Abstract
Covalent labeling along with mass spectrometry is finding more use as a means of studying the higher order structure of proteins and protein complexes. Diethylpyrocarbonate (DEPC) is an increasingly used reagent for these labeling experiments because it is capable of modifying multiple residues at the same time. Pinpointing DEPC-labeled sites on proteins is typically needed to obtain more resolved structural information, and tandem mass spectrometry after protein proteolysis is often used for this purpose. In this work we demonstrate that, in certain instances, scrambling of the DEPC label from one residue to another can occur during collision-induced dissociation (CID) of labeled peptide ions, resulting in ambiguity in label site identity. From a preliminary study of over 30 labeled peptides, we find that scrambling occurs in about 25% of the peptides and most commonly occurs when histidine residues are labeled. Moreover, this scrambling appears to occur more readily under non-mobile proton conditions, meaning that low-charge state peptide ions are more prone to this reaction. For all peptides, we find that scrambling does not occur during electron transfer dissociation, which suggests that this dissociation technique is a safe alternative to CID for correct label site identification.
Introduction
Covalent labeling along with mass spectrometry is being increasingly used to study higher order protein structure and protein-protein complexes [1-9]. Covalent labels that are typically used either label specific amino acid residues (e.g. succinimides for lysines) or a range of amino acid residues (e.g. hydroxyl radicals). We have recently shown that diethylpyrocarbonate (DEPC) is a promising reagent molecule because of its ability to modify a wide range of amino acids [1, 10-14]. This potential to modify numerous amino acids enables DEPC to probe approximately 30% of the average protein [14]. Usually, proteins that are labeled with DEPC are then subjected to proteolysis so that the modification sites can be pinpointed to individual residues, thereby improving the resolution of this method.
While DEPC has been successfully used to study the structures of proteins and protein complexes, this labeling reagent introduces an electrophilic site into the side chains of the residues it modifies, opening up the possibility for some unwanted chemistry. For example, we recently demonstrated that, in solution, cysteine residues have the ability to capture carbethoxy groups from other residues that were modified by DEPC. This unwanted label transfer can be eliminated by deactivating cysteine’s strong nucleophilic character via alkylation just after DEPC labeling of the protein is finished [15]. The presence of a new electrophilic site might also affect the gas-phase chemistry of DEPC-labeled peptides especially when subjected to slow collisional activation in a quadrupole ion trap mass spectrometer. Indeed, Reid and co-workers and others have reported that phosphorylated peptide ions can have their phosphate group transferred from one amino acid to another upon CID [16-18]. The result is the incorrect assignment of the phosphorylation site on the peptide. In addition to the scrambling of phosphate groups in peptide ions, methyl [19], acetyl, and formyl groups [20] have also been documented to undergo transfer from one amino acid to another during CID in the gas phase.
With the known chemistry of DEPC and these previous studies in mind, we set out to investigate whether DEPC label scrambling can occur during CID of DEPC labeled peptide ions. Through the study of numerous peptides, we find that label transfer can occur in DEPC-labeled peptides, and it occurs with similar characteristics to phosphate group transfer in phosphorylated peptides.
Methods
Materials
Diethylpyrocarbonate (DEPC), imidazole, iodoacetamide, and tris(2-carboxyethyl)phosphine (TCEP) were obtained from Sigma Aldrich (St. Louis, MO). Angiotensin 1-10 (DRVYIHPFHL) , angiotensin 1-13 (DRVYIHPFHLVIH), apelin 13 (QRPRLSHKGPMPA), β amyloid (YEVHHQKLVFF), semastatin (SPWTKCSATCGGGHYMRTR), neuromedin-C (GNHWAVGHLM ) and adrenocorticotropic hormone (ACTH) 1-13 (SYSMEHFRWGKPV) were obtained from the American Peptide Company (Sunnyvale, CA). Human β-2-microglobulin (β2m) was obtained from Lee Biosolutions (St. Louis, MO). Immobilized chymotrypsin and triethylamine acetate (pH 8.0) were obtained from Princeton Separations (Adelphia, NJ). Ammonium acetate, methanol, formic acid, acetonitrile, and water were purchased from Fisher Scientific (Fair lawn, NJ). Centricon molecular weight cutoff (MWCO) filters were obtained from Millipore (Burlington, MA).
DEPC Labeling Reactions
Peptide solutions with a concentration of 100 μM in 10 mM ammonium acetate were reacted with DEPC at either a 1:1 or 1:4 (peptide:DEPC) ratio for 2-5 min at 37°C. A 6 mM stock solution of DEPC in acetonitrile was added to obtain the final DEPC concentration. The final DEPC concentration resulted in a total acetonitrile concentration that was less than 2% (v/v) of the reaction mixture. The reaction time and ratio were chosen to ensure that the unmodified and the singly modified peaks were the most prominent products. The protein β2m was reacted with DEPC at a 1:4 (protein:DEPC) ratio for 1 min at 37°C. In all cases, the DEPC reactions were quenched by the addition of 10 mM imidazole [11].
Proteolytic Digestion
Prior to proteolytic digestion of β2m, the quenching agent, imidazole, was removed using a 10,000 MWCO filter, resulting in a 40 μL solution of 250 μM β2m. β2m was then incubated in a buffer solution (100 mM triethylamine at pH 8.0) with 10% (v/v) acetonitrile at 50 °C for 45 min. Next β2m was reacted with TCEP (1:40 ratio) and iodoacetamide (1:80 ratio) in the dark for 30 min to reduce its disulfide bond and alkylate the resulting free thiols. Immobilized chymotrypsin was then added to achieve a 1:10 ratio of enzyme to substrate. The digestion reaction was allowed to proceed at 37 °C for 2 hours.
HPLC Separation
HPLC separations were performed using a Supelco Discovery C18 column (15 cm × 2.1 mm, 5 μm particle size, St. Louis, MO) on an HP1100 HPLC system (Agilent, Wilmington, DE). To achieve sufficient separation of DEPC-labeled peptide isomers, an isocratic elution was used (40% acetonitrile in water with 0.1% formic acid) for 20 min. This was followed by a 0.5%/min increase of acetonitrile for an additional 20 min. A flow rate of 0.35 mL/min was used, and the effluent from the column was split by four prior to introduction into the mass spectrometer. The proteolytic digests were eluted using a linear gradient of methanol ranging from 10 – 100% methanol over 30 min. This mobile phase contained 0.1% acetic acid and had a flow rate of 0.25 mL/min.
Mass Spectrometry
Mass analysis was performed using either a Bruker Esquire-LC or a Bruker AmaZon (Billerica,MA) quadrupole ion trap mass spectrometer. These instruments are equipped with electrospray ionization sources, and the needle voltage was typically set to 3000 V. The capillary temperature was set to 300°C. Both CID and ETD were used to obtain tandem mass spectra. The ion isolation width for both methods was set to 1.0 Da. CID voltages were typically between 0.65 and 1.1 V and were chosen to achieve optimal dissociation efficiency. For ETD experiments, the low m/z cutoff was typically set to 135, and the reaction time was typically set to 150 ms.
