Abstract
In mammals, the vast majority of transcripts expressed are noncoding RNAs, ranging from short RNAs (including microRNAs) to long RNAs spanning up to hundreds of kb. While the actions of microRNAs as destabilizers and repressors of the translation of protein-coding transcripts (mRNAs) have been studied in detail, the influence of microRNAs on long noncoding RNA (lncRNA) function is only now coming into view. Conversely, the influence of lncRNAs upon microRNA function is also rapidly emerging. In some cases, lncRNA stability is reduced through the interaction of specific miRNAs. In other cases, lncRNAs can act as microRNA decoys, with the sequestration of microRNAs favoring expression of repressed target mRNAs. Other lncRNAs derepress gene expression by competing with miRNAs for interaction with shared target mRNAs. Finally, some lncRNAs can produce miRNAs, leading to repression of target mRNAs. These microRNA-lncRNA regulatory paradigms modulate gene expression patterns that drive major cellular processes (such as cell differentiation, proliferation, and cell death) which are central to mammalian physiologic and pathologic processes. We review and summarize the types of microRNA-lncRNA crosstalk identified to-date and discuss their influence on gene expression programs.
1. Introduction
In mammalian cells, the vast majority of transcribed RNAs are noncoding [1–4]. Many of them are processed to generate small RNAs, including microRNAs, the best-known class of small RNAs, but also small interfering (si)RNAs, Piwi-interacting (pi)RNAs, transcription initiation (ti)RNAs, tRNA-derived stress-induced RNAs, and small nucleolar (sno)RNAs [5,6]. Other transcripts (long noncoding RNAs or lncRNAs) remain larger than 200 nucleotides in their mature form [7–9]. Through their interaction with DNA, RNA, and proteins, ncRNAs affect all levels of gene regulation, including chromatin remodeling, transcription, pre-mRNA splicing, mRNA turnover, mRNA translation, and protein stability [10–13]. It has recently become apparent that there is interesting cross-regulation between microRNAs and lncRNAs. In this review, we describe and discuss the mutual regulatory influence of mammalian microRNAs and lncRNAs.
1.1. MicroRNA biogenesis and function
The expression of microRNAs begins through transcription of a primary miRNA (pri-miRNA) that harbors the microRNA. The pri-miRNA is processed by a nuclear complex known as a microprocessor, which includes the DiGeorge syndrome critical region 8 (DGCR8) and the ribonuclease Drosha, to generate a miRNA precursor (pre-miRNA). After pre-miRNAs are exported to the cytoplasm, they are cleaved by the ribonuclease Dicer, yielding ~22-nt duplex RNAs; one strand of each duplex is loaded into the miRNA-containing ribonucleoprotein (RNP) complex RISC (RNA-induced silencing complex), which contains Argonaute (Ago) proteins [5,14]. Each of these complexes then targets a specific mRNA, generally (but not exclusively) at the mRNA 3′-untranslated region (UTR), forming a partial hybrid with the miRNA ‘seed’ region (nucleotides 2–7) or other regions of the miRNA [10,15,16]. MicroRNAs regulate the pattern of expressed proteins via a number of proposed mechanisms, most of them repressive. They can modulate gene transcription rates, inhibit the initiation and elongation of translation of target mRNAs, promote target mRNA decay, and reduce the stability of proteins newly synthesized from target mRNAs. They can also sequester target mRNAs in cytoplasmic regions (P-bodies) where translation and stability are diminished [11,17–21].
1.2. LncRNA biogenesis and function
The expression of lncRNAs is largely regulated like that of coding RNAs. LncRNAs are subject to transcriptional regulation, and some lncRNAs (although not all) are further regulated via splicing, processing at the 5′ and 3′ ends, and export to the cytoplasm [22,23]. Some lncRNAs can produce small peptides, but the vast majority of lncRNAs are not translated [24,25]. Their function has been tied to their modular structure, which permits them to interact specifically with proteins, DNA, and RNA [26]. Studies on the influence of lncRNAs on the patterns of expressed genes have revealed increasing richness and complexity of mechanisms through which lncRNAs control gene expression, at both transcriptional and post-transcriptional levels, as explained below.
