Abstract
Aims
Recently, microRNAs (miRNAs) have been implicated in control of Edn1 mRNA in several tissues. Here we examined the role of miRNA action on Edn1 mRNA expression in renal distal collecting duct cells.
Main methods
A microarray study was conducted to provide a comprehensive assessment of miRNAs present in a murine inner medullary collecting duct (mIMCD-3) cell line. The experiment was designed as a comparison between mIMCD-3 cells grown in the presence and absence of aldosterone. Argonaute (Ago) immunoprecipitation experiments were used to investigate binding of the RNA induced silencing complex (RISC) to Edn1 mRNA.
Key findings
Thirty-four miRNAs were detected in very high abundance in mIMCD-3 cells, and a large number of others were present at lower levels. The microarray experiments were validated by quantitative PCR analysis of selected miRNAs. The microarray data, in combination with in silico examination of the Edn1 3’ UTR provided a panel of candidate miRNAs that could act upon the Edn1 expression. Edn1 mRNA was co-immunoprecipitated with an Argonaute protein antibody, and this interaction was blocked by anti-miR-709 oligonucleotides.
Significance
These results define the miRNA landscape of the mIMCD-3 cell line. Moreover, Edn1 was shown to interact with Argonaute protein suggesting that it is a target of the RNA induced silencing complex (RISC).
Keywords: miRNA, kidney, endothelin-1
INTRODUCTION
The kidney is responsible for maintaining acid-base homeostasis and ion balance in the blood stream. Aldosterone is the major hormone involved in stimulation of sodium retention in the kidney collecting duct, where its primary mode of action is to stimulate transcription of genes favoring sodium retention. In contrast, endothelin favors sodium loss with the urine, thus aldosterone and endothelin-1 (ET-1) apparently promote opposing physiological responses (Kohan et al. 2011). Collecting duct-specific knockout of Edn1 yielded a hypertensive phenotype in mice (Ahn et al. 2004). Recently, inhibition of net sodium reabsorption in the collecting duct was shown to be dependent on both endothelin A and B receptors (Lynch et al. 2013). Paradoxically, the endothelin-1 (Edn1 in mouse) gene is among the most highly induced aldosterone-response genes in collecting duct cells (Gumz et al. 2003; Stow et al. 2009).
Circulating ET-1 accumulates in the kidney (Johnström etal. 2005), and the renal collecting duct shows the highest level of ET-1 expression in the entire body (Moridaira et al. 2003). Regulation of Edn1 occurs predominantly at the level of transcription (Stow et al. 2011; Welch et al. 2013), but it is also increasingly apparent that Edn1 mRNA is regulated at the post-transcriptional level (Jacobs et al. 2013). Edn1 mRNA is unstable, and the mechanisms responsible for this instability center on the 3` untranslated region (UTR) of the Edn1 mRNA. In mammals, the Edn1 3’UTR represents more than half of the total mRNA length. Alignment of Edn1 3’ UTRs of mammals yields roughly 80% sequence identity. Mammalian Edn1 3’ UTRs are typically equipped with AU-rich elements (AREs) thought contribute to Edn1 RNA turnover via the AUF1-proteosome pathway (Mawji et al. 2004; Reimunde et al. 2005). MicroRNAs (miRNAs) offer an alternative mechanism for control of Edn1 expression. Endogenous miRNAs are small, noncoding RNAs that facilitate binding of the RNA induced silencing complex (RISC) to the 3’ UTR. Translation of RISC-bound mRNAs is impaired and degradation increases. Changes in the miRNA landscape are known to occur in pathogenic states including polycystic kidney disease, renal cell carcinoma, and diabetic nephropathy (Pandey et al. 2008; Juan et al. 2012; Krupa et al. 2010). However, the change in the miRNA landscape in response to physiological stimuli remains unclear.
Our working hypothesis is that the miRNA content in the inner medullary collecting duct is an important regulatory mechanism for Edn1 expression and consequently sodium reabsorption. Here we report the miRNA landscape in cells derived from the murine renal inner medullary collecting duct (mIMCD-3) using miRNA microarray analysis. Comparison of the microarray data with an in silico analysis of the mRNA yielded a list of candidate miRNAs predicted to act on the Edn1 3’ UTR. The RISC-miR-709 complex was shown to target murine Edn1 mRNA in mIMCD-3 cells.