Results and Discussion
CID for Label Site Identification
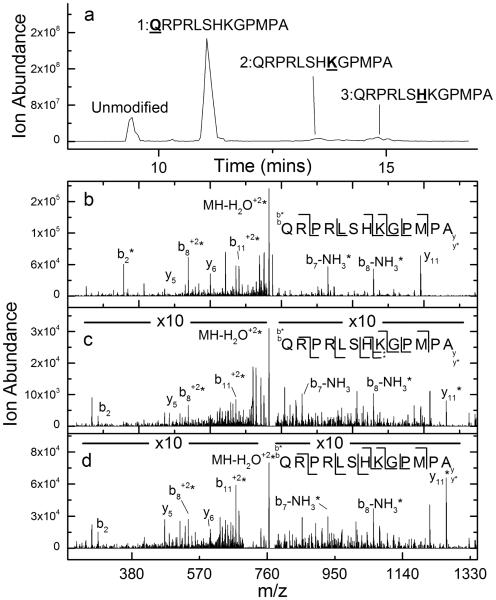
CID is commonly used to determine label locations in covalently labeled peptides and is typically successful. As an example, CID of the peptide apelin 13 (QRPRLSHKGPMPA) can readily enable the identification of the DEPC-labeled residues upon a 2 minute reaction with this reagent (Figure 1). LC/MS analysis of the labeled peptide suggests that there are three labeled sites as indicated by the chromatogram in Figure 1a. By examining the sequence ions upon CID of each peak in the chromatogram, the specific amino acid residues labeled by DEPC can be assigned. For DEPC-labeled apelin 13, the CID spectrum of the second chromatographic peak shows a series of b ions that are all labeled, and several y ions that are not labeled, indicating that the N-terminus is the labeled site (Figure 1b). The CID spectrum of the third chromatographic peak is consistent with a label on Lys8, as suggested by labeled y6, y7, y9, and y11 product ions, unlabeled y4 and y5 ions, labeled b8 and b11 ions, and unlabeled b2 and b7 ions (Figure 1c). The final chromatographic peak corresponds to a labeled His7 product. This assignment is evident from the labeled y7, y9, and y11 product ions, unlabeled y4, y5, and y6 ions, labeled b7, b8 and b11 ions, and an unlabeled b2 ion (Figure 1d).
Figure 1.
(a) Total ion chromatogram after HPLC separation of DEPC-labeled apelin 13. (b) CID spectrum of the (M+2H)2+ ion of the second chromatographic peak from the DEPC-labeled sample of apelin 13, indicating that the N-terminus is modified. (c) CID spectrum of the (M+2H)2+ ion of the third chromatographic peak from the DEPC-labeled sample of apelin 13, indicating that Lys8 is modified. *Interfering ions make assignment of the labeled and unlabeled versions of the y6 ion somewhat ambiguous. (d) CID spectrum of the (M+2H)2+ ion of the fourth chromatographic peak from the DEPC-labeled sample of apelin 13, indicating that His7 is modified.
Ambiguous Label Assignment by CID
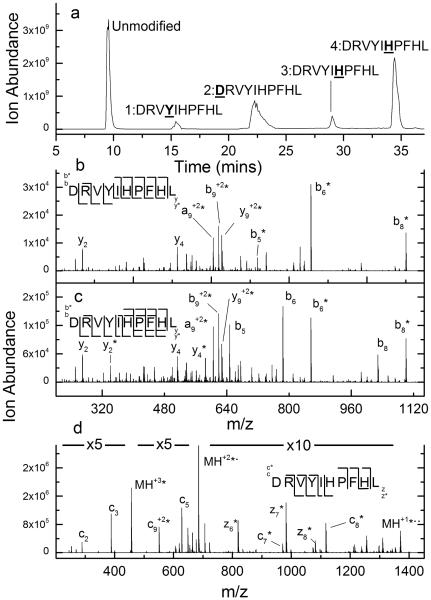
While CID often provides correct labeling site information, as exemplified by the data for apelin 13, we have also found evidence that covalent labels can be scrambled from one site to another during the CID process. The CID data of DEPC labeled-angiotensin I illustrates this phenomenon. Upon labeling this peptide with DEPC for 5 min, on average a single modification site per peptide is measured, resulting in four isomers as indicated by the chromatogram in Figure 2a. Each peak in the chromatogram corresponds to a chemically distinct structure, and indeed the CID data for the first (Figure 2b) and second (Figure S1 in the Supplemental Information) labeled peaks are consistent with this assertion, as Tyr4 and the N-terminus, respectively, are identified as the modified sites.
Figure 2.