The roles of lncRNAs in regulating gene transcription have been studied extensively. Since the early finding that chromatin was purified with extensive amount of RNAs [27], many lncRNAs have been found to interact with chromatin-modifying enzymes and to modulate the transcriptional activation or silencing of genes [28]. For example, the lncRNA XIST, expressed from one X chromosome, inactivates the other X chromosome via mechanisms that include, but are not restricted to, recruitment of PRC2 (Polycomb repressive complex) [29]. In mammalian cells, the lncRNA HOTAIR represses target gene expression by associating with PRC2 and LSD1 and triggering genome-wide histone H3 lysine-27 trimethylation (H3K27me3) and lysine 4 demethylation (H3K4me2), while a lncRNA transcribed from the CEBPA (CCAAT/enhancer binding protein α) locus activates CEBPA mRNA transcription by inhibiting DNMT1 (DNA cytosine-5-methyltransferase 1) [30–34]. In addition, p53-regulated lncRNAs (lincRNA-p21, PANDA, lincRNA-PINT) suppress target gene transcription by interacting with transcription factors (hnRNPK and NF-YA) or PRC2 [35–37]. The complete spectrum of mechanisms of through which lncRNAs affect transcription is not fully known.
Some lncRNAs have also been found to regulate gene expression post-transcriptionally [13]. Examples of this regulation include the lncRNA MALAT1 (metastasis-associated long adenocarcinoma transcript 1), which sequesters SR (Serine/Arginine) proteins to modulate pre-mRNA alternative splicing [38], and lncRNAs 1/2-sbsRNAs (half-Staufen 1-binding site RNAs), which promote target mRNA degradation by partial complementarity with target mRNAs and subsequent recruitment of Staufen 1 [39]. In contrast, perfect complementarity between an mRNA and its antisense lncRNA [β amyloid-cleaving enzyme (BACE)1 mRNA and BACE1AS (BACE1 antisense)] protects BACE1 mRNA from RNase cleavage and degradation [40]. LncRNAs have also been found to repress translation globally in neurons and germ cells (e.g., lncRNA BC1 [41]) or that of specific target mRNAs (lincRNA-p21, [42]). A translation-enhancing function was also reported for AS Uchl1 (ubiquitin carboxyl-terminal esterase L1 ubiquitin thiolesterase) through its complementarity with target Uchl1 mRNA and embedded SINEB2 repeat [43]. The post-transcriptional functions of lncRNAs are quickly coming to light.
A number of studies over the past decade have also begun to uncover the interaction among mammalian lncRNAs and miRNAs. LncRNAs are targeted by miRNAs to reduce lncRNA stability (section 2). LncRNAs can also function as molecular decoys or sponges of microRNAs, as proposed by the Pandolfi Group [44] and reported in a few circular RNAs studies (section 3). In addition, lncRNAs can also compete with miRNAs for binding to shared target mRNAs (section 4) and are precursors for the generation of miRNAs to silence target mRNAs (section 5). In this review, we summarize the recent progress in understanding the interplay between lncRNAs and miRNAs.
2. microRNA-triggered lncRNA decay
The abundance of numerous lncRNAs is controlled by microRNAs (Figure 1A). Since many of these lncRNAs affect different cell functions (proliferation, differentiation, senescence, apoptosis, transformation), changes in their abundance directly alter the cellular response in physiologic and pathologic processes.
