Abstract
Purpose. To investigate the association between CpG island methylator phenotype (CIMP) and the overall survival of sporadic colorectal cancer (CRC) in Northeast China. Methods. 282 sporadic CRC patients were recruited in this study. We selected MLH1, MGMT, p16, APC, MINT1, MINT31, and RUNX3 as the CIMP panel markers. The promoter methylation was assessed by methylation sensitive high resolution melting (MS-HRM). Proportional hazards-regression models were fitted with computing hazard ratios (HR) and the corresponding 95% confidence intervals (95% CI). Results. 12.77% (36/282) of patients were CIMP-0, 74.1% (209/282) of patients were CIMP-L, and 13.12% (37/282) of patients were CIMP-H. The five-year survival of the 282 CRC patients was 58%. There was significant association between APC gene promoter methylation and CRC overall survival (HR = 1.61; 95% CI: 1.05–2.46; P = 0.03). CIMP-H was significantly associated with worse prognosis compared to CIMP-0 (HR = 3.06; 95% CI: 1.19–7.89; P = 0.02) and CIMP-L (HR = 1.97; 95% CI: 1.11–3.48; P = 0.02), respectively. While comparing with the combine of CIMP-L and CIMP-0 (CIMP-L/0), CIMP-H also presented a worse prognosis (HR = 2.31; 95% CI: 1.02–5.24; P = 0.04). Conclusion. CIMP-H may be a predictor of a poor prognosis of CRC in Northeast China patients.
1. Introduction
Colorectal cancer (CRC) is one of the most common malignancies, representing the third most common cancer in men and the second in women worldwide [1]. In 2012, about 694,000 deaths from CRC were estimated in the world, which account for 8.5% of all cancer deaths, making it the fourth most common cause of death from cancer [2]. Although the relative 5-year survival of CRC increased in Europe during 1995–2007 [3], it was only about 30%–65% worldwide [4]. Anatomic and pathological staging are still the most accurate predictors of CRC prognosis until now. Therefore, novel molecular prognostic markers for colorectal cancer are needed for the accurate prediction of prognosis.
DNA methylation of tumor suppressor genes leading to transcriptional inactivation has been identified as an important mechanism in human carcinogenesis [5, 6]. CpG island methylation phenotype (CIMP), characterized by the extensive hypermethylation of multiple CpG islands, is currently recognized as one of the major mechanisms in the colorectal carcinogenesis [7, 8]. Compared to CIMP-low/negative CRCs, CIMP-high/positive CRCs have distinct clinicopathological and molecular profiles such as older age, female gender, proximal tumor location, poorly differentiated or mucinous histology, and high rates of MSI and BRAF mutation [9, 10]. Most of these clinicopathