Abstract
The interaction of myosin with actin filaments is the central feature of muscle contraction and cargo movement along actin filaments of the cytoskeleton. Myosin converts the chemical energy stored in ATP into force and movement along actin filaments. Myosin binding to actin induces conformational changes that are coupled to the nucleotide-binding pocket and amplified by a specialized region of the motor domain for efficient force generation. Tropomyosin plays a key role in regulating the productive interaction between myosins and actin. Here, we report the 8 Å resolution structure of the actin-tropomyosin-myosin complex determined by cryo electron microscopy. The pseudo-atomic model of the complex obtained from fitting crystal structures into the map defines the large actin-myosin-tropomyosin interface and the molecular interactions between the proteins in detail and allows us to propose a structural model for tropomyosin dependent myosin binding to actin and actin-induced nucleotide release from myosin.
Keywords: actin, myosin, tropomyosin, cryo electron microscopy
Introduction
Muscle contraction and many other motile processes such as cell motility, endocytosis or cytokinesis are mediated by the interaction of members of the myosin superfamily with actin filaments. ATP binding and hydrolysis by myosin trigger a sequence of conformational changes that are associated with an approximately 10.000-fold change in actin affinity. Actin binding in turn promotes the successive release of Pi and ADP. The movement of myosin along actin filaments is thought to be predominately caused by the large conformational changes associated with the release of the hydrolysis products. Rapid rebinding of ATP leads to myosin dissociation from the filament and starts a new cycle1–3.
This mechanochemical reaction cycle, first described by Lymn and Taylor1, has only been partially characterized at the structural level. Crystal structures of myosin in different states provide detailed insights into states in which myosin is dissociated from the actin filament4,5. However, since the complex of filamentous (F-)actin with myosin is refractory to crystallization, structural information on the actin-myosin complex was only obtained at medium resolution from X-ray fiber diffraction6,7 and cryo electron microscopic (EM) studies8–10. Due to its helical symmetry, F-actin decorated with myosin motor domains in the absence of ATP is an ideal subject for cryo-EM. The highest resolution obtained so far for a three-dimensional cryo-EM reconstruction was 14 Å and allowed the rigid-body fitting of actin and myosin crystal structures10. Along with spectroscopic studies and crystal structures of myosin II and Va in the absence of nucleotide11–13, this reconstruction strongly suggests that myosin closes its so-called 50-kDa cleft upon binding to F-actin. This conformational change is coupled to the opening of the nucleotide-binding pocket in a manner that is not yet fully understood.
A better mechanistic description of the interaction of actin and myosin and its regulation requires a spatial resolution at which secondary structure elements of proteins can be identified directly. Unfortunately, F-actin filaments decorated with myosin motor domains are flexible, which disrupts the long-range order of the proteins. As it is challenging to restore bent filaments a posteriori14, we searched for filaments optimized for EM (fully decorated, straight, stiff and not bundled) by screening many combinations of F-actins and myosin motor domains. The combination of rabbit skeletal muscle F-actin with the motor domain of Dictyostelium discoideum myosin-IE15 (myoE), a fast single-headed molecular motor that is involved in the phagocytic uptake of solid particles and cells16, proved to be the best specimen for this study. The addition of skeletal muscle tropomyosin, which wraps around the F-actin filament and in conjunction with troponin regulates the Ca2+-dependent contraction or relaxation of muscles17, improved the rigidity of the filaments even further. Finally, we collected high-quality data using an electron microscope equipped with an Omega in column filter (Jeol JEM 3200 FSC) to achieve maximal resolution.
Structure determination and overall architecture
We observed actin filaments to be either fully decorated with myosin and tropomyosin or not to bind the proteins at all (Fig. 1). This indicates that tropomyosin and myosin have a higher affinity to the F-actin-myosin and F-actin-tropomyosin complex, respectively, than to F-actin alone. These findings are in good agreement with previous studies showing that the myosin motor domain subfragment 1 (S1) binds to F-actin-tropomyosin filaments with a 7-fold higher affinity than to undecorated F-actin18 and that myosin increases the actin affinity of tropomyosin19.
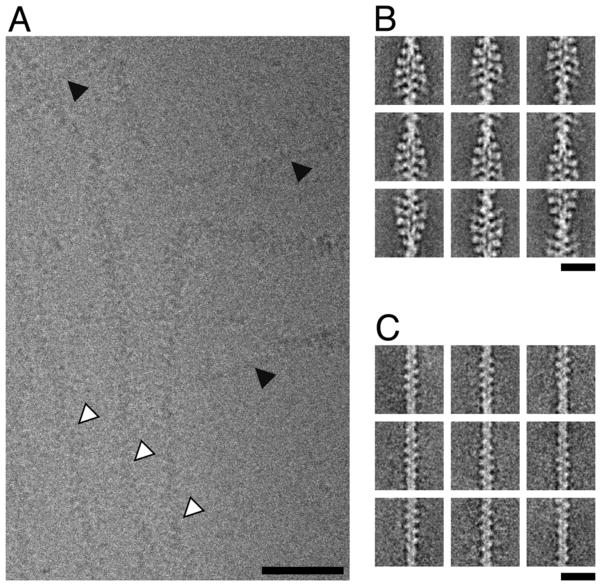
Figure 1. Cryo electron microscopy of F-actin (undecorated actin) and the complex of F-actin, myosin-IE motor domain and tropomyosin (decorated actin).