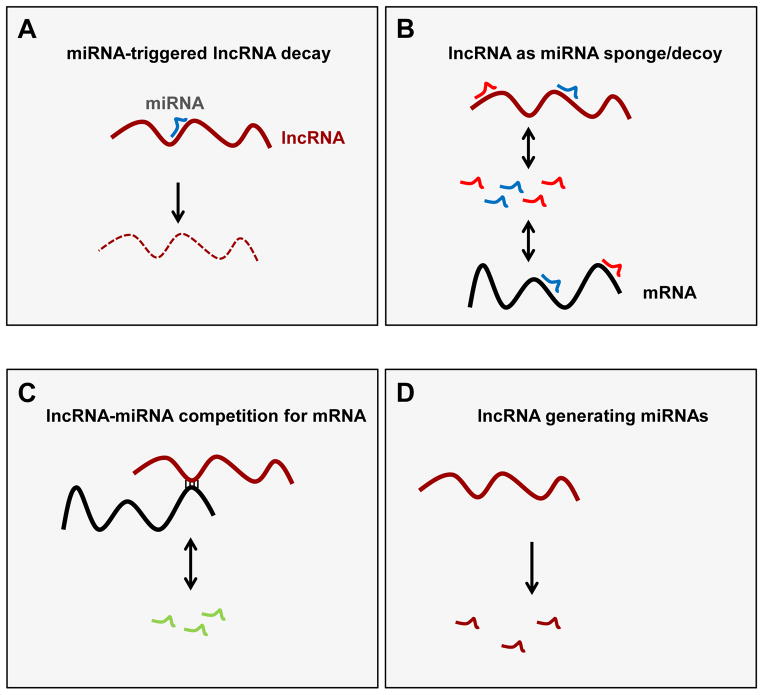
FIGURE 1. Modes of direct post-transcriptional interaction among lncRNAs and miRNAs.
Schematic of the major forms of interplay among lncRNAs and miRNAs. (A) Some lncRNAs are degraded by miRNAs, as described for lincRNA-p21 (degraded by let-7), HOTAIR (by let-7), MALAT1 (by miR-9), LOC285194 (by miR-211), PTCSC3 (by miR-574-5p), H19 (by let-7), and lincRNA-RoR (by miR145-5p and miR-181a-5p). (B) Other lncRNAs can serve as sponges/decoys for microRNAs, as described for linc-MD1 (sequestering miR-133 and miR-135), circular RNAs (sequestering miR-7, although many more examples are predicted to occur), lincRNA-RoR (miR-145-5p), and H19 (let-7). (C) A few examples of lncRNAs that compete with miRNAs for binding to mRNAs have also been reported, including BACE1 AS, which competes with miR-485-5p for binding to BACE1 mRNA, and ncNRFR, which competes with let-7 to derepress let-7 site-bearing mRNAs to promote carcinogenesis. (D) Several lncRNAs also produce microRNAs and other small RNAs, as shown for linc-MD1, which generates miR-206 and miR-133b, H19 (generates miR-675), and RMRP (generates miRNAs RMRP-S1 and RMRP-S2). See text for details.
LincRNA-p21
A recent report uncovered that the stability of lincRNA-p21, a lncRNA transcriptionally activated by p53, was regulated by changes in its turnover rate. The RNA-binding proteins (RBP) HuR and Ago2 (Argonaute 2, a component of the RISC), and the miRNA let-7b, all contributed to lowering lincRNA-p21 stability in human cervical carcinoma cells. HuR depletion stabilized lincRNA-p21, whereas let-7b overexpression facilitated lincRNA-p21 degradation; moreover, the HuR-mediated lowering of lincRNA-p21 half-life required the recruitment of let-7/Ago2 to the lncRNA, suggesting a cooperative mechanism of lincRNA-p21 repression by HuR and let-7b. The accumulation of lincRNA-p21 under reduced HuR or let-7b levels enhanced the formation of partially complementary hybrids with JUNB and CTNNB mRNAs, causing the repression of JunB and β-catenin translation [42].
HOTAIR
The recruitment of let-7 by HuR also caused decreased stability of another lncRNA, HOTAIR [45], suggesting that the mechanism of lncRNA decay through HuR-enhanced microRNA interactions may be widespread. This recruitment model remains to be elucidated in detail, but it may be linked to the ability of HuR to binds several miRNAs, including members of the let-7 family, and to promote the interaction between Ago2 and let-7b [45,46]. Under conditions of high abundance, HOTAIR functioned as a platform to foment the interaction of associated RBPs, as shown for two pairs of proteins: Ataxin-1 and Snurportin-1, the substrates of E3 ligases Dzip3 and Mex3b. With HOTAIR serving as a scaffold, the increased interaction of these pairs of proteins led to enhanced ubiquitination of Ataxin-1 and Snurportin-1. In addition, microRNA miR-34a was also shown to lower the stability of HOTAIR in the human prostate cancer cell lines PC3 and DU145 [47].
