ELAV1) generally stabilizes ARE-containing mRNAs [87]. HuR and other ELAV-like proteins can concurrently bind ARE sites and poly(A) tails >70 nt long on target mRNAs [93]. Unexpectedly, HuR binding to poly(A) tails does not appear to prevent removal of the poly(A) tail, but it does delay mRNA decay [94]. TTP and the related zinc finger protein Brf-1 bind to ARE sites and recruit the enzymes responsible for decapping, deadenylation and exonucleolytic activity, leading to mRNA decay [86]. A possible explanation of how TTP family proteins contribute to shortening of the poly(A) tails on ARE-containing mRNAs is through their ability to stimulate PARN in cell-free systems to enhance deadenylation [95] or by interacting with CCR4-NOT [96]. Multiple short-lived mRNAs contain GU-rich elements in their 3′ UTRs, which serve as docking sites for CUG-binding protein 1 (CUGBP1 or CELF1), a member of the CELF RNA-binding protein family. CUGBP-1/CELF1 can specifically target mRNAs containing GREs to mediate poly(A) tail shortening via recruitment of PARN and mRNA decay [97,98].
MicroRNAs are small endogenous RNAs that target mRNAs containing specific miRNA seed sequences and leading to gene silencing, generally by inducing mRNA decay, previously reviewed in [71]. Once bound to the target transcripts, miRNAs recruit the RNA-induced silencing complex (RISC). Argonaute, one of the RISC proteins, can induce endonucleolytic cleavage to cause degradation, but this pathway is not commonly utilized in animal cells [99]. Instead, miRNAs in animal cells are associated with rapid deadenylation of their target mRNAs through the CAF1-CCR4-NOT1 complex, thus triggering mRNA decay [100,101]. Other components of RISC, the GW182 proteins (called TNRC proteins in mammalian cells), directly interact with PAN3 of the PAN2-PAN3 deadenylation complex and NOT1 to induce poly(A) tail shortening of miRNA targets [102]. The GW182 proteins also appear to cause PABP to dissociate from mRNA targets to disrupt the translation complex and enhance deadenylation [103]. These results indicate that deadenylation is not a side-effect of miRNA binding, but that members of the RISC complex actually recruit factors involved in removal of the poly(A) tail [99]. Notably, miRNAs can also induce gene silencing independent of a poly(A) tail and the eIF4E/eIF4G/PABP translation complex, which would suggest that miRNAs could also repress mRNAs with shortened (A) tails or alternative 3′ end modifications [101,103].
The poly-C binding proteins (PCBPs) are trans-acting factors that stabilize mRNAs in the cytoplasm through the poly(A) tail. Two representatives of this group of proteins are αCP1 and αCP2, which contribute to a ribonucleoprotein complex that enhances the stability of α-globin mRNA. The removal of the αCP ribonucleoprotein complex from α-globin mRNA enhanced deadenylation and mRNA decay. In addition, the αCPs are associated with PABP, which may account for the prevention of deadenylation and the increased mRNA stability [76].
In oocytes, embryonic cells, and neurons, a long poly(A) tail is initially added to mRNAs during 3′ end processing, but is removed from mRNAs in the cytoplasm as a means of controlling translation [64]. The cytoplasmic polyadenylation element binding protein (CPEB) is the main factor involved in the various stages polyadenylation regulated translation in the cytoplasm. The CPEB is a zinc-finger containing protein with an RNA recognition motif (RRM) that binds to the CPE consensus sequence in the 3′UTR. Once bound to its target mRNA, CPEB forms a complex with symplekin, CPSF, the poly(A) ribonuclease (PARN) deadenylase, and germ-line development factor 2 (Gld2), a poly(A) polymerase. PARN removes the A-residues added by Gld2 to maintain the short (A) tail, until the activation of a cell-signaling pathway that phosphorylates CPEB leading to release of PARN from the complex. Without PARN, Gld2 is able to elongate the poly(A) tail, thereby stimulating translation [104]. Interestingly, CPEB has recently been implicated as a major factor in cancer, senescence, and neuronal functions.