(a) Total ion chromatogram after HPLC separation of DEPC-labeled angiotensin. (b) Example CID spectrum of the (M+2H)2+ ion of the second chromatographic peak from the DEPC-labeled sample of angiotensin I, indicating that Tyr4 is modified. Unlabeled y2 to y6 ions, labeled y7 to y9 ions, labeled b5 to b9 ions, and an unlabeled b2 ion indicate that Tyr4 is labeled. (c) CID spectrum of the (M+2H)2+ ion of the final chromatographic peak from the DEPC-labeled sample of angiotensin I. The presence of both labeled and unlabeled b6, b8, y4, and y2 product ions causes the labeled site to be ambiguous. (d) ETD spectrum of the (M+3H)3+ ion of the final chromatographic peak from the DEPC-labeled sample of angiotensin I, indicating that His6 is labeled.
The identity of the labeled sites for the modified peptides associated with the final two chromatographic peaks, however, are not as clear. This fact is most evident from the presence of both labeled and unlabeled versions of the b6, b8, y4, and y2 product ions in both spectra (Figures 2c and S2). One possible explanation for this is that these chromatographic peaks correspond to two unseparated isomers – one modified at His6 and the other at His9. This possibility is unlikely considering the great care taken to identify LC conditions to separate any isomers present. Moreover, the ETD spectra of these peaks (Figures 2d and S3) indicate that only His6 is modified with no evidence for the modification of His9. This conclusion comes from the series of c and z product ions in which ions that include His6 are labeled and all that do not are unlabeled. Incidentally, the ETD spectra of peaks 1 and 2 lead to the same conclusions about modification sites as the CID data (see Figures S4 and S5). Another possible explanation is that the DEPC label is lost as a neutral. This would, for example, explain the presence of both labeled and unlabeled b6 and b8 product ions in the CID spectrum; however, the absence of an unlabeled b9 ion and no evidence for label loss during the CID of other DEPC-labeled peptides (e.g. apelin 13 in Figure 1) suggests that label loss is not the cause. Moreover, label loss cannot explain the presence of labeled y2 and y4 ions. These latter ions can only arise from the label being transferred to His9.
Upon considering the data for DEPC-labeled angiotensin I, we propose that the CID data for the final two chromatographic peaks of modified angiotensin I arise via a collision-induced transfer of the DEPC label from one His residue to another. Such a gas-phase rearrangement of a side chain modification has been seen before [16-20] during CID analyses, as indicated in the introduction. The migration of the DEPC label from His6 to His9 during collisional activation of angiotensin I can explain the existence of all observed ions and is consistent with differences in dissociation mechanisms between ETD and CID. Unlike label loss, transfer of DEPC from His6 to His9 explains, for example, why the b9 product ion is only observed as a labeled species. The fact that label migration does not take place during ETD makes sense due to the non-ergodic nature of its dissociation mechanism. We suspect that the scrambling of the label is driven by the fact that addition of DEPC to a histidine side chain results in an electrophilic site that is vulnerable to attack by other nucleophilic sites on the peptide. In the case of angiotensin, this nucleophilic site could be the other, unlabeled histidine residue. A possible mechanism is illustrated in Figure 3.
Figure 3.
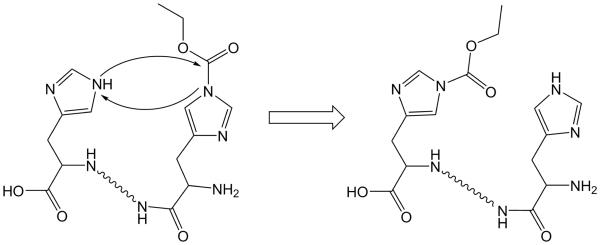
Possible mechanism of rearrangement, where the DEPC label on one histidine side chain is transferred to another via a nucleophilic attack.