MALAT1
MALAT1 is also targeted by microRNA-Ago2-RISC in the human primary glioblastoma cell line U87MG and the human Hodgkin cell line L428 [48]. Silencing Ago2 or antagonizing miR-9 increased the steady state level of MALAT1, whereas miR-9 overexpression decreased it. Interestingly, the lowering of MALAT1 stability by miR-9 was mainly elicited in the nucleus, highlighting a nuclear decay-promoting function for this miRNA.
LOC285194
One of the p53-inducible lncRNAs, the tumor suppressor transcript LOC285194 is the target of Ago2 and miR-211 in the colon cancer cell line HCT-116 [49]. The downregulation of LOC285196 in colon cancer specimens was proposed to be triggered by miR-211, which shows enhanced levels in colon cancer. A reciprocal negative regulation of LOC285194 upon the levels of mature miR-211 was also hypothesized, perhaps with LOC285194 acting as a ‘sponge’ (below).
PTCSC3
Another cancer-related lncRNA, the tumor suppressor PTCSC3, is targeted by miR-574-5p. In thyroid cancer cell lines (BCPAP, FTC133, and 8505C), miR-574-5p overexpression reduced PTCSC3 expression levels [50]. Given that PTCSC3 overexpression arrests cells in G0/G1 and induces apoptosis, reduction in PTCSC3 levels by miRNAs might directly affect cell proliferation and tumorigenesis.
H19
One of the classic lncRNAs, H19 is predicted to have 4 let-7 target sites (let7a, let-7b, let-7g, let-7i) [51]. The findings that overexpressing let-7 in C2C12 myoblasts promoted myotube formation while depleting H19 increased it suggested that let-7 and H19 have opposite functions in myoblast differentiation [51].
LincRNA-RoR
LincRNA-RoR is also targeted by many miRNAs in human embryonic stem cells [52]. Among the putative regulators, overexpression of miR145-5p, miR-181a-5p or miR-99b-3p specifically decreased the activity of a reporter containing lincRNA-RoR sequences [52].
3. lncRNAs as miRNA sponges/decoys
The reciprocal influence, lncRNAs affecting the levels and function of microRNAs (Figure 1B), has also emerged in recent years. In particular, it has been proposed that the concentration of microRNAs in the cytoplasm (and likely the nucleus) can be titrated by the abundant lncRNAs that harbor similar microRNA target sequences and hence can sequester microRNAs away from mRNAs, as first described in plants [54]; these lncRNAs are known as competing endogenous (ce)RNAs [44,53,55]. As a group, mammalian lncRNAs antagonize miRNA function by titrating miRNAs for target mRNA binding during processes that include muscle differentiation and stem cell self-renewal.
PTENP1
The first example of ceRNAs was described by Poliseno and colleagues [56], who found that expression of the tumor suppressor PTEN (phosphatase and tensin homolog) was dependent on the expression levels of the transcript encoded by pseudogene PTENP1. The PTENP1 RNA elicited this influence by acting as a decoy for PTEN mRNA-targeting microRNAs. Accordingly, the PTENP1 locus is lost in many human cancers.
Linc-MD1
A muscle-specific lncRNA, linc-MD1 modulates muscle differentiation by competing with miRNAs for target mRNA binding [57]. Overexpression of linc-MD1 in mouse accelerated muscle differentiation whereas its depletion prevented it. Moreover, in keeping with the ceRNA mechanism of regulation, linc-MD1 enhanced expression of MAML1 and MEF2C mRNAs by reducing the availability of miR-133 and miR-135, respectively, from binding to target mRNAs in mouse and human myoblasts. Recently, a feed-forward regulatory loop was described for the RNA-binding protein HuR, a target of repression by miR-133. High levels of linc-MD1 in early stages of myogenesis sponges miR-133 away from HuR, allowing HuR to accumulate in cells and establish a myogenic gene expression program [58].