(A) Subarea of an unprocessed image of the vitrified sample recorded at a defocus of 1 μm. The presence of a mixture of filament types is evident (decorated filaments marked by white arrows, undecorated filaments marked by black arrows). Filaments appear to be either decorated or undecorated over their complete length. Scale bar, 50 nm. (B) Representative class sums of the decorated filaments (35,374 segments). No classes with partial decoration were identified. Scale bar, 20 nm. (C) Representative class sums of the undecorated filaments (4,629 segments). No classes with partial decoration were identified. Scale bar, 20 nm.
After separation of the data into two sets for either decorated or non-decorated filaments (Fig. 1), we determined three-dimensional (3D) cryo-EM structures of both F-actin and the F-actin-tropomyosin-myoE (ATM) complexes (Supplementary Fig. 1A). The F-actin data set contained only ~ 4,600 segments but nevertheless resulted in a 3D map at a resolution of 8.9 Å (Fig. 2A) (also see Methods and Supplementary Fig. 2). The resolution of the ATM complex, which was determined using almost ten times more segments yielded only similar resolution and had artifactual density distribution with myoE underrepresented in comparison to F-actin and tropomyosin. In addition, the regions most distant from the helical axis were not as well resolved as central regions. By comparing three independent reconstructions with the same data to estimate the confidence of the map, we found that the reproducibility of the outer regions of myoE showed significant variability in comparison with those of F-actin and tropomyosin. This indicated a structural heterogeneity of myoE. Therefore, we adapted the codimensional principal component analysis method20 to structures with helical symmetry (Behrmann et al., accepted) and identified the presence of five subpopulations in the data set.
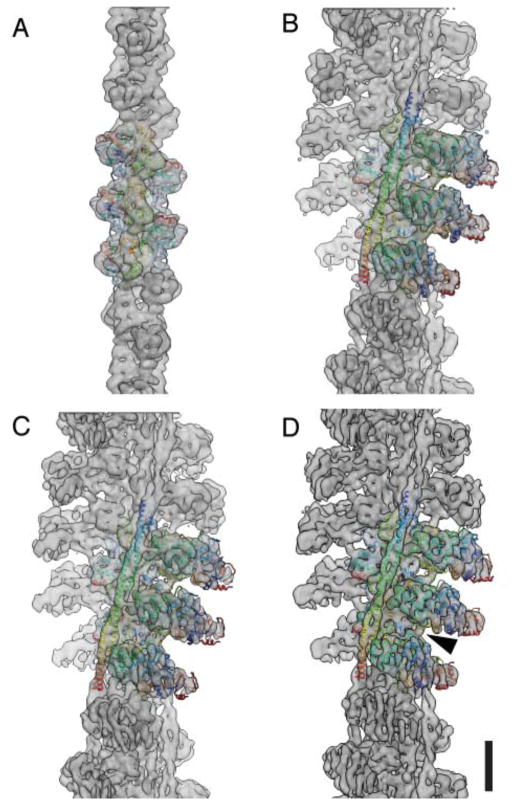
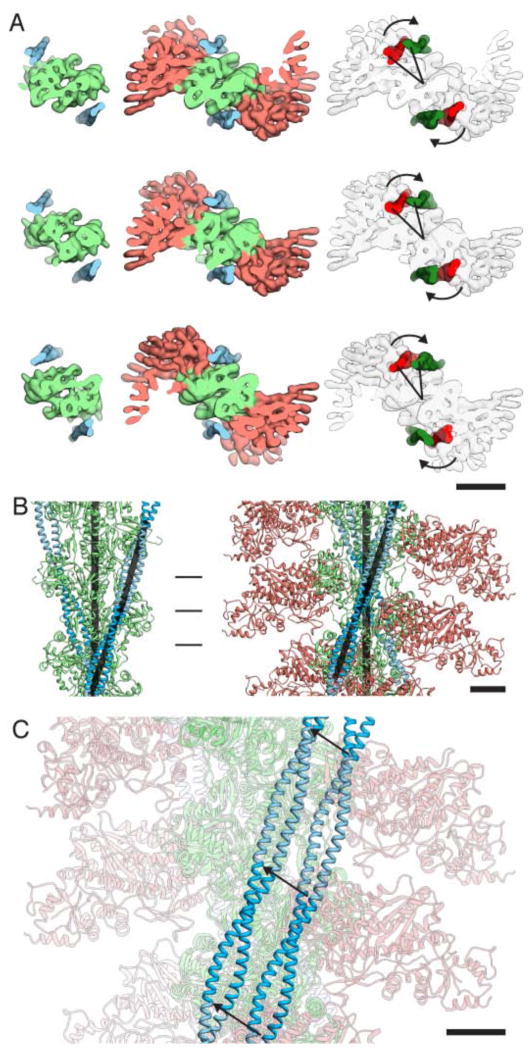
Figure 2. Fit of pseudo-atomic models into the electron density maps of the undecorated F-actin filament and three ATM complex conformers.
Electron density maps for (A) the undecorated filament and (B–D) the three decorated filaments (group 1–3). Central subunits are depicted as ribbon traces of the Cα coordinates colored with a rainbow gradient from blue (amino terminus) to red (carboxy terminus). Arrow indicates the Milligan contact. Scale bar, 5 nm.