PABPC4 is a minor cytoplasmic poly(A)-binding protein isoform that has dissimilar binding specificity and function than the far more prevalent PABPC1. Reduction of PABPC4 levels in mouse eythroleukemia (MEL) cells, used as a model of erythroid differentiation, altered the steady-state expression of mRNAs that contain an AU-rich region within the 3′UTR. Affected transcripts included those involved in cell growth, metabolism and erythroid differentiation. In addition, PABPC4 depletion by shRNA altered the ratio of long to short poly(A) tails, leading to a decreased abundance of mRNAs with tails having fewer than 30 A-residues. These results suggest that PABPC4 may protect mRNAs with critically short tails from mRNA decay [105].
In summary, trans-acting factors, including proteins and microRNAs, are critical in regulating the poly(A) tail and its role in the mRNA life cycle. While there are numerous proteins involved in polyadenylation, deadenylation and the regulatory steps in between that have already been described, more trans-acting factors that interact with the poly(A) tail will undoubtedly be uncovered. As with the sequences, the number and complexity of the transacting factors that regulate the poly(A) tail suggest that it is a key aspect of post-transcriptional regulation. The poly(A) tail is seemingly subject to major and minor changes in length that may ultimately have significant effects on post-transcriptional regulation of gene expression.
Some Final Thoughts on the Dynamics of Poly(A) Tail Size
Interestingly, if PABPN1 is knocked down in myoblasts, poly(A) tails are shortened and proliferation and differentiation are decreased in these cells [78]. A key point in understanding poly(A) tail dynamics, however, is that bigger is not always better. Hyperadenylation, as seen in viral infection or NPM-1 depletion, leads to nuclear retention of mRNAs, which would reduce the ability of these mRNAs to be translated and could have other negative effects on nuclear activity [10,11,78,82]. Hyperadenylation is also associated with increased mRNA turnover rates [10]. Curiously, increased PAP activity has been noted in multiple tumor types and can be associated with a worse prognosis in certain cancers [106]. This implies that longer poly(A) tails could result from increased enzymatic activity and will dysregulate normal cellular functions.
A second key point is that reduction of poly(A) tail length can have beneficial and very significant functions in cell biology. In embryos, short poly(A) tails are necessary to repress translation until the appropriate stage of development is reached [9]. In addition, short poly(A) tails appear to denote mRNAs that are critical for early development, and may be a way to regulate translation in a dose and time-dependent manner [107]. Thus poly(A) tail length control and development go hand in hand. Interestingly, cordycepin and other inhibitors of PAP activity can reduce the induction of inflammatory genes, presumably by decreasing polyadenylation efficiency [108]. Polyadenylation inhibitors can also decrease cell proliferation and increase apoptosis in cancer cells [109,110]. Clearly, proper regulation of poly(A) tail length is important for maintaining appropriate biological behavior in cells, but whether tails need to be shorter or longer appears to be transcript-specific.
Based on the findings from multiple genome wide sequencing analyses, a significant population of mRNAs (other than those encoding histones) have very short or absent poly(A) tails, and not merely as a result of deadenylation. Thus a third key point is that we need a clearer understanding of the sequence elements that coordinate poly(A) tail length. While there is evidence of sequence and protein-mediated generation of mRNA populations with short poly(A) tails, there are still many unknowns as to how and why some mRNAs avoid typical nuclear polyadenylation. The PLE has been found in several transcripts with short poly(A) tails and continued searching could reveal its presence in many other mRNAs. It is also entirely possible that there are other regulatory sequence elements that act in coordination with or in place of the PLE. Protection from decay could be a motivation for an mRNA to maintain a short poly(A) tail. Perhaps cytoplasmic transcripts with short enough (A) tails are not detected by the deadenylases and thus completely avoid deadenylation-mediated decay. In addition, RNA-binding proteins that are recruited by a PLE-like sequence may protect the transcript from mRNA decay [56,57]. There could also be structural features of the mRNA that resist exonucleases, such as circularization or secondary folding structures in the 5′ or 3′ UTRs. Terminal uridylation, for example, has recently been implicated in stabilizing short poly(A) tails [58]. Finally, perhaps the short poly(A) tail specifies transport to a particular subcellular localization where these mRNAs are essentially in limbo and are protected from degradation and are not undergoing translation.