MATERIALS AND METHODS
Cell Culture and Biological Reagents
mIMCD-3 cells were obtained from American Type Culture Collection. mIMCD-3 cells between passages 13 and 23 were used for all experiments. mIMCD-3 cells were grown in DMEM-F-12 (Genesee Scientific) supplemented with 10% fetal bovine serum (Invitrogen), and 50 μg/ml of gentamycin in T-75 flasks (Genesee Scientific) at 37 °C in 95% air and 5% CO2. Total RNA isolated from murine inner medullary tissue was a kind gift of Dr. Michelle Gumz.
miRNA Microarray Approximately 70,000 mIMCD-3 cells were plated in Corning® CoStar® transwell dishes to induce polarity. Cells were grown to confluence, at which point the medium was replaced by hormone-free media for 24 hours. The cells were treated with either vehicle (ethanol) or 100nM aldosterone. After 24 hours of aldosterone treatment, total cellular RNA was isolated using TRIzol® (Invitrogen). Cells were washed twice with 1mL PBS, and 1 mL of TRIzol® was added to each well. The cells were incubated at room temperature for 5 minutes. Cell lysates were gently resuspended by pipetting up and down. The cells were incubated in a 1.5mL test tube for an additional 3 minutes to obtain complete lysis. Chloroform (200 μL) was added to the tube, and the tube was mixed by shaking for 15 seconds. The samples were centrifuged at 12,000 rpm at 4°C for 20 minutes. The aqueous phase was collected and 800 μL of isopropanol was added to each sample. Samples were incubated at −80°C overnight, thawed on ice, and centrifuged at 12,000 rpm at 4°C for 30 minutes. The RNA pellet was washed with 70% ethanol, centrifuged 12,000 rpm at 4°C for 10 minutes, air dried for 10 minutes, and resuspended in RNase-free water. To obtain the amount of RNA required for the miRNA microarray, it was necessary to concentrate the samples. Six transwells for each passage were pooled into one 150 μL total RNA sample. RNA integrity was analyzed by an Agilent 2100 Bioanalyzer (UF ICBR), using a Small RNA Bioanalyzer Chip (Agilent). Samples were then shipped to Toray Industries, Inc. For each sample, 500 ng of total RNA was labeled using miRCURY LNA™ microRNA Power Labeling Kits Hy5 (Exiqon). The labeled samples were individually hybridized onto the DNA chip surface, and were incubated at 42°C for 16 hours. The washed and dried DNA chip was scanned using a ProScanArray™ microarray scanner (PerkinElmer Inc.). The obtained microarray images were analyzed using Genepix Pro™ 4.0 software (Molecular Devices). In this study, the median values of the foreground signal minus the local background were represented as the feature intensities. The microarray data was deposited in the GEO data base (accession number GSE47901).
Treatment with anti-miRNA inhibitors
mIMCD-3 cells were transfected with 10nM anti-miR™ miRNA inhibitors (Life Technologies) using DharmaFECT4 (ThermoScientific) according to the manufacturer’s protocol. 24 hours post-transfection 100nM aldosterone or vehicle was added to the media, and cells were cultured for an additional 24 hours.
RNA Quantification by Real-Time PCR
Total RNA (miRNA and mRNA) was isolated from mIMCD-3 cells using TRIzol® (Invitrogen). For mRNA quantification, RNA was reverse transcribed using the High Capacity cDNA synthesis kit (Applied Biosytems) followed by real-time analysis using sequence – specific primers and TaqMan master mix (Life Technologies) with a BioRad DNA Engine Opticon® 2 PCR system. For miRNA quantification, five nanograms of total cellular RNA was used for a reverse transcriptase reaction (TaqMan® MicroRNA Reverse Transcription Kit, Life Technologies) using miRNA-specific primers, followed by real-time PCR analysis using TaqMan® miRNA probes.
Argonaute Pull-Down Assay
The Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore) was used with an anti-pan-Ago antibody (MABE56, EMD Millipore) for the Argonaute immunoprecipitation experiments. The assay was performed according to manufacturer’s instructions. RNA concentration was normalized to percent of input using relative standard curve.
RESULTS
The miRNA landscape of mIMCD-3 cells
In order to determine the miRNA content of an inner medullary collecting duct cell line, total RNA was prepared from mIMCD-3 cells. Five independent cell cultures were treated with 100 nM aldosterone and five cell cultures were exposed to vehicle (ethanol) only. The quality of all RNA samples was examined using an Agilent 2100 Bioanalyzer, and only samples with a RNA Integrity Number (RIN) of 10 were used (Figure 1). Each independent RNA sample was applied to the Toray 3D-Gene™ Mouse miRNA Oligo chip (16v1.0.0) carrying probes for 1079 miRNAs. The microarray analysis identified 41 highly expressed miRNAs in mIMCD-3 cells (Table 1). In addition, 78 moderately expressed miRNAs, and a large number of miRNAs were detected at low levels in at least 9 out of 10 samples (GEO ascension number GSE47901). Representative miRNAs from the highly, moderately, and lowly expressed miRNAs were selected and studied using qRT-PCR to verify levels seen in the microarray. The relative abundance measured by qRT-PCR coincided with the values seen in the microarray experiment (Table 2). Applying the most rigorous statistical considerations suggested that five lowly expressed miRNAs were unambiguously upregulated more than two-fold in the presence of aldosterone (Table 3). Changes in higher level miRNAs were less convincing.