We determined structures for three of the five subpopulations (see also Methods). The resolutions of the maps were 7.7 – 8.1 Å with an even density distribution and reproducible myoE densities (Fig. 2B–D, Supplementary Fig. 1). The pseudo-atomic model of F-actin obtained from a previously published cryo-EM study21 and the atomic model of myoE15 were then fit into the electron densities (Fig. 2) (see also Methods and Supplementary Information). Since no crystal structure of the full tropomyosin complex has been obtained, we used a previously published tropomyosin model that was created based on crystal structures of subfragments and its optimal electrostatic fit to actin22. In this way we obtained a pseudo-atomic model of the complete ATM complex that defines the molecular interactions between the partner proteins.
Conformational changes in F-actin
We first compared the F-actin models obtained from our four structures (Fig. 2A–D) with each other and with previously published F-actin models21,23. In order to characterize differences between the models we superposed them on an invariable region identified as the β-sheets of subdomain (SD) 3, calculated the root mean-square deviations (RMSD) of all alpha carbon positions and, in order to determine major correlated variability between the models, which can be interpreted as conformational variability of structures, we performed eigenanalysis of the distribution of alpha carbon atoms within the family of models (see also Methods). The low RMSDs showed that the F-actin models obtained from the three ATM reconstructions (Fig. 2B–D) differed only marginally (Supplementary Fig. 2A–F). We identified the highest variability in the DNaseI binding loop and the region next to the N-terminus upon comparison of our F-actin model (Fig. 2A) with the previously published models21,23. The remaining regions of actin were almost invariable (Supplementary Fig. 2 G–L). The positions of the variable regions agree with those identified by Splettstoesser et al. applying molecular dynamics simulations on different published actin structures24.
To identify conformational changes in F-actin upon formation of the ATM complex, we compared our F-actin model with that of the ATM complex (Fig. 3A–C). While the low RMSDs between the carbon-alpha positions of the models indicate that conformational changes are minimal and distributed over the whole structure (Fig. 3A/B), the highest differences were localized again in the region of the DNaseI binding loop and near the N-terminus (Fig. 3C). It is tempting to speculate that this variability indicates conformational changes upon myosin binding, especially since the regions of highest variability are directly involved in complex formation (Fig. 4). Although our sample contained both decorated and undecorated F-actin and we can exclude structural changes due to specimen preparation such as ice thickness and fluid flow during blotting, we cannot rule out that the observed differences are caused just by a higher flexibility of these F-actin regions, as was described for F-actin24,25.
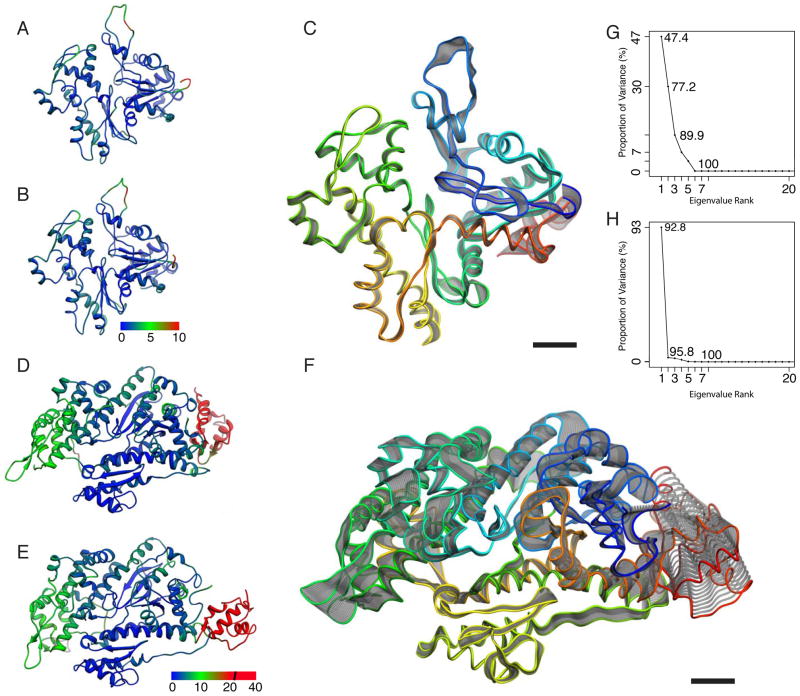
Figure 3. Analysis of conformational changes induced by complex formation.
(A–C) Analysis of the variability of the F-actin models between the undecorated and with myoE and tropomyosin decorated state. (A–B) Atomic models of an undecorated and decorated (group 3) F-actin subunit, respectively. Models are color coded by Cα RMSD values ranging from 0 Å (blue) to 10 Å (red). Major differences are found in carboxy terminal region of the DNase I binding loop (residues 39–52) and the amino terminal region (residues 1–5). (C) Visualization of the first eigenvector obtained after PCA as a trajectory of standard deviation scaled displacements from the average structure. No clear pattern of displacements is discernable. Actin is colored with a rainbow gradient from blue (amino terminus) to red (carboxy terminus). (D–F) Analysis of the variability of the myoE models between pre-power stroke and rigor state. (D–E) Atomic models of pre-power stroke (PDB ID: 1LKX chain C) and rigor state (group 3 myosin) color coded by Cα RMSD values ranging from 0 Å (blue) to 20 Å (red). In the converter domain, displacements ranged up to 40 Å. The second highest displacement locates to the UD50. (F) Visualization of the first eigenvector for myoE. While the LD50, especially the helix-loop-helix motif, is almost invariable, clear changes are seen in all other domains indicating closure of the 50-kDa-cleft and rotation of the converter domain. Myosin is colored with a rainbow gradient from blue (amino terminus) to red (carboxy terminus). (G–H) Eigenvalue spectra for actin PCA and myoE PCA, respectively. While changes upon complex formation cannot easily be accounted for in the case of actin, one eigenvector is enough to account for over 92 % of observed variance in the case of myosin. Scale bar, 1 nm.
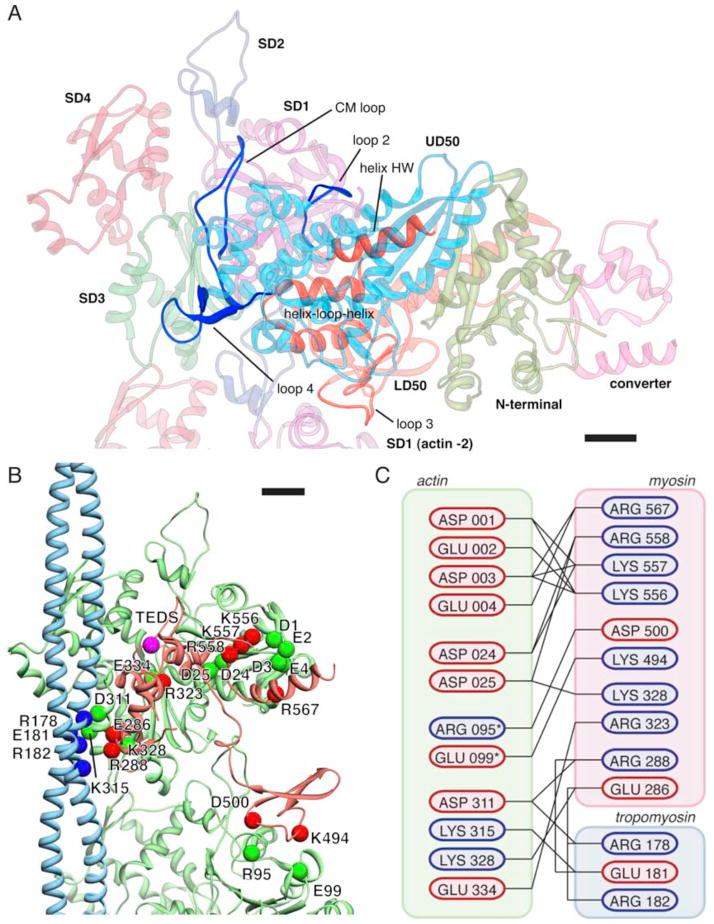
Figure 4. The binding interface with potential key electrostatic interactions between myosin, tropomyosin and actin(0) and (−2).
(A) Overview of the binding interface between myosin and actin. Regions on myosin involved in actin binding are highlighted and labeled. (B) Pseudo-atomic model of the complete binding interface. Potential interaction partners with complimentary charges in close proximity are depicted as colored spheres. In addition, the TEDS site (which is not part of the interface) is depicted as a pink sphere. The interface between actin(0) and myosin extends over 1,450 Å2, the interface between actin(−2) and myosin over 370 Å2, the interface between actin(0) and tropomyosin over 210 Å2 and the interface between myosin and tropomyosin over 300 Å2. (C) Cartoon representation of the interface. Residues are colored by charge at pH 7.4. Asterisk denotes residues that are part of the actin (−2) interface. Scale bars, 1 nm.
From pre-power stroke to rigor state
We then compared the myoE models obtained from our three reconstructions (Fig. 2B–D) with each other and with the four previously published conformers of the myoE pre-power-stroke crystal structure15. We identified an invariable core region in the lower 50 kDa domain (LD 50), superposed all models on this region, determined RMSDs and we performed eigenanalysis of the distribution of alpha carbon atoms within the family of different myosin models as well as within the family of actin models (see also Methods). Major differences in the upper 50 kDa domain (UD50) and especially in the converter domain become obvious when the myoE models obtained from the three ATM reconstructions (Fig. 2B–D) are compared (Supplementary Figs. 3 A–F and 4A). This suggests that myosin displays conformational flexibility even when it is tightly bound to F-actin (Supplementary Movie 1A–B), as previously reported by Klein et al.26. Interestingly, high variability was also observed in the region of the converter domain when the four conformers of the myoE pre-power-stroke crystal structure were compared (Supplementary Fig. 3G–L)15. This indicates that flexibility of the converter domain is an intrinsic property of myoE.
To identify conformational changes in myosin upon binding to F-actin, we compared the pre-power stroke with the rigor state conformations of myoE, identifying large conformational changes in the UD50, the N-terminal and the converter domain (Fig. 3D–F). The UD50 rotates towards actin resulting in the closure of the 50-kDa cleft (Supplementary Movies 2 and 3A–D). The converter domain rotates in the opposite direction and swings the lever arm by ~70°.