Circular RNAs
This class of long noncoding RNAs was first identified in adult mouse testis [59] but their function was unknown. Recent advances in high-throughput RNA sequencing (RNA-seq) technology characterized a variety of circular RNAs with the same sequence information in many model organisms and cell types. Circular RNAs are mainly generated as by-products of pre-mRNA splicing intermediates, although their biogenesis is not limited to protein-coding RNAs [60–63]. Once generated, these transcripts are strongly resistant to exonuclease digestion and to microRNA-mediated decay, suggesting that they collectively have very high stabilities, although circRNAs can be degraded by the presence of a perfectly matched miRNA target site [64]. One of the functions currently proposed for circular RNAs is the sequestration of miRNAs away from target mRNAs, thereby derepressing mRNAs that bear miRNA target sequences. Such function was reported for the human circRNA CDR1as (antisense to the cerebellar degeneration-related protein 1 transcript), which harbors a strikingly high number (greater than 70) of conserved binding sites for miR-7 [62], while the testis-specific circRNA Sry (sex-determining region Y) serves as a sponge for miR-138, prompting the hypothesis that circular RNAs might serve as efficient miRNA sponges [59]. Other potential functions of circular RNAs, as proposed by Hentze and Preiss [65], include serving as vehicles for miRNAs or RBPs, RBP sponges, assembly platforms for RBPs that have shared functions, allosteric regulators of RBPs, modulators of mRNA expression, and templates for translation.
LincRNA-RoR
In human embryonic stem cells, lincRNA-RoR was downregulated under differentiation conditions like the triggering of embryonic body formation, the removal of fibroblast growth factor (bFGF), and treatment with bone morphogenetic protein (BMP) 4 [52]. Under these conditions, lincRNA-RoR is not present in enough abundance to sequester miR-145-5p, a microRNA that targets mRNAs encoding the pluripotency proteins Nanog, Oct4, and Sox2. Interestingly, Nanog, Oct4, and Sox2 activate lincRNA-RoR transcription, so that human embryonic stem cells maintain their pluripotency through this self-regulatory loop [52].
H19
As indicated above, H19 also functions as an antagonist of let-7 during muscle differentiation [51]. H19 displays sequences partially complementary with let-7e, let-7g, and let-7i and is downregulated by these miRNAs. During differentiation of the mouse myoblast cell line C2C12, the steady state levels of H19 and let-7 increase along with those of myogenic factors MHC (myosin heavy chain) and Myogenin. Interestingly, H19 depletion or let-7 overexpression increased the levels of MHC mRNA, Myog mRNA, and Igf2, in turn promoting myotube formation.
4. lncRNAs competing with miRNAs for interaction with mRNAs
In addition to functioning as ceRNAs, lncRNAs can also compete with miRNAs for binding to target mRNAs (Figure 1C).
BACE1AS
The lncRNA BACE1AS, partially antisense to BACE1 mRNA (encoding the β-site amyloid precursor protein cleaving enzyme 1) promoted the stabilization of BACE1, which bears a miR-485-5p site [66]. In the human embryonic kidney HEK-293 cells, miR-485-5p overexpression reduced BACE1 mRNA levels; this decline was rescued by overexpressing BACE1AS, which is complementary to BACE1 mRNA in a region that includes the BACE1 mRNA miR-485-5p site. This finding suggests that BACE1AS protects BACE1 mRNA by preventing access of miR-485-5p for miRISC-mediated degradation.
ncNRFR
The tumor-promoting lncRNA ncNRFR (non-coding Nras functional RNA) contains a 22-nt sequence which is identical to let-7a and differs with other microRNAs (let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, and miR-98) by 1–4 nucleotides [67]. Overexpression of ncNRFR in the colonic epithelial cell line YAMC increased the activity of a heterologous reporter bearing a let-7 target site, suggesting that ncNRFR lowered let-7 function. However, the authors did not elucidate the underlying mechanism, nor did they find evidence that ncNRFR affected Nras mRNA abundance. Interestingly, YAMC cells overexpressing ncNRFR displayed higher levels of let-7 target mRNAs and were highly invasive when injected into nude mice, further suggesting that the tumorigenic function of ncNRFR was linked to its ability to suppress let-7 actions upon endogenous target mRNAs [67].