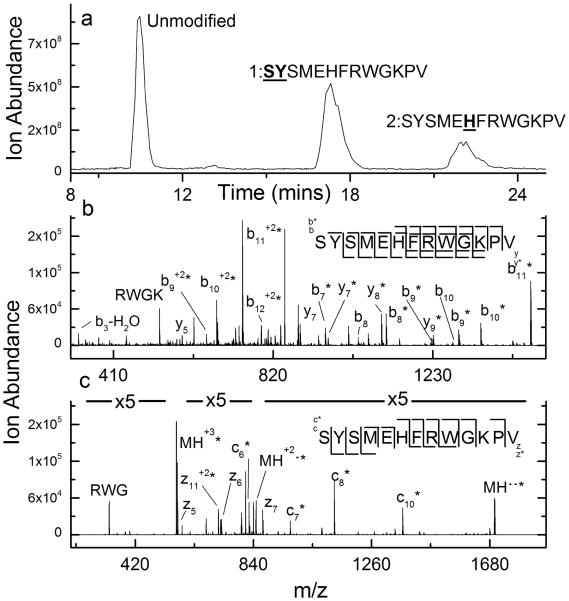
Another peptide that appears to undergo label scrambling during CID is the peptide adrenocorticotropic hormone (ACTH). LC/MS analysis of DEPC labeled ACTH indicates that two amino acids are labeled in this peptide (Figure 4a). In this case, the label sites associated with both chromatographic peaks from the labeled peptide are ambiguous when CID is used. The tandem mass spectrum of the first labeled peak suggests scrambling between the N-terminus, His6, and Lys11 (Figure S6) as indicated by the presence of labeled and unlabeled versions of the b2 - b10 series of product ions and the y6 - y11 series of product ions. The fact that the labeling site is clearly identified as the N-terminus when dissociated by ETD (Figure S7) suggests that unseparated isomers are not the source of this ambiguity. CID of the second labeled chromatographic peak suggests both His6 and Lys11 as the labeled sites based on the existence of labeled and unlabeled versions of the b6 - b10 series of product ions and y3 - y7 series of product ions (Figure 4b). In contrast, the ETD spectrum of this labeled peptide readily identifies His6 as the labeled site (Figure 4c).
Figure 4.
(a) Total ion chromatogram after HPLC separation of DEPC-labeled ACTH. (b) CID spectrum of the (M+2H)2+ ion of the first labeled chromatographic peak from the DEPC-labeled sample of ACTH. The presence of both labeled and unlabeled b10, b9, b8, and y7 product ions suggests label scrambling. (c) ETD spectrum of the (M+3H)3+ ion of the second labeled chromatographic peak from the DEPC-labeled sample of ACTH, indicating that His6 is labeled.
Effect of Charge State on Labeling Scrambling
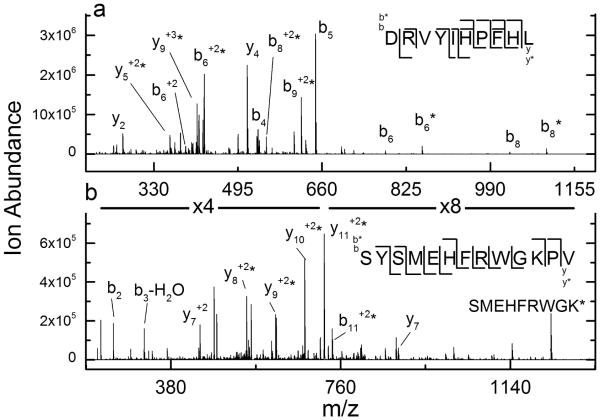
Previous work has shown that the gas-phase rearrangement of phosphate and methyl groups in peptide ions is dependent on the precursor ion’s charge state and the subsequent proton mobility within the ion [16,19]. Phosphorylated and methylated peptide ions with non-mobile and partially mobile protons were found to have a higher propensity to rearrange. Proton mobility in this context refers to the number of net protons on the peptide ion as compared to the number of basic residues in the peptide. Non-mobile protons are present when the number of net protons is lower than the number of basic residues in the peptide. To test whether proton mobility influences DEPC label scrambling, we compared the CID spectra of the [M+2H+DEPC]2+ and [M+3H+DEPC]3+ ions of the fifth chromatographic peak of labeled angiotensin (i.e. peaks in Figure 2a). Interestingly, the different charge states result in drastically different dissociation patterns with regard to label scrambling. There are no longer any labeled y2 or y4 product ions observed upon dissociation of the +3 charge state (Figure 5a); these ions were key indicators of label scrambling for the +2 ion (see Figure 2c). In addition, the unlabeled b6 and b8 ions, which also implied label scrambling in the +2 ion, are substantially reduced in relative abundance.