Actin-Myosin interface
Our pseudo-atomic models clearly show that myosin interacts with two adjacent actin molecules via several loops forming a large contact surface (1,820 Å2). Whereas loop2 (547–561), and helix HW (563–577), loop4 (278–298) and the cardiomyopathy (CM) loop (322–342) of myoE form large contacts to SD 1 and 3 of the neighboring actin molecule, loop3 (482–501) attaches to SD 1 of the next lower actin along the long pitch helix. In addition, a helix-loop-helix motif of the LD50 domain (445–481) protrudes into a cavity, which is formed by SD1 and 3 of the upper and by the SD2 domain of the lower actin, thereby interacting with both of them (Fig. 4 and Supplementary Fig. 5A). As becomes obvious from examining hydrophobic surface potentials, both the helix-loop-helix motif and the cavity are predominantly hydrophobic (Supplementary Fig. 5B). In addition, two clusters of hydrophobic residues on the CM-loop (residues 327–330 and 335–341) interact with hydrophobic regions on actin (residues 29–31 and 329–334). As shown before, these hydrophobic stretches are important to maintain the myosin strong binding state27.
In contrast, the contacts between other regions are mostly mediated by electrostatic interactions and potentially involve the formation of salt bridges (Fig. 4, Supplementary Fig. 5C). Salt bridge formation is probably arranged in a double sandwich composed of the highly negatively charged N-terminus of actin (D1, E2, D3, E4), a conserved positively charged region in myo-IE loop2 and on helix HW (K556, K557, R558, R567), a negatively charged loop from actin SD1 (residue 20–28 (D24, D25)), and conserved positively charged residues on the myo-IE CM-loop (R323, K331, R 332) (Supplementary Figs. 5D and 6).
The importance of the involved residues has been previously shown. MyoE residue R332 corresponds to human β-cardiac myosin R403, whose mutation to glutamine reduces the ATPase activity of myosin in the presence of actin and is associated with familial hypertrophic cardiomyopathy28.
The two conserved lysines at the C-terminal end of loop2 (K652/K653 in smooth muscle myosin-II and K556/K557 in myoE) are necessary for triggering actin activation. The net charge and charge density of loop2 greatly affect actin affinity and for non-processive myosins actin-activated ATPase activity29–32. Loop2 sits not only in the middle of the described double sandwich but also at the center of the major actin-myosin interface. Our results shed light on the contribution of the actin N-terminus to the actin-myosin interaction. It is well known that negative charges at the N-terminus of actin are of critical importance for the ATP-dependent actin-myosin interaction33, especially for the weakly bound actomyosin states34. The N-terminus is one of the rare regions of actin that is not highly conserved (Supplementary Fig. 5D). It is therefore tempting to speculate that changes in this area affect not only binding affinity but also serve as a means for discriminating between different myosin isoforms.
The conserved TEDS site (S334, in our structure mutated to E334 to mimic phosphor-serine) located in the CM loop is not oriented towards actin (Fig. 4A–B). As becomes obvious from our model, its negative charge stabilizes the CM loop through interaction with the neighboring residue K332 as was also suggested from comparing the unphosphorylated myoE crystal structure, where the CM loop was disordered, to structures of other myosins with a glutamate or aspartate residue at this position mimicking either phospho-serine or -threonine15.
It was shown that the residues DEA460-462 on the myosin helix-loop-helix motif are important for the myosin-actin interaction35,36. Although these conserved residues are at the actin-myosin interface in our model there are no obvious complementary charges on the actin surface (Supplementary Figs. 5C,D). It is therefore more likely that the negative charges in this region are important for stabilizing myosin loop 2 (R567) by electrostatic interactions. This would also explain the observed effect of charge-modifying mutations on actin affinity36.
Schröder et al. and Rayment et al. suggested an additional stabilization of the actomyosin complex by interaction of loop3 with the SD2 of the adjacent actin8,37,38. Using our cryo-EM structure, we are now able to prove that this contact exists (Fig. 2D) and based on our model we can demonstrate that it is most likely formed by potential salt bridges (myosin D500 – actin R95 and myosin K494 – actin E99) (Fig. 4). However, its influence on either the formation of a high-affinity actin-myosin interface or modulation of actomyosin ATPase activity is not supported by biochemical experiments, which moreover indicate that loop3 mediated contacts are not a feature shared between all myosin isoforms39.
Tropomyosin in the M(open) -state
The ~40 nm long tropomyosin dimer winds itself as a α-super helix around the actin filament. Overlaps between the N- and C-termini of neighboring tropomyosins occur at every seventh actin, leading to the formation of a continuous rope-like structure. Tropomyosin is divided into seven pseudo-repeating units, each of which binds to a successive actin monomer along F-actin. Since these units are very similar and not distinguishable at a resolution of ~8 Å, we applied the helical symmetry of actomyosin in order to achieve maximal resolution but at the same time accepting that the overlap region of the N- and C-termini will not be visible in the maps.