5. lncRNAs generating miRNAs
Finally, some lncRNAs are processed to generate miRNAs (Figure 1D). Linc-MD1 generates miR-206 and miR-133b from an intron and an exon, respectively, indicating additional regulatory mechanisms by linc-MD1 in muscle differentiation and muscle dystrophy [57]. LncRNA H19 can also generate miR-675, a process that is repressed by HuR [68], in turn suppressing translation of the Insulin Growth Factor Receptor (Igf1r) mRNA in mouse. Very recently, H19 knockdown in myoblasts was reported to decrease myogenesis; interestingly, this inhibition was attributed to the loss of miR-675, since ectopic restoration of miR-675 rescued muscle cell differentiation [69]. In addition, the RNA component of RNase MRP (RMRP), a lncRNA localized in mitochondria, also produces two miRNAs, RMRP-S1 and RMRP-S2; these microRNAs are putative regulators of PTCH2 and SOX4 mRNAs, encoding proteins that are linked to the human disease Cartilage Hair Hypoplasia (CHH) [70].
In sum, an expanding body of evidence reveals that lncRNAs and miRNAs work jointly to control gene expression via a number of complex post-transcriptional mechanisms (Figure 1). More examples of lncRNAs eliciting post-transcriptional actions by competing or cooperating with small RNAs are expected to emerge. Together, microRNAs and lncRNAs contribute to a robust and dynamic control of expressed proteins.
6. Concluding remarks and perspectives
MicroRNAs and lncRNAs regulate gene expression on all levels – transcriptional, post-transcriptional, and post-translational. Through this multi-leveled influence on protein expression patterns, these vast families of noncoding RNAs affect all aspects of cell metabolism, including cell division, senescence, differentiation, stress response, immune activation, and apoptosis. Here, we have reviewed the regulation of lncRNA expression and function by microRNAs, and vice versa. The examples described underscore the rich and complex regulation of gene expression resulting from the reciprocal regulation of ncRNAs.
The effects of microRNAs on lncRNAs (for example, in reducing lncRNA stability) were not entirely unexpected, since lncRNAs resemble mRNAs in many respects. Whether microRNAs also modulate the transcription or splicing of lncRNAs has not been reported. In addition, while lncRNAs are believed not to be translated, they are often found associated with polysomes [25,42,71,72]; whether or not microRNAs affect these interactions also warrants further study.
Further investigation of the effects of lncRNAs on microRNA levels, localization, and function promises to be particularly exciting. In addition to the mechanisms already recognized (e.g., lncRNA sponging microRNAs, serving as microRNA precursors, and competing with microRNAs for binding to mRNAs), one can envision many other functions for lncRNAs as modulators of microRNA biology. For example, lncRNAs could serve as platforms for presentation of microRNAs to RBPs or mRNAs, as means of intracellular microRNA transport, and even as vehicles for transport outside of the cell or between cells. In this regard, it is important to be reminded that some lncRNA functions are tied to their great stability (circular RNAs, for example) and in some cases their high abundance.
Another immediate consequence of the microRNA-lncRNA interplay is that one must consider additional layers of regulation by lncRNAs and microRNAs. Besides their direct actions upon target complementary mRNAs, pre-mRNAs, and DNA, indirect actions need to be carefully evaluated. For example, a microRNA that appears to repress transcriptionally the levels of an mRNA lacking a microRNA target site may in fact be lowering the abundance of a lncRNA required for the transcriptional activation of the mRNA in question. In another scenario, a lncRNA that appears to enhance the stability of a given mRNA may function indirectly by sponging a microRNA that would otherwise accelerate the decay of that mRNA. As many other mechanisms of indirect impact are possible, we should use extreme care and rigor when interpreting the consequences of lncRNA and microRNA actions on gene expression.
A full elucidation of the complex ncRNA regulatory circuits will require a great deal of additional work. Future studies must focus on lncRNA-microRNA interactions in animal models, on the joint consideration of lncRNA-microRNA when establishing linkages to disease, as well as on the analysis of other lncRNAs (e.g., very long lncRNAs, pseudogenes, antisense RNAs) and other small RNAs (e.g., piRNAs, siRNAs, snoRNAs). Given the unexpected consequences from lncRNA-miRNA interactions identified to-date, future studies of crossregulation among ncRNAs promises to be a thrilling journey.