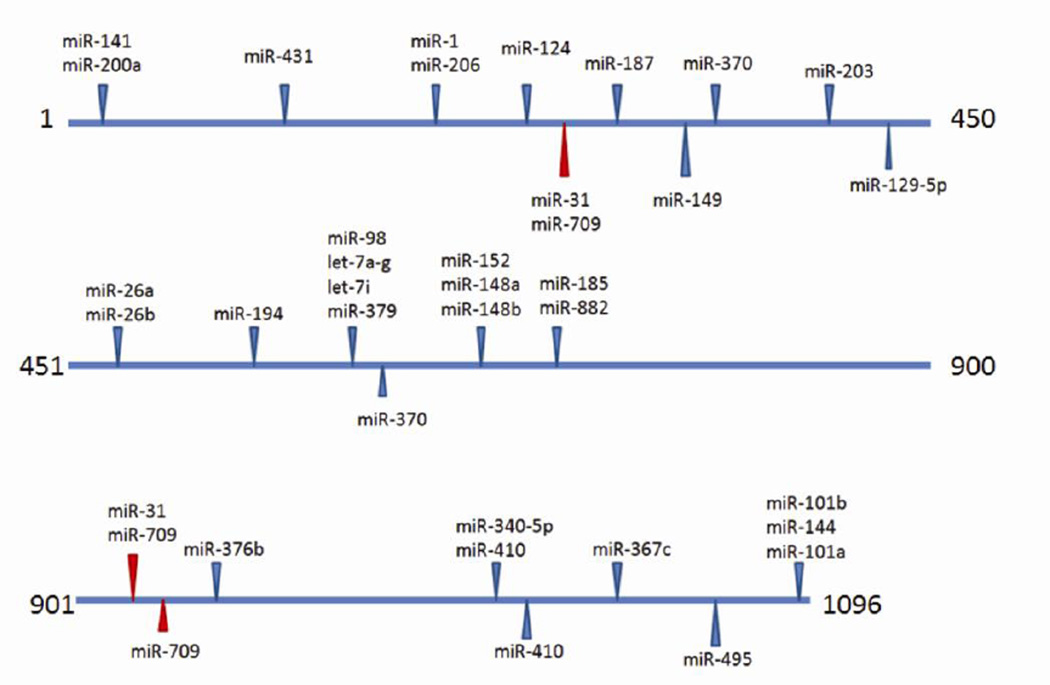
Figure 1. Predicted miRNA binding sites in the Edn1 3’ UTR.
Predicted miRNA target sites in the murine Edn1 3′UTR (NM_010104) were determined using microRNA.org (Betel et al., 2008). Blue arrows indicate predicted target sites of miRNAs with high probability mirSVR scores., Red arrows show the putative miR-709 binding sites.
TABLE 1.
Highly expressed miRNAs in the mIMCD-3 cell line.
| miRNA | Microarraya | Accession Number | |
|---|---|---|---|
| Vehicle | Aldosterone | ||
| miR-690 | 11.60 ± .44 | 11.94 ± .31 | MIMAT0003469 |
| miR-1937b | 11.43 ± .77 | 11.62 ± .31 | MIMAT0009414 |
| miR-1937a | 11.28 ± .42 | 11.36 ± .21 | MIMAT0009401 |
| miR-1937c | 11.28 ± .42 | 11.36 ± .21 | MIMAT0009429 |
| miR-709 | 10.52 ± .60 | 10.26 ± .54 | MIMAT0003499 |
| miR-2137 | 10.14 ± .43 | 10.37 ± .41 | MIMAT0011213 |
| miR-21 | 9.88 ± 1.05 | 10.75 ±.54 | MIMAT0000530 |
| miR-2861 | 9.33 ± .30 | 9.40 ± .28 | MIMAT0013803 |
| let-7a | 9.32 ± .54 | 9.50 ± .38 | MIMAT0000521 |
| let-7c | 8.97 ± .52 | 8.95 ± .13 | MIMAT0000523 |
| miR-30c | 8.86 ± .35 | 8.90 ± .23 | MIMAT0000514 |
| let-7d | 8.83 ± .38 | 8.83 ± .26 | MIMAT0000383 |
| miR-762 | 8.81 ± .31 | 8.73 ± .35 | MIMAT0003892 |
| miR-29a | 8.58 ± .33 | 8.86 ± .25 | MIMAT0000535 |
| mir-23a | 8.41 ± .57 | 8.76 ± .05 | MIMAT0000532 |
| let-7b | 8.27 ± .35 | 8.95 ± .17 | MIMAT0000522 |
| let-7f | 8.20 ± 1.11 | 8.67 ± .38 | MIMAT0000525 |
| miR-30a | 8.15 ± .65 | 8.76 ± .65 | MIMAT0000128 |
| miR-16 | 8.10 ± .76 | 8.51 ± .27 | MIMAT0000527 |
| miR-23b | 8.08 ± .39 | 8.36 ± .12 | MIMAT0000125 |
| miR-10a | 8.08 ± .53 | 8.19 ± .31 | MIMAT0000648 |
| miR-15b | 8.07 ± .38 | 8.20 ± .22 | MIMAT0000124 |
| miR-30d | 7.63 ± .48 | 8.06 ± .47 | MIMAT0000515 |
| miR-93 | 7.62 ± .41 | 7.83 ± .34 | MIMAT0000540 |
| miR-1944 | 7.53 ± .13 | 7.60 ± .35 | MIMAT0009409 |
| miR-103 | 7.50 ± .30 | 7.63 ± .16 | MIMAT0000546 |
| miR-130a | 7.49 ± .52 | 7.82 ± .24 | MIMAT0000141 |
| miR-17 | 7.47 ± .74 | 7.78 ± .29 | MIMAT0000649 |
| miR-31 | 7.42 ± .20 | 7.65 ± .22 | MIMAT0000538 |
| miR-25 | 7.37 ± .51 | 7.61 ± .28 | MIMAT0000652 |
| miR-24 | 7.35 ± .50 | 7.72 ± .07 | MIMAT0000219 |
| miR-677* | 7.35 ± .50 | 7.44 ± .26 | MIMAT0017246 |
| miR-107 | 7.21 ± .32 | 7.47 ± .18 | MIMAT0000647 |
| miR-106b | 7.09 ± .97 | 7.63 ± .40 | MIMAT0000386 |
Toray 3D-Gene Data is expressed as the mean of at least five trials and standard deviation.