Our electron densities show clearly two separated rod-like features corresponding to the α-helices of tropomyosin winding around the actomyosin complex (Fig. 2B–D). It becomes obvious from our structure and model that tropomyosin fits nicely into the groove between actin and myosin (Supplementary Movie 4). This shape complementarity, referred to as Gestalt-binding40, was described by Holmes and Lehman as a necessary prerequisite for the specific actin-tropomyosin interaction. In addition, the complex is stabilized by electrostatic interactions between myosin and tropomyosin. Myosin loop4 (285–292) acts as a central linker interacting both with actin and tropomyosin via potential salt bridges (Fig. 4). In muscle thin filaments, three states of tropomyosin exist. At low Ca2+ concentrations troponin holds tropomyosin in the B (blocked) position. When Ca2+ levels increase tropomyosin can move by ~25° to adopt the C (closed) state that allows initial myosin binding to actin. Myosin binding induces a further ~10° shift to the M (open)-position41,42. Li et al. identified a position of tropomyosin on actin with optimal electrostatic complementarity and postulated this to be its preferred position in the absence of troponin or other constraints43 (Fig. 5). Since this position covers the myosin-binding sites on actin SD3 and part of actin SD2 (Supplementary Movie 5A–B), it corresponds rather to the B- than to the C-state of tropomyosin when troponin is present. In order to move from this position to the M-state, the position of tropomyosin in our cryo-EM structure, tropomyosin needs to be slightly rotated and shifted by ~23 Å (Fig. 5, Supplementary Movie 5A–B). This corresponds to an azimuthal rotation of ~31.5° and results in an overall upward shift of tropomyosin along the filament (Fig. 5, Supplementary Movie 6). In comparison to the model of the C-state, the torsion angle of tropomyosin does not significantly change, but its radius is slightly decreased (Fig. 5).
Figure 5. Tropomyosin in the B and M state.
(A) Top views along the filament axis at the three positions indicated by vertical markers in (B). Upon myosin binding, the position of tropomyosin is rotated azimuthally by ca. 30° as indicated by the arrows. The density of F-actin with tropomyosin was calculated at 8 Å resolution based on the tropomyosin model in its B-state43. In addition to the rotation, the radial position of tropomyosin is reduced from 43 Å to 40 Å. Scale bar, 5 nm. (B) Side view orthogonal to the filament axis. The vectors defined by the positions of the Cα atoms of two pseudorepeats are depicted as black bars. The angle between the tropomyosin filament and the actin filament remains unchanged upon myosin binding at ~20°. Scale bar, 2.5 nm (C) Overlay of the two states depicted in (B) with actin and myosin faded. Displacement of tropomyosin can be described as a shift along the surface of the actin filament with additional lateral movement. MyoE, tropomyosin and actin are salmon, light green and blue, respectively. Scale bar, 2.5 nm.
The large azimuthal rotation suggests that we observe a shift from the B- to the M-state rather than from the C- to the M-state, using the nomenclature adopted for troponin-decorated actin filaments. It has long been deliberated whether tropomyosin slides or rolls over actin when changing between the different states22. Our results show that tropomyosin undergoes mainly sliding movements on F-actin.
Model for myosin binding and the actin-catalyzed power stroke
Our pseudo-atomic model provides important insights into the actin-myosin-tropomyosin interaction. Although important intermediates are still missing to explain the complete process of myosin binding, actin-catalyzed ATP hydrolysis, and the power stroke, we can use the information provided by our models of myoE in the pre-power stroke and rigor state to suggest the following mechanism for it.
Since our models show that the LD50 domain and loop4 are the most invariant regions between myosin in the pre-power stroke and the rigor state (Fig. 3D–E), we propose that myosin initially binds mainly via this region to actin. In contrast to the other binding sites on actin, the ones for the LD50 domain and loop3 only exist on F-actin and not on G-actin since a contact site between two different actins forms them. Initial binding in this region might therefore also serve as specificity control.
In order for myosin to bind to tropomyosin-decorated actin in the absence of troponin, it would have to be azimuthally rotated by 20° away from its final binding position. This mode of binding is consistent with previously described interactions of loop 2 with actin29,44. A subsequent rotation and closure of myosin’s 50-kDa cleft induced by strong interactions between actin and myosin will sterically push tropomyosin further aside by about 23 Å (Supplementary Movie 7). At the same time tropomyosin diffusion from the actomyosin complex is prevented by Gestalt-binding and electrostatic interactions. In this model, an oscillation of tropomyosin between the M- and B-state as described by McKillop and Geeves41 is not necessary. However, if myosin cannot rotate in this manner, its binding to actin will be sterically hindered as long as tropomyosin remains in the B-state and oscillation of tropomyosin is required (Supplementary Movie 8).
Whereas actin does not change its conformation significantly during the interaction with myosin, myosin undergoes large changes. A rotation of the UD50 towards actin closes the 50-kDa cleft (Fig. 3D,E, Supplementary Movies 2,3). During this movement, helix HK (241–256) squeezes with its hydrophobic residues between helix HG (127–146) and helix HH (342–372), thereby replacing their hydrophobic interactions and pushing the HG helix aside (Supplementary Fig. 4B). This displacement is then directly transmitted to switch I and loop1. Pi and ADP binding by switch I is destabilized and the backdoor for Pi is opened. Loop1 transmits the force to the N-terminal domain (Supplementary Fig. 4C). Induced by strong binding of loop 2 to actin, the HW helix (562–579) of the LD50 is pulled towards actin, transmitting the force to the fifth β-strand (581–589) (Supplementary Fig. 4D).