Lastly, from a functional standpoint, the length of the poly(A) tail could identify a portion of specific mRNAs that may not be used to encode proteins, but rather these transcripts could have an alternate function in the cell. As long as their 3′ ends were stabilized in some fashion (triple helices, terminal uridylation, etc), such mRNAs would not necessarily need a poly(A) tail of significant size to carry out their regulatory functions. For example, competing endogenous RNAs (ceRNAs) can act as microRNA sponges to prevent miRNA-mediation post-transcriptional regulation [111,112]. Specific mRNAs destined to serve as primary-miRNA precursors in the nucleus that are subject to DROSHA processing also may not necessarily need a significant poly(A) tail. Other possible functions of mRNAs with short poly(A) tails include acting as RNA scaffolds, targeting other transcripts for trans-regulation by staufen-mediated decay [113] and other processes [114] and by behaving as RNA-based protein sponges [115]. Clearly there may be a large potential for novel insights into gene regulation by additional experimentation in the area of poly(A) tail dynamics.
Highlights.
mRNA poly(A) tails naturally occur in a variety of lengths
Accurately defining poly(A) lengths for individual mRNAs has been challenging
Numerous sequence elements and factors determine poly(A) size in mammalian cells
Poly(A) length dynamics may play a key role in several aspects of cell biology
Acknowledgements
Studies on RNA stability and polyadenylation in the Wilusz laboratory have been funded by the NIH (GM072481) and NSF (MCB-1301983).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. doi:10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Natalizio BJ, Wente SR. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 2013;23:365–373. doi: 10.1016/j.tcb.2013.03.006. doi:10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fuke H, Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2007;36:1037–1049. doi: 10.1093/nar/gkm1120. doi:10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. Rna New York N. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Richter JD. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. doi:10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- [6].Wu H-Y, Ke T-Y, Liao W-Y, Chang N-Y. Regulation of Coronaviral Poly(A) Tail Length during Infection. Plos One. 2013;8:e70548. doi: 10.1371/journal.pone.0070548. doi:10.1371/journal.pone.0070548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Svitkin YV, Yanagiya A, Karetnikov AE, Alain T, Fabian MR, Khoutorsky A, et al. Control of Translation and miRNA-Dependent Repression by a Novel Poly(A) Binding Protein, hnRNP-Q. Plos Biol. 2013;11:e1001564. doi: 10.1371/journal.pbio.1001564. doi:10.1371/journal.pbio.1001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rutledge CE, Lau H-T, Mangan H, Hardy LL, Sunnotel O, Guo F, et al. Efficient Translation of Dnmt1 Requires Cytoplasmic Polyadenylation and Musashi Binding Elements. Plos One. 2014;9:e88385. doi: 10.1371/journal.pone.0088385. doi:10.1371/journal.pone.0088385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. doi:10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bresson SM, Conrad NK. The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay. Plos Genet. 2013;9:e1003893. doi: 10.1371/journal.pgen.1003893. doi:10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kumar GR, Glaunsinger BA. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol. Cell. Biol. 2010;30:4996–5008. doi: 10.1128/MCB.00600-10. doi:10.1128/MCB.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen C-YA, Shyu A-B. Mechanisms of deadenylation-dependent decay, Wiley Interdiscip. Rev. Rna. 2011;2:167–183. doi: 10.1002/wrna.40. doi:10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Darnell JE, Philipson L, Wall R, Adesnik M. Polyadenylic acid sequences: role in conversion of nuclear RNA into messenger RNA. Science. 1971;174:507–510. doi: 10.1126/science.174.4008.507. [DOI] [PubMed] [Google Scholar]
- [14].Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. doi:10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. doi:10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang L, Duff MO, Graveley BR, Carmichael GG, Chen L-L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. doi:10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chang H, Lim J, Ha M, Kim VN. TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications. Mol. Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. doi:10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- [18].Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell Cycle Georget. Tex. 2010;9:4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. doi:10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation: Factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev. Rna. 2013;4:437–461. doi: 10.1002/wrna.1171. doi:10.1002/wrna.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. doi:10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions: Poly(A) RNA-binding proteins and polyadenosine RNA. Wiley Interdiscip. Rev. Rna. 2014 doi: 10.1002/wrna.1233. n/a–n/a. doi:10.1002/wrna.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schmidt K, Butler JS. Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip. Rev. Rna. 2013;4:217–231. doi: 10.1002/wrna.1155. doi:10.1002/wrna.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. doi:10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- [25].Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. Embo J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. doi:10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chowdhury A, Raju KK, Kalurupalle S, Tharun S. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1-7-Pat1 complex. Rna New York N. 2012;18:936–944. doi: 10.1261/rna.029876.111. doi:10.1261/rna.029876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012 doi: 10.1038/nrg3160. doi:10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- [29].Hilleren P, Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. Rna New York N. 2001;7:753–764. doi: 10.1017/s1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wilusz JE, JnBaptiste CK, Lu LY, Kuhn C-D, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. doi:10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. doi:10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Conrad NK. The emerging role of triple helices in RNA biology: The emerging role of RNA triple helices. Wiley Interdiscip. Rev. Rna. 2014;5:15–29. doi: 10.1002/wrna.1194. doi:10.1002/wrna.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Broitman SL, Im DD, Fresco JR. Formation of the triple-stranded polynucleotide helix, poly(A.A.U) Proc. Natl. Acad. Sci. U. S. A. 1987;84:5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sippel AE, Stavrianopoulos JG, Schutz G, Feigelson P. Translational properties of rabbit globin mRNA after specific removal of poly(A) with ribonuclease H. Proc. Natl. Acad. Sci. U. S. A. 1974;71:4635–4639. doi: 10.1073/pnas.71.11.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sallés FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods San Diego Calif. 1999;17:38–45. doi: 10.1006/meth.1998.0705. doi:10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- [36].Murray EL, Schoenberg DR. Methods Enzymol. Elsevier; 2008. Chapter 24 Assays for Determining Poly(A) Tail Length and the Polarity of mRNA Decay in Mammalian Cells; pp. 483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. ePAT: A simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA. 2012;18:1289–1295. doi: 10.1261/rna.031898.111. doi:10.1261/rna.031898.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Patil DP, Bakthavachalu B, Schoenberg DR. Poly(A) polymerase-based poly(A) length assay. Methods Mol. Biol. Clifton Nj. 2014;1125:13–23. doi: 10.1007/978-1-62703-971-0_2. doi:10.1007/978-1-62703-971-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Beilharz TH, Preiss T. Transcriptome-wide measurement of mRNA polyadenylation state. Methods. 2009;48:294–300. doi: 10.1016/j.ymeth.2009.02.003. doi:10.1016/j.ymeth.2009.02.003. [DOI] [PubMed] [Google Scholar]
- [40].Beilharz TH, Preiss T. Polyadenylation state microarray (PASTA) analysis. Methods Mol. Biol. Clifton Nj. 2011;759:133–148. doi: 10.1007/978-1-61779-173-4_9. doi:10.1007/978-1-61779-173-4_9. [DOI] [PubMed] [Google Scholar]
- [41].Zheng D, Tian B. Sizing up the poly(A) tail: insights from deep sequencing. Trends Biochem. Sci. 2014 doi: 10.1016/j.tibs.2014.04.002. doi:10.1016/j.tibs.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. Rna. 2011;2:348–361. doi: 10.1002/wrna.56. doi:10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- [43].Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. Rna. 2012;3:385–396. doi: 10.1002/wrna.116. doi:10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. Mmbr. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. doi:10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen F, Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. doi:10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Beaudoing E. Patterns of Variant Polyadenylation Signal Usage in Human Genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. doi:10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tian B. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. doi:10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kamasawa M, Horiuchi J. Identification and characterization of polyadenylation signal (PAS) variants in human genomic sequences based on modified EST clustering. Silico Biol. 2008;8:347–361. [PubMed] [Google Scholar]
- [51].Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. Embo J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. doi:10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. doi:10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- [53].Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. doi:10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rao M. Regulated nuclear polyadenylation of Xenopus albumin pre-mRNA. Nucleic Acids Res. 1996;24:4078–4083. doi: 10.1093/nar/24.20.4078. doi:10.1093/nar/24.20.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Das Gupta J, Gu H, Chernokalskaya E, Gao X, Schoenberg DR. Identification of two cis-acting elements that independently regulate the length of poly(A) on Xenopus albumin pre-mRNA. Rna New York N. 1998;4:766–776. doi: 10.1017/s1355838298971837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gu H, Das Gupta J, Schoenberg DR. The poly(A)-limiting element is a conserved cis-acting sequence that regulates poly(A) tail length on nuclear pre-mRNAs. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8943–8948. doi: 10.1073/pnas.96.16.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gu H. U2AF modulates poly(A) length control by the poly(A)-limiting element. Nucleic Acids Res. 2003;31:6264–6271. doi: 10.1093/nar/gkg823. doi:10.1093/nar/gkg823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon J-M, et al. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41:7115–7127. doi: 10.1093/nar/gkt465. doi:10.1093/nar/gkt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Fox CA, Sheets MD, Wickens MP. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- [60].Simon R, Richter JD. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol. Cell. Biol. 1994;14:7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Paillard L, Maniey D, Lachaume P, Legagneux V, Osborne HB. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in Xenopus embryos. Mech. Dev. 2000;93:117–125. doi: 10.1016/s0925-4773(00)00279-3. doi:10.1016/S0925-4773(00)00279-3. [DOI] [PubMed] [Google Scholar]
- [62].Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. doi:10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Charlesworth A, Welk J, MacNicol AM. The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′-untranslated region. Dev. Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. doi:10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- [64].Weill L, Belloc E, Bava F-A, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. doi:10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- [65].Barreau C. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. doi:10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bakheet T. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. doi:10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Vlasova-St Louis I, Bohjanen PR. Post-transcriptional regulation of cytokine signaling by AU-rich and GU-rich elements. J. Interf. Cytokine Res. Off. J. Int. Soc. Interf. Cytokine Res. 2014;34:233–241. doi: 10.1089/jir.2013.0108. doi:10.1089/jir.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mukherjee D. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. Embo J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. doi:10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ma W-J, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. doi:10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Su Y-L, Wang S-C, Chiang P-Y, Lin N-Y, Shen Y-F, Chang G-D, et al. Tristetraprolin Inhibits Poly(A)-Tail Synthesis in Nuclear mRNA that Contains AU-Rich Elements by Interacting with Poly(A)-Binding Protein Nuclear 1. Plos One. 2012;7:e41313. doi: 10.1371/journal.pone.0041313. doi:10.1371/journal.pone.0041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. doi:10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Djuranovic S, Nahvi A, Green R. miRNA-Mediated Gene Silencing by Translational Repression Followed by mRNA Deadenylation and Decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. doi:10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. doi:10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- [74].Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. doi:10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- [75].Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. Rna New York N. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Morales J, Russell JE, Liebhaber SA. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- [78].Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 2010;19:1058–1065. doi: 10.1093/hmg/ddp569. doi:10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA. 2014;20:681–688. doi: 10.1261/rna.043984.113. doi:10.1261/rna.043984.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) Tail Length Is Controlled by the Nuclear Poly(A)-binding Protein Regulating the Interaction between Poly(A) Polymerase and the Cleavage and Polyadenylation Specificity Factor. J. Biol. Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. doi:10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Palaniswamy V, Moraes KCM, Wilusz CJ, Wilusz J. Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat. Struct. Mol. Biol. 2006;13:429–435. doi: 10.1038/nsmb1080. doi:10.1038/nsmb1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. Embo J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. doi:10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Soucek S, Corbett AH, Fasken MB. The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim. Biophys. Acta Bba - Gene Regul. Mech. 2012;1819:546–554. doi: 10.1016/j.bbagrm.2012.03.006. doi:10.1016/j.bbagrm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bear DG, Fomproix N, Soop T, Björkroth B, Masich S, Daneholt B. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 2003;286:332–344. doi: 10.1016/s0014-4827(03)00123-x. doi:10.1016/S0014-4827(03)00123-X. [DOI] [PubMed] [Google Scholar]