TABLE 2.
Relative abundance of select miRNAs as measured by qRT-PCR
| miRNA | Average CT ± SEa | Level of miRNA detected in microarray |
|---|---|---|
| Total RNA isolated from mIMCD-3 cells | ||
| snoRNA-202 | 22.6 ± 0.5 | (control) |
| let-7c | 25.7 ± 0.1 | High |
| let-7f | 25.7 ± 0.1 | High |
| miR-98 | 31.8 ± 0.1 | Moderate |
| miR-199 | 38.8 ± 0.2 | Low |
| Total RNA isolated from the inner medulla | ||
| miR-709 | 19.3 ± 0.2 | NAb |
| miR-98 | 29.3 ± 0.3 | NA |
CT values < 30 are considered to indicate high abundance, CT values in the range of 30–37 are moderate abundance, and > 37 suggest low abundance.
Not applicable.
TABLE 3.
Aldosterone-responsive miRNAs determined by miRNA microarray
| miRNA | Average fold Change ± SDa |
|---|---|
| miR-200c | 2.68 ±0.58 |
| miR-467b* | 2.68 ±0.44 |
| miR-297a* | 2.02 ±0.87 |
| miR-327 | 2.10 ±0.46 |
| miR-1966 | 2.02 ± 0.66 |
Toray 3D-Gene Data is expressed as the mean of at least five trials and standard deviation.
Candidate miRNAs targeting Edn1 expression
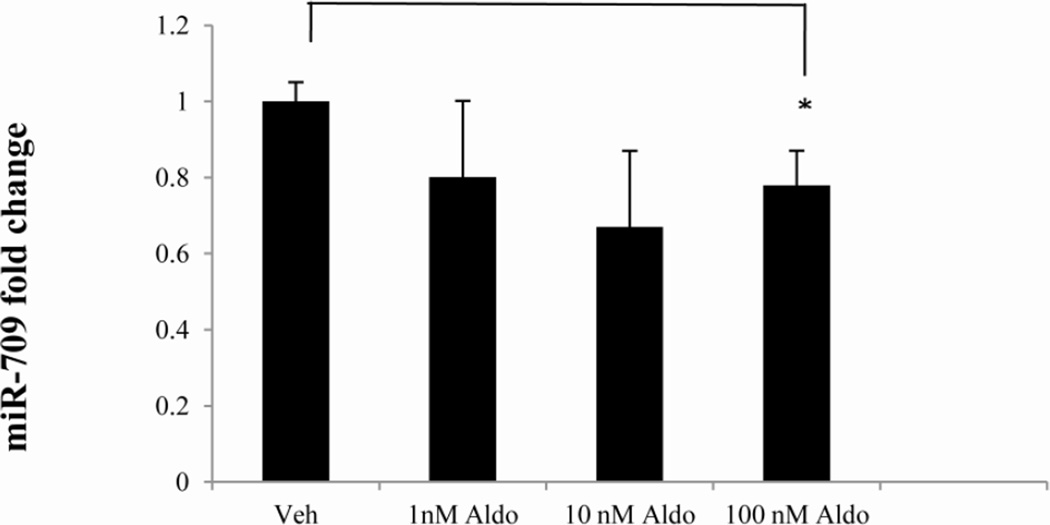
The murine Edn1 3’ UTR was examined using two separate algorithms (Targetscan.org and microRNA.org) to look for putative miRNA binding sites for highly and moderately expressed miRNAs identified by the microarray (Figure 1). Comparison of the miRNAs detected in the microarray experiment with those which had putative binding sites provided a list of candidate miRNAs that might reasonably be expected to act on Edn1 expression. Approximately a dozen miRNAs were highly to moderately expressed and predicted by both algorithms to target the Edn1 3’ UTR (Table 4). We elected to focus on miR-709. miR-709 was very abundant, had three putative binding sites in the Edn1 mRNA 3’ UTR, and tended to be reduced in cells treated with 100 nM aldosterone (Figure 2). At higher levels miR-709 returned to the control value.