As a result, pushing by loop1 and pulling by the HW loop compress the N-terminal domain like a spring, which relaxes by rotation only after Pi is released. This is the mechanistic basis for the amplification of movement and release of energy obtained from ATP hydrolysis by interaction with myosin at the LD50 and UD50 domain. The P-loop can then be moved upwards, destabilizing Mg2+-ADP binding. According to the model, rotation of the N-terminal domain subsequently pulls on the SH-1 and SH-2 helix, which induces a rotation of the converter domain accompanied by a stretching of the kinked relay helix and a large swing of the lever arm (Supplementary Fig. 4E). The power stroke can only be fully completed after Mg2+-ADP has been released from the nucleotide-binding pocket.
However, in order to support our model for myosin binding and the actin-catalyzed power stroke additional structures of intermediate states, such as the weakly and strongly bound ADP state are needed. We are currently collecting relevant EM data, also including data sets of additional actomysin complexes meant as a control.
Methods Summary
Filament preparation
F-actin was prepared from rabbit skeletal muscle. Recombinant tropomyosin and myo-IE were purified from E. coli and Dictyostelium discoideum cells, respectively. Thin filaments were prepared by mixing F-actin (0.1 mg ml−1) with tropomyosin at a molar ratio of 1:7 in 5 mM HEPES-OH, pH 7.2, 100 mM KCl, 2 mM MgCl2, 50 mM glutamine, 50 mM arginine. The full complex was prepared by additionally adding myoE at slight molar excess to the thin filaments. The resulting filament suspension was used for cryo-EM within 15 min to minimize bundling.
Electron microscopy
For cryo-EM, the sample was diluted 10-fold, applied to C-Flat R2/1 holey carbon grids (Protochips) and vitrified. Filaments were imaged with a JEM-3200FSC electron microscope (JEOL) at an acceleration voltage of 200 kV operated at liquid nitrogen temperature. An in-column omega energy filter was used to improve image contrast by zero-loss filtering with a slit-width of 12 eV. Micrographs were recorded at 169,644x magnification with an 8k × 8k TemCam-F816 CMOS camera (TVIPS) under minimal dose conditions (15–20 e−/Å2) resulting in an effective pixel size of 0.92 Å on the specimen scale.
Image processing
The SPARX software45 was used for all image-processing steps, with the exception of the initial defocus determination, which was done using CTFFIND346. These initial defocus settings were subsequently refined as described previously47. Decorated filaments were identified using K-means clustering and processed according to the SPARX implementation of the iterative helical real-space refinement scheme14. Structural heterogeneity found in myosin was separated using a co-dimensional PCA modified to work with structures having helical symmetry (Behrmann et al., accepted).
Fitting
Atomic models of F-actin21 (PDB ID, 3MFP), myoE15 (PDB ID, 1LKX) and tropomyosin43 were fit into the EM volumes first by rigid-body using Chimera48 and then by flexible fitting with DireX49.
Supplementary Material
Acknowledgments
We are grateful to R.S. Goody for continuous support and useful comments on the manuscript. We thank Oliver Hofnagel for assistance at the electron microscope and Ingrid Vetter for help with the installation of SPARX. We are grateful to Gunnar Schröder for helpful comments on the use of DireX and William Lehman for providing the tropomyosin model in the B-state. We thank Krishna Chinthalapudi, Ralph Diensthuber, Sarah M. Heissler and Manuel Taft for providing purified proteins. This work was supported by the ‘Deutsche Forschungsgemeinschaft’ Grants RA 1781/1-1 (to S.R.), MA 807/17-1 (to H.G.M), MA 1081/16-2 (to. D.J.M), the ‘Fonds der chemischen Industrie’ Grant 684052 (to E.B.), funding from the European Union Seventh Framework Programme (FP7/2007-2011) under grant agreement n° 228971 (Molecular Motors-based Nano Devices –MONAD) (to D.J.M.), the NIH Grants GM_U54_094598 and GM R01 60635 (to P.A.P) and the Max Planck Society (to S.R. and E.B.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
M.M. and. H.G.M. purified proteins, E.B. and H.G.M. screened samples and collected EM data, E.B., P.A.P and S.R. processed, refined and analyzed EM data; E.B. and S.R. wrote the paper. D.J.M. and S.R. designed the study. D.J.M. co-wrote the paper. All authors discussed the results and commented on the manuscript.