Figure 5.
(a) CID spectrum of the (M+3H)3+ ion of the final labeled chromatographic peak from the DEPC-labeled sample of angiotensin I. The absence of labeled y2 and y4 product ions indicates label scrambling is not occurring. (b) CID spectrum of the (M+3H)3+ ion of the second labeled chromatographic peak from the DEPC-labeled sample of ACTH. The absence of unlabeled b6, labeled y3, and y7 product ions in this spectrum indicates label scrambling is not occurring.
A similar influence of proton mobility on label scrambling is also observed for ACTH. As shown earlier in Figure 4b, the +2 charge state of third chromatographic peak exhibits scrambling upon activation with CID. When the +3 charge state of this same labeled peptide is dissociated using CID, no evidence for scrambling is observed (Figure 5b). The absence of unlabeled b6, labeled y3, and y7 product ions in this spectrum removes all ambiguity about the label assignment. The dramatic influence of charge state on the dissociation behavior of these DEPC-modified peptides further supports the notion that label scrambling is occurring.
Prevalence of Label Scrambling
To test the pervasiveness of label scrambling, we examined several other labeled peptides, including ones from a proteolytic digest of the protein β-2-microglobulin that had been labeled with DEPC for 1 minute. Each collection of peptides was separated by LC and then subjected to both CID and ETD. In all, 34 different labeled sites were probed (Table 1). The criteria for concluding that label scrambling had occurred were: (i) unlabeled and labeled versions of product ions were observed during CID and (ii) CID and ETD identified different labeled sites in a given peptide. Of these 34 labeled peptides, nine showed evidence for scrambling. Interestingly, all nine had a histidine residue as either the originally labeled site or the site to which the labeled was transferred, indicating the lability of labeled histidine residues. Another interesting observation from this limited data set is that an acidic residue is present in all the peptides that undergo scrambling. Because of the effect of proton mobility on the prevalence of this scrambling reaction (see Figure 5), it is possible that intramolecular interactions between charged groups may be necessary to facilitate this reaction. Further work is clearly needed, though, to more fully understand how amino acid sequence affects this scrambling reaction.
Table 1.
DEPC-labeled sites on peptides as identified by ETD and CID and whether scrambling has occurred.