TABLE 4.
Candidate miRNAs possibly targeting the murine Edn1 3’ UTR
| miRNA | Microarraya | ||||
|---|---|---|---|---|---|
| Highly expressed | Aldosterone | Vehicle | Total Context Scoreb | miRANDAc | Sitesd |
| miR-709 | 10.26 ± .14 | 10.52 ± .60 | −0.31 | −0.52 | 212^,610,668 |
| let-7a | 9.50 ± .38 | 9.32 ± .54 | −0.34 | −0.99 | 563 |
| let-7c | 8.95 ± .13 | 8.97 ± .52 | −0.34 | −0.99 | 563 |
| let-7d | 8.83 ± .26 | 8.83 ± .38 | −0.34 | −1.01 | 563 |
| let-7f | 8.67 ± .38 | 8.20 ±1.11 | −0.34 | −0.99 | 563 |
| let-7b | 8.95 ± .17 | 8.27 ± .35 | −0.34 | −0.99 | 563 |
| miR-31 | 7.65 ± .22 | 7.42 ± .20 | −0.20 | −1.01 | 220,665^ |
| Moderately expressed | |||||
| let-7i | 7.15 ± .30 | 6.69 ± .71 | −0.34 | −0.99 | 560 |
| miR-200a | 6.68 ± .55 | 5.95 ± .88 | −0.09 | −0.75 | 1^,345 |
| let-7e | 6.66 ± .33 | 6.27 ± .85 | −0.34 | −1.01 | 563 |
| let-7g | 6.12 ± .37 | 5.43 ±1.32 | −0.34 | −0.99 | 561 |
| miR-425 | 4.13 ± .26 | 3.91 ± .56 | −0.08 | −0.88 | 578,802^ |
| miR-185 | 4.40 ± .40 | 3.76 ± .79 | −0.27 | −0.59 | 54,618^ |
Toray 3-D Gene data is expressed as the mean of at least four trials and standard deviation.
Total context score as computed by Targetscan.org. The total context score represents the sum of the contribution of six features: site-type contribution, 3' pairing contribution, local AU contribution, position contribution, target site abundance contribution, and seed-pairing stability contribution.
mirSRV score as computed by microRNA.org.
Position of the first base of a seed sequence within the 3’UTR. ^ indicates the major seed sequence(s) contributing to the mirSRV score.
Figure 2. Effect of aldosterone on miR-709 expression.
mIMCD-3 cells were treated with aldosterone or vehicle for 24 hours. RNA was isolated and miR-709 expression was determined using aTaqMan miRNA assay. miR-709 levels were normalized to an endogenous small, nucleolar RNA control (snoRNA-202). *, p < 0.05
In silico examination of the Edn1 3’ UTR putative miR-709 binding sites (position 212, 610, and 668) provided a prediction of the minimum free energy of secondary structures surrounding these sites. Using mFold (http://mfold.rna.albany.edu) to examine the free energy of the 70 nucleotides flanking the putative miRNA binding sites in the Edn1 3’UTR, the sites at positions 610 and 668 have a minimum free energies that are higher than randomly expected (ΔG = −13.4 kcal/mol) (Martin et al. 2007). This suggests that these sites are accessible to RISC binding. Importantly, miR-709 was readily detectable in inner medulla tissue dissected from mice (Table 2).