The coordinates and structure factors for the actin-myosin-tropomyosin EM reconstructions and the atomic models have been deposited in the EM Data Bank and the RCSB Protein Data Bank under accession codes EMD-1987-1990 and 4a7n, 4a7l, 4a7h, 4a7f, respectively.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 2.Mannherz HG, Leigh JB, Holmes KC, Rosenbaum G. Identification of the transitory complex myosin-ATP by the use of, -methylene-ATP. Nat New Biol. 1973;241:226–229. doi: 10.1038/newbio241226a0. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney HL, Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 4.Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. Embo J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huxley HE, Faruqi AR, Bordas J, Koch MH, Milch JR. The use of synchrotron radiation in time-resolved X-ray diffraction studies of myosin layer-line reflections during muscle contraction. Nature. 1980;284:140–143. doi: 10.1038/284140a0. [DOI] [PubMed] [Google Scholar]
- 7.Irving M, et al. Conformation of the myosin motor during force generation in skeletal muscle. Nat Struct Biol. 2000;7:482–485. doi: 10.1038/75890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rayment I, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 9.Volkmann N, et al. Myosin isoforms show unique conformations in the actin-bound state. Proc Natl Acad Sci U S A. 2003;100:3227–3232. doi: 10.1073/pnas.0536510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 11.Coureux PD, et al. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 12.Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin-induced nucleotide release in myosin. Nat Struct Biol. 2003;10:826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
- 13.Conibear PB, Bagshaw CR, Fajer PG, Kovacs M, Malnasi-Csizmadia A. Myosin cleft movement and its coupling to actomyosin dissociation. Nat Struct Biol. 2003;10:831–835. doi: 10.1038/nsb986. [DOI] [PubMed] [Google Scholar]
- 14.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 15.Kollmar M, Durrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. Embo J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durrwang U, et al. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci. 2006;119:550–558. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- 17.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251:191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- 18.Geeves MA, Halsall DJ. The dynamics of the interaction between myosin subfragment 1 and pyrene-labelled thin filaments, from rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1986;229:85–95. doi: 10.1098/rspb.1986.0076. [DOI] [PubMed] [Google Scholar]
- 19.Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science. 1976;192:1337–1339. doi: 10.1126/science.131972. [DOI] [PubMed] [Google Scholar]
- 20.Penczek PA, Kimmel M, Spahn CM. Identifying Conformational States of Macromolecules by Eigen-Analysis of Resampled Cryo-EM Images. Structure. 2011;19:1582–1590. doi: 10.1016/j.str.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 22.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. Journal of structural biology. 2010;170:313–318. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 24.Splettstoesser T, Holmes KC, Noe F, Smith JC. Structural modeling and molecular dynamics simulation of the actin filament. Proteins. 2011;79:2033–2043. doi: 10.1002/prot.23017. [DOI] [PubMed] [Google Scholar]
- 25.Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein JC, et al. Actin-binding cleft closure in myosin II probed by site-directed spin labeling and pulsed EPR. Proc Natl Acad Sci U S A. 2008;105:12867–12872. doi: 10.1073/pnas.0802286105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki N, Asukagawa H, Yasuda R, Hiratsuka T, Sutoh K. Deletion of the myopathy loop of Dictyostelium myosin II and its impact on motor functions. J Biol Chem. 1999;274:37840–37844. doi: 10.1074/jbc.274.53.37840. [DOI] [PubMed] [Google Scholar]
- 28.Geisterfer-Lowrance AA, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 29.Furch M, Geeves MA, Manstein DJ. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 1998;37:6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- 30.Knetsch ML, Uyeda TQ, Manstein DJ. Disturbed communication between actin- and nucleotide-binding sites in a myosin II with truncated 50/20- kDa junction. J Biol Chem. 1999;274:20133–20138. doi: 10.1074/jbc.274.29.20133. [DOI] [PubMed] [Google Scholar]
- 31.Van Dijk J, et al. Differences in the ionic interaction of actin with the motor domains of nonmuscle and muscle myosin II. Eur J Biochem. 1999;260:672–683. doi: 10.1046/j.1432-1327.1999.00172.x. [DOI] [PubMed] [Google Scholar]
- 32.Joel PB, Trybus KM, Sweeney HL. Two conserved lysines at the 50/20- kDa junction of myosin are necessary for triggering actin activation. J Biol Chem. 2001;276:2998–3003. doi: 10.1074/jbc.M006930200. [DOI] [PubMed] [Google Scholar]
- 33.Sutoh K, Ando M, Toyoshima YY. Site-directed mutations of Dictyostelium actin: disruption of a negative charge cluster at the N terminus. Proc Natl Acad Sci U S A. 1991;88:7711–7714. doi: 10.1073/pnas.88.17.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen JE, Marner J, Pavlov D, Rubenstein PA, Reisler E. Structural transition at actin’s N-terminus in the actomyosin cross-bridge cycle. Biochemistry. 2000;39:1792–1799. doi: 10.1021/bi991873c. [DOI] [PubMed] [Google Scholar]
- 35.Giese KC, Spudich JA. Phenotypically selected mutations in myosin’s actin binding domain demonstrate intermolecular contacts important for motor function. Biochemistry. 1997;36:8465–8473. doi: 10.1021/bi963141f. [DOI] [PubMed] [Google Scholar]
- 36.Furch M, Remmel B, Geeves MA, Manstein DJ. Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry. 2000;39:11602–11608. doi: 10.1021/bi000985x. [DOI] [PubMed] [Google Scholar]
- 37.Schroder RR, et al. Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature. 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- 38.Milligan RA. Protein-protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci U S A. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Dijk J, Furch M, Lafont C, Manstein DJ, Chaussepied P. Functional characterization of the secondary actin binding site of myosin II. Biochemistry. 1999;38:15078–15085. doi: 10.1021/bi991595h. [DOI] [PubMed] [Google Scholar]
- 40.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 41.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 43.Li XE, et al. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy CT, Spudich JA. The sequence of the myosin 50–20K loop affects Myosin’s affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry. 1999;38:3785–3792. doi: 10.1021/bi9826815. [DOI] [PubMed] [Google Scholar]
- 45.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 47.Mouche F, Boisset N, Penczek PA. Lumbricus terrestris hemoglobin--the architecture of linker chains and structural variation of the central toroid. J Struct Biol. 2001;133:176–192. doi: 10.1006/jsbi.2001.4362. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Schroder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.