Table 1. Examples of direct cross-regulation among lncRNAs and miRNAs.
LncRNAs (column 1) affecting the levels and/or activity of microRNAs (column 2) and vice versa are summarized (column 3); if known, the consequences on gene expression are also indicated (column 4).
| lncRNA | microRNA | Interplay | Consequences | References |
|---|---|---|---|---|
| lincRNA-p21 | let-7b | lncRNA decay | Translational repression of JUNB and CTNNB mRNAs | [42] |
| HOTAIR | let-7i | lncRNA decay | Ataxin-1 and Snurportin-1 ubiquitination | [45] |
| miR-34a | lncRNA decay | Tumorigenesis | [47] | |
| MALAT1 | miR-9 | lncRNA decay | Repression of MALAT1 | [48] |
| LOC285194 | miR-211 | lncRNA decay | Tumorigenesis | [49] |
| PTCSC3 | miR-574-5p | lncRNA decay | Tumorigenesis Proliferation | [50] |
| H19 | let7a,b,g,i | lncRNA decay, decoy | Decay of let-7 target mRNAs in muscle cells | [51] |
| miR-675 | miRNA production | Proliferation, myogenesis | [67,69] | |
| lincRNA-RoR | miR145-5p miR-181a-5p miR-99b-3p |
lncRNA decay miRNA competition lncRNA decay lncRNA decay |
Stability of Nanog, Oct4, and Sox2 mRNAs | 52] |
| Linc-MD1 | miR-133 miR-135 miR-206 miR-133b |
miRNA competition miRNA competition miRNA production miRNA production |
Muscle differentiation, and abundance of MAML1 and MEF2C mRNAs | [57] |
| CDR1as/ciRS-7 | miR-7 | miRNA sponge | Neuronal function | [60] |
| Sry | miR-138 | miRNA sponge | [59] | |
| BACE1AS | miR-485-5p | miRNA competition | Brain | [66] |
| ncNRFR | let-7b,c,d,e,f,g,i miR-98 |
miRNA competition miRNA competition |
Tumorigenesis | [67] |
| RMRP | RMRP-S1 RMRP-S2 |
miRNA production miRNA production |
CHH (Cartilage-hair hypoplasia) disease | [70] |
Highlights.
LncRNA stability and function can be potently regulated by miRNAs
LncRNAs can function as miRNA decoys, derepressing miRNA target mRNAs
LncRNAs can compete with miRNAs for degradation of target mRNAs
LncRNAs can give rise to miRNA
Acknowledgments
JHY, KA, and MG were supported by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Abbreviations
- ceRNA
competing endogenous RNAs
- lncRNA
long noncoding RNA
- lincRNA
large intergenic noncoding RNA
- miRNA
micro RNA
- RBP
RNA-binding protein
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermüller J, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17:852–64. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–32. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 11.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–22. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 13.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–30. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, et al. iR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′ UTR microRNA recognition elements. Mol Cell. 2009;35:610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 18.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 19.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 21.Morozova N, Zinovyev A, Nonne N, Pritchard LL, Gorban AN, Harel-Bellan A. Kinetic signatures of microRNA modes of action. RNA. 2012;18:1635–55. doi: 10.1261/rna.032284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 28.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 30.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–6. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CU, Tiedge H. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171:811–21. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 44.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long noncoding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, et al. PLoS One. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–87. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5:1143–6. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol Cell. 2013;52:101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Nat Genet. 2007;39:1033–7. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 55.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506–14. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–30. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 60.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 61.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 64.Hansen TB1, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–22. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–5. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Franklin JL, Rankin CR, Levy S, Snoddy JR, Zhang B, Washington MK, et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem Biophys Res Commun. 2013;440:99–104. doi: 10.1016/j.bbrc.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–65. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dey BK1, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet. 2014;23:368–82. doi: 10.1093/hmg/ddt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]