Anti-miR Inhibitor Impact on RNA Levels
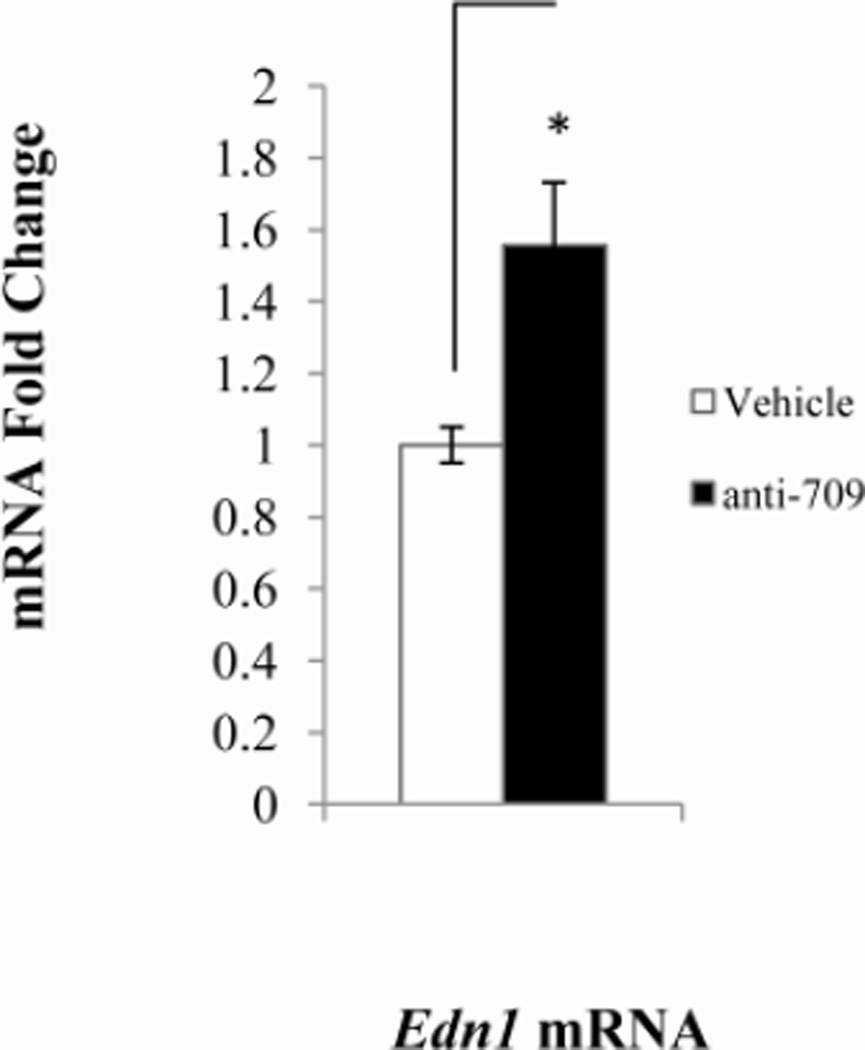
The levels of Edn1 mRNA in mICMCD-3 cells rises as much as three-fold in a dose-dependent manner in response to aldosterone (Stow et al. 2009). In order to determine if miR-709 was indeed targeting Edn1, miR709 action was blocked using anti-miR-709 inhibitors. Transfection of anti-miR-709 resulted in approximately 60% increase in Edn1 mRNA levels (Figure 3).
Figure 3. Effect of anti-miR inhibitors on target mRNA levels.
Levels of mRNAs were determined by qPCR. Probes for qPCR were as follows: beta actin, ACTGAGCTGCGTTTTACACCCTTTC; Edn1, ACTGAGCTGCGTTTTACACCCTTTC; Sgk1, CTCTACGGCCTGCCCCCGTTTTATA, Scn2a1, GATCTTTCCGACTGCTTAGAGTCTT; Atp1b1, GCCCCGCCAGGATTGACACAGATTC. Panels: A, mIMCD-3 cells were transfected with anti-miR-709 for 48 hours; B, anti-miR-1898 for 48 hours; C, anti- 135a-3p for 48 hours. mRNA expression levels were normalized to a β actin control. *, p < 0.05, ‡, p < 0.08, **, p < 0.01
Argonaute-Immunoprecipitation
Although a number of miRNAs have been previously shown to affect Edn1 mRNAs in various cell lines, direct binding of RISC to Edn1 mRNA has not been reported. An Argonaute-immunoprecipitation experiment was used to detect the direct binding of the central RISC subunit to Edn1 mRNA. Isolation of RNA from immunoprecipitates generated using a pan-Argonaute protein antibody demonstrated that Edn1 mRNA was readily detectable by qPCR. In a follow-up experiment, mIMCD-3 cells were treated with anti-miR-709 inhibitors. Edn1 mRNA was barely detectable above background levels in Argonaute immunoprecipitates from these cells. The result suggested that miR-709 likely directs RISC to the Edn1 mRNA.
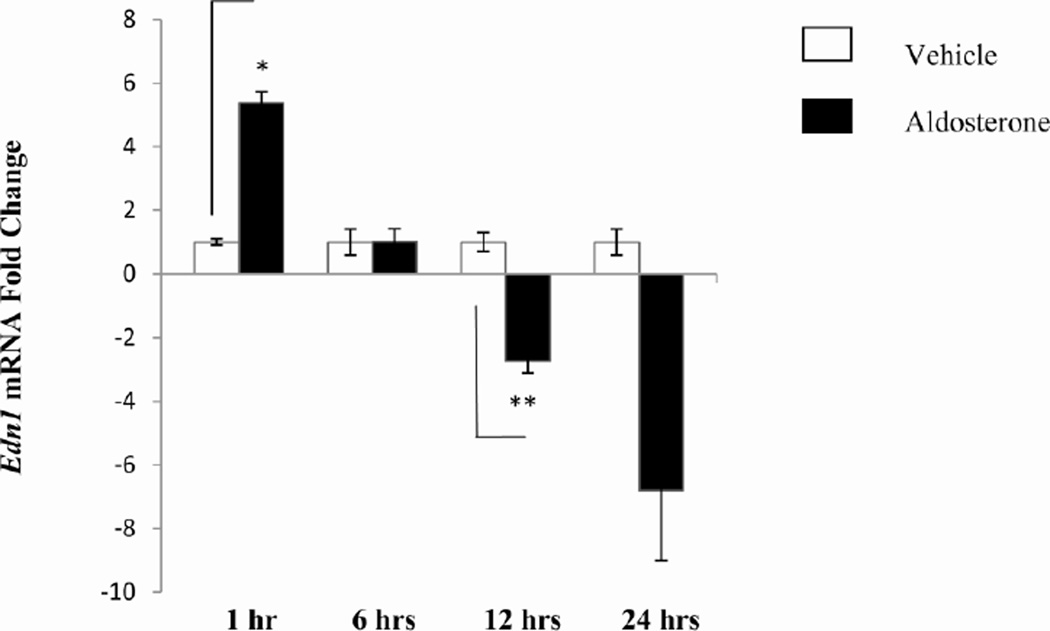
The Argonaute-immunoprecipitation technology was also used to consider the influence of miRNAs on the aldosterone-response in mIMCD-3 cells. Argonaute immunoprecipitation assays were performed on whole cell lysates from cells treated with aldosterone or vehicle over a period of up to 24 hours. After one hour of aldosterone treatment, immunoprecipitation with an anti-Argonaute antibody yielded a five- fold more efficient Edn1 mRNA pulldown (Figure 4). There was a dramatic decrease in the amount of Edn1 mRNA interacting with RISC at later time points. After 24 hours of aldosterone treatment the amount of Edn1 mRNA dropped by eight-fold. These results suggest that decreased miRNA action on Edn1 mRNA may contribute to the build-up of ET-1 seen in the inner medulla in response to aldosterone.