| Sequence | ETD of (M+3H)3+ |
CID of (M+3H)3+ |
Scrambling in (M+3H)3+? |
CID of (M+2H)2+ |
Scrambling in (M+3H)2+? |
|---|---|---|---|---|---|
| DRVYIHPFHL peak 1 | Y4 | Y4 | No | Y4 | No |
| DRVYIHPFHL peak 2 | N-term | N-term | No | N-Term | No |
| DRVYIHPFHL peak 3 | H6 | H6 | No | H6, H9 | Yes |
| DRVYIHPFHL peak 4 | H6 | H6 | No | H6, H9 | Yes |
|
| |||||
| GNHWAVGHLM peak 1 | N-Term | N-Term | No | N-Term | No |
| GNHWAVGHLM peak 2 | H8 | H8 | No | H8 | No |
| GNHWAVGHLM peak 3 | H3 | H3 | No | H3 | No |
| GNHWAVGHLM peak 4 | H8 | H8 | No | H8 | No |
| GNHWAVGHLM peak 5 | H3 | H3 | No | H3 | No |
|
| |||||
| DRVYIHPFHLVIH peak 1 | Y4 | No | Y4 | No | |
| DRVYIHPFHLVIH peak 2 | N-term | No | N-term | No | |
| DRVYIHPFHLVIH peak 3 | H9 | No | H6, H9, H13 |
Yes | |
| DRVYIHPFHLVIH peak 4 | H9 | No | H6, H9 | Yes | |
| DRVYIHPFHLVIH peak 5 | H13 | No | H9, H13 | Yes | |
| DRVYIHPFHLVIH peak 6 | H6 | No | H6, H9 | Yes | |
|
| |||||
| SYSMEHFRWGKPV peak 1 | N-term | N-term | No | N-term, H6, K11 |
Yes |
| SYSMEHFRWGKPV peak 2 | H6 | H6 | No | H6, K11 | Yes |
|
| |||||
| YEVHHQKLVFF peak 1 | H4 | H4, H5 | Yes | H4, H5 | Yes |
| YEVHHQKLVFF peak 2 | N-term | N-Term | No | N-term | No |
| YEVHHQKLVFF peak 3 | H4 | H4 | No | H4 | No |
| YEVHHQKLVFF peak 4 | H5 | H5 | No | H5 | No |
| YEVHHQKLVFF peak 5 | K7 | K7 | No | K7 | No |
|
| |||||
| SPWTKCSATCGGGHYMRTR peak 1 | Y15 | No | Y15/H14* | No | |
| SPWTKCSATCGGGHYMRTR peak 2 | H14 | No | H14 | No | |
| SPWTKCSATCGGGHYMRTR peak 3 | Y15/H14* | No | Y15/H14* | No | |
| SPWTKCSATCGGGHYMRTR peak 4 | N-term | No | N-Term | No | |
| SPWTKCSATCGGGHYMRTR peak 5 | K5 | No | K5 | No | |
|
| |||||
| SRHPAENGKSNF | S1/R2 | No | N-term | No | |
| SRHPAENGKSNF | H3 | No | H3 | No | |
|
| |||||
| IQRTPKIQVY | K6 | No | Y3 | No | |
| IQRTPKIQVY | N-term | No | N-term | No | |
|
| |||||
| KNGERIEKVEHSDL | K1 | No | K1 | No | |
|
| |||||
| SQPKIVKW | K4 | No | K4 | No | |
|
| |||||
| SRHPAENGKSNFLNCY | C15 | No | C15 | No | |
Tandem mass spectrometry is unable to identify which of the listed residues is labeled due to the absence of the appropriate product ions.
Conclusions
Covalent labeling is increasingly used to study the surface structure of proteins and protein complexes, and DEPC is a labeling reagent that has shown great promise in these experiments because of its ability to label multiple residues simultaneously. Identifying the labeled sites is often done using CID after proteolytic digestion of the protein, but in this work, we show that scrambling of the DEPC label to another site on a peptide can occasionally occur during CID, resulting in ambiguous labeling site assignments. Like previous studies that reported the scrambling of functional groups (e.g. phosphate groups) on modified peptides, this scrambling occurs most readily under low proton mobility conditions. Also, scrambling does not appear to occur when ETD is used to dissociate the labeled peptides; this is consistent with the known mechanistic differences between CID and ETD. Based on the analysis of over 30 labeled peptides, we find evidence for scrambling about 25% of the time. In all the cases where scrambling is observed, histidine residues are involved. The increased tendency of histidine residues to undergo scrambling has important consequences as this residue is the most reactive amino acid with DEPC. A solution to this problem may be to produce larger peptides upon proteolysis so that the labeled peptides have higher charge states and are thus more amenable to ETD. Alternatively, supercharging agents could be used to produce higher peptide charge states to avoid the low proton mobility conditions that foster label scrambling during CID.