- [85].Millevoi S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. doi:10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. doi:10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Damgaard CK, Lykke-Andersen J. Regulation of ARE-mRNA Stability by Cellular Signaling: Implications for Human Cancer. In: Wu JY, editor. Rna Cancer. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. pp. 153–180. [DOI] [PubMed] [Google Scholar]
- [88].Brooks SA, Blackshear PJ. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta Bba - Gene Regul. Mech. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. doi:10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhu H, Zhou H-L, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. doi:10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- [90].Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- [91].Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. Embo J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. doi:10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ma W-J, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. doi:10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Peng SS-Y. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. Embo J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. doi:10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and Its Family Members Can Promote the Cell-Free Deadenylation of AU-Rich Element-Containing mRNAs by Poly(A) Ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. doi:10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, et al. Structural basis for the recruitment of the human CCR4–NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. doi:10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Moraes KCM, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. Rna New York N. 2006;12:1084–1091. doi: 10.1261/rna.59606. doi:10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St. Louis-Vlasova IA, et al. Analysis of CUGBP1 Targets Identifies GU-Repeat Sequences That Mediate Rapid mRNA Decay. Mol. Cell. Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. doi:10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. doi:10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [100].Wu L. From the Cover: MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. doi:10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2008;15:21–32. doi: 10.1261/rna.1399509. doi:10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. doi:10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- [103].Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. doi:10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. doi:10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kini HK, Kong J, Liebhaber SA. Cytoplasmic Poly(A) Binding Protein C4 Serves a Critical Role in Erythroid Differentiation. Mol. Cell. Biol. 2014;34:1300–1309. doi: 10.1128/MCB.01683-13. doi:10.1128/MCB.01683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Scorilas A. Polyadenylate polymerase (PAP) and 3′ end pre-mRNA processing: function, assays, and association with disease. Crit. Rev. Clin. Lab. Sci. 2002;39:193–224. doi: 10.1080/10408360290795510. doi:10.1080/10408360290795510. [DOI] [PubMed] [Google Scholar]
- [107].Gohin M, Fournier E, Dufort I, Sirard M-A. Discovery, identification and sequence analysis of RNAs selected for very short or long poly A tail in immature bovine oocytes. Mol. Hum. Reprod. 2013 doi: 10.1093/molehr/gat080. doi:10.1093/molehr/gat080. [DOI] [PubMed] [Google Scholar]
- [108].Kondrashov A, Meijer HA, Barthet-Barateig A, Parker HN, Khurshid A, Tessier S, et al. Inhibition of polyadenylation reduces inflammatory gene induction. RNA. 2012;18:2236–2250. doi: 10.1261/rna.032391.112. doi:10.1261/rna.032391.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Thomadaki H, Scorilas A, Tsiapalis CM, Havredaki M. The role of cordycepin in cancer treatment via induction or inhibition of apoptosis: implication of polyadenylation in a cell type specific manner, Cancer Chemother. Pharmacol. 2008;61:251–265. doi: 10.1007/s00280-007-0467-y. doi:10.1007/s00280-007-0467-y. [DOI] [PubMed] [Google Scholar]
- [110].Imesch P, Hornung R, Fink D, Fedier A. Cordycepin (3′-deoxyadenosine), an inhibitor of mRNA polyadenylation, suppresses proliferation and activates apoptosis in human epithelial endometriotic cells in vitro. Gynecol. Obstet. Invest. 2011;72:43–49. doi: 10.1159/000322395. doi:10.1159/000322395. [DOI] [PubMed] [Google Scholar]
- [111].Ebert MS, Sharp PA. Emerging Roles for Natural MicroRNA Sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. doi:10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. doi:10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. Rna. 2013;4:423–435. doi: 10.1002/wrna.1168. doi:10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Reveal B, Yan N, Snee MJ, Pai C-I, Gim Y, Macdonald PM. BREs mediate both repression and activation of oskar mRNA translation and act in trans. Dev. Cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. doi:10.1016/j.devcel.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. Changes in Cellular mRNA Stability, Splicing, and Polyadenylation through HuR Protein Sequestration by a Cytoplasmic RNA Virus. Cell Reports. 2013;5:909–917. doi: 10.1016/j.celrep.2013.10.012. doi:10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]