Figure 4. Argonaute Immunoprecipitation of Edn1 mRNA.
A pan-Argonaute antibody was conjugated to magnetic beads. Whole cell lysate isolated from mIMCD-3 cells treated with aldosterone for 1,6,12, or 24 hours was incubated with the argonaute-magnetic bead complex. RNA was isolated from the bound complexes and analyzed using qRT-PCR. *p < 0.006, **p < 0.01, n = 3.
DISCUSSION
In this paper we report the miRNA landscape of mIMCD-3 cells by microarray analysis. The microarray quantitatively measured miRNA abundance for 1079 different murine miRNAs. Interestingly, expression of the vast majority of the miRNAs did not significantly change in response to aldosterone. However, the levels of a select few miRNAs present in moderate to low abundance were altered. The miRNAs identified in the microarray provide a list of candidate miRNAs that can by employed to look for miRNAs that play a role in regulating transcripts involved in sodium reabsorption by the collecting duct. One of the high abundance miRNAs, miR-709, was shown to alter the efficiency of co-immunoprecipitation of Edn1 mRNA with Argonaute proteins. These experiments suggest direct binding of RISC to Edn1 mRNA.
Several miRNAs have been implicated in regulation of ET-1 expression in cell lines derived from various tissues (Jacobs et al. 2013). Yeligar et al. (2009) found miR-199 regulates ethanol-induced ET-1 expression in both rat liver sinusoidal cells and human microvascular endothelial cells. In human umbilical vein endothelial cells, EDN1 mRNA and ET-1 levels increased in the presence of glucose when cells were transfected with a miR-320 mimic (Feng and Chakrabarti, 2012). Additionally, miR-1 expression has been observed in human cardiac and skeletal muscle and its expression was inversely correlated with ET-1 expression (Li et al. 2012). These studies share the use of either miRNA overexpression, introduction of miRNA mimics or antisense miRNAs to detect an effect on Edn1 RNA. These approaches are established and valid, but one of the challenges in examining miRNA functions is to determine the difference between a direct binding of RISC to a target mRNA and a change observed due to off target effects. The Argonaute immunoprecipitation method used here provides direct evidence of a physical interaction between endogenous RISC and Edn1 mRNA.
The human genome does not contain an obvious homologue of miR-709. However, alignment of the Edn1 3’UTRs of 18 mammalian species to the apparent murine miR-709 showed that both sites at positions 610 and 668 are highly conserved. For example, the murine Edn1 3’ UTR around position 610 is identical to the human sequence at 15 of the 16 nucleotides. There are some notable mammalian outliers, both Otolemur galago and Echinops telfairi lack the target site. Although the sequence surrounding position 668 appears to be more divergent, the human and mouse Edn1 3’ UTRs are identical for 15 of 17 nucleotides. It seems reasonable to suspect that humans and other mammals have a miRNA that serves a comparable purpose to the murine miR-709.
Edn1 mRNA levels in mIMCD-3 cells increase in the presence of aldosterone (Gumz et al. 2003). Clearly one mechanism of this increase is due to induction of transcription of the Edn1 gene (Stow, 2011). Activated mineralocorticoid and glucocorticoid receptors mediate the response by binding to a response element located in the 5’ UTR of the Edn1 gene (Stow et al. 2009; Stow et al. 2012). An increase in ET-1 in the inner medullary collecting duct results in sodium retention (Kohan et al. 2011). The time dependent change in the efficiency of Argonaute–pulldown of Edn1 mRNA suggests that miRNA action likely contributes to increased Edn1 levels and consequently sodium retention. A limitation to the Argonaute immunoprecipitation approach is that while we do detect a stable interaction between Argonaute and Edn1 mRNA, we are not able to directly detect the specific miRNA directing RISC to the Edn1 message. The experiment is not designed as a direct measure of changes in the absolute levels of ET-1 production. While inhibition of miR-709 did prevent the binding of Argonaute to Edn1 mRNA, there may be other miRNAs that could act on the Edn1 mRNA at different times in response to either acute or chronic aldosterone treatment. It remains to be seen if changes in miR-709 or other miRNAs are specifically mediating this response, or alternatively, whether changes seen in the pulldown efficiency reflect secondary effects on RISC action.
ACKNOWLEDGEMENTS
This work was supported by Public Health Service Grant 1RO1DK02680 to CSW and BDC, and a predoctoral training grant 5T32DK076541 supported MEJ. Additional support for microarray studies was provided by Toray Industries, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn D, Ge Y, Stricklett PK, Gill P, Taylor D, Hughes AK, Yanagisawa M, Miller L, Nelson RD, Kohan DE. Collecting duct-specific knockout of endothelin-1 causes hypertension and sodium retention. J Clin Invest. 2004;114:504–511. doi: 10.1172/JCI21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Chakrabarti S. miR-320 regulates glucose-induced gene expression in diabetes. ISRN Endocrinol. 2012:549875. doi: 10.5402/2012/549875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumz ML, Popp MP, Wingo CS, Cain BD. Early transcriptional effects of aldosterone in a mouse inner medullary collecting duct cell line. Am J Physiol. 2003;285:F664–F673. doi: 10.1152/ajprenal.00353.2002. [DOI] [PubMed] [Google Scholar]
- Jacobs ME, Wingo CS, Cain BD. An emerging role for microRNA in the regulation of endothelin-1. Front Physiol. 2013;4:22. doi: 10.3389/fphys.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnström P, Fryer TD, Richards HK, Harris NG, Barret O, Clark JC, Pickard JD, Davenport AP. Positron emission tomography using 18F-labelled endothelin-1 reveals prevention of binding to cardiac receptors owing to tissue-specific clearance by ET B receptors in vivo. Br J Pharmacol. 2005;144:115–122. doi: 10.1038/sj.bjp.0706064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2012;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Kohan DE, Rossi NF, Inscho EW, Pollock DM. Regulation of blood pressure and salt homeostasis by endothelin. Physiol Rev. 2011;91:1–77. doi: 10.1152/physrev.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, He B, Zhang H, Shan SF, Liang Q, Yuan WJ, Ren AJ. The inhibitory effect of miRNA-1 on ET-1 gene expression. FEBS Lett. 2012;586:1014–1021. doi: 10.1016/j.febslet.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Lynch IJ, Welch AK, Kohan DE, Cain BD, Wingo CS. Endothelin-1 inhibits sodium reabsorption by ETA and ETB receptors in the mouse cortical collecting duct. Am J Physiol Renal Physiol. 2013;305:F568–F573. doi: 10.1152/ajprenal.00613.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mawji IA, Robb GB, Tai SC, Marsden PA. Role of the 3'-untranslated region of human endothelin-1 in vascular endothelial cells. Contribution to transcript lability and the cellular heat shock response. J Biol Chem. 2004;279:8655–8667. doi: 10.1074/jbc.M312190200. [DOI] [PubMed] [Google Scholar]
- Moridaira K, Nodera M, Sato G, Yanagisawa H. Detection of prepro-ET-1 mRNA in normal rat kidney by in situ RT-PCR. Nephron Exp Nephrol. 2003;95:e55–e61. doi: 10.1159/000073672. [DOI] [PubMed] [Google Scholar]
- Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics. 2008;23:624. doi: 10.1186/1471-2164-9-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimunde FM, Castañares C, Redondo-Horcajo M, Lamas S, Rodríguez-Pascual F. Endothelin-1 expression is strongly repressed by AU-rich elements in the 3'-untranslated region of the gene. Biochem J. 2005;387:763–772. doi: 10.1042/BJ20041687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow LR, Gumz ML, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Wingo CS. Aldosterone modulates steroid receptor binding to the endothelin-1 gene (edn1) J Biol Chem. 2009;284:30087–30096. doi: 10.1074/jbc.M109.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow LR, Voren GE, Gumz ML, Wingo CS, Cain BD. Dexamethasone stimulates endothelin-1 gene expression in renal collecting duct cells. Steroids. 2012;77:360–366. doi: 10.1016/j.steroids.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow LR, Jacobs ME, Wingo CS, Cain BD. Endothelin-1 gene regulation. FASEB J. 2011;25:16–28. doi: 10.1096/fj.10-161612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch AK, Jacobs ME, Wingo CS, Cain BD. Early progress in epigenetic regulation of endothelin pathway genes. Br J Pharmacol. 2013;168:327–334. doi: 10.1111/j.1476-5381.2012.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microRNA-199. J Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]