Supplementary Material
Acknowledgments
This work was supported by NIH grant RO1 GM015092. NB was partially supporting by a NIH training grant (T32 GM008515).
References
- 1.Mendoza VL, Vachet RW. Probing Protein Structure by Amino Acid-Specific Covalent Labeling and Mass Spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konermann L, Stocks BB, Pan Y, Tong X. Mass Spectrometry Combined with Oxidative Labeling for Exploring Protein Structure and Folding. Mass. Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 3.Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 4.Hambly D, Gross M. Laser Flash Photochemical Oxidation to Locate Heme Binding and Conformational Changes in Myoglobin. Int. J. Mass. Spectrom. 2007;259:124–129. [Google Scholar]
- 5.Hambly DM, Gross ML. Laser Flash Photolysis of Hydrogen Peroxide to Oxidize Protein Solvent-Accessible Residues on the Microsecond Timescale. J. Am. Soc. Mass. Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Anal. Chem. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Rempel DL, Gross ML. Temperature jump and fast photochemical oxidation probe submillisecond protein folding. J. Am. Chem. Soc. 2010;132:15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konermann L, Pan Y. Exploring membrane protein structural features by oxidative labeling and mass spectrometry. Expert Rev. Proteomics. 2012;9:497–504. doi: 10.1586/epr.12.42. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Rempel DL, Gau BC, Gross ML. Fast Photochemical Oxidation of Proteins and Mass Spectrometry Follow Submillisecond Protein Folding at the Amino-Acid Level. J. Am. Chem. Soc. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendoza VL, Antwi K, Barón-Rodríguez MA, Blanco C, Vachet RW. Structure of the Pre-amyloid Dimer of β-2-microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza VL, Vachet RW. Protein Surface Mapping using Diethylpyrocarbonate with Mass Spectrometric Detection. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper Binding to Beta-2-Microglobulin and its Pre-Amyloid Oligomers. Biochemistry. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza VL, Barón-Rodríguez MA, Blanco C, Vachet RW. Structural Insights into the Pre-Amyloid Tetramer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Vachet RW. Increased Protein Structural Resolution from Diethylpyrocarbonate-Based Covalent Labeling and Mass Spectrometric Detection. J. Am. Soc. Mass. Spectrom. 2012;23:708–717. doi: 10.1007/s13361-011-0332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Vachet RW. Diethylpyrocarbonate Labeling for the Structural Analysis of Proteins: Label Scrambling in Solution and How to Avoid it. J. Am. Soc. Mass. Spectrom. 2012;23:899–907. doi: 10.1007/s13361-012-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palumbo AM, Reid GE. Evaluation of Gas-Phase Rearrangement and Competing Fragmentation Reactions on Protein Phosphorylation Site Assignment using Collision Induced Dissociation-MS/MS and MS3. Anal. Chem. 2008;80:9735–9747. doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Reid GE. Examining Factors that Influence Erroneous Phosphorylation Site Localization Via Competing Fragmentation and Rearrangement Reactions During Ion Trap CID-MS/MS and - MS(3) Proteomics. 2013;13:964–973. doi: 10.1002/pmic.201200384. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Sanchez MB, Lanucara F, Hardman GE, Eyers CE. Gas-Phase Intermolecular Phosphate Transfer Within a Phosphohistidine Phosphopeptide Dimer. Int. J. Mass. Spectrom. 2014;367:28–34. doi: 10.1016/j.ijms.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong L, Ping L, Yuan B, Wang Y. Methyl Group Migration During the Fragmentation of Singly Charged Ions of Trimethyllysine-Containing Peptides: Precaution of using MS/MS of Singly Charged Ions for Interrogating Peptide Methylation. J. Am. Soc. Mass. Spectrom. 2009;20:1172–1181. doi: 10.1016/j.jasms.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs R, Budzikiewicz H. Rearrangement Reactions in the Electrospray Ionization Mass Spectra of Pyoverdins. Int. J. Mass. Spectrom. 2001;210/211:603–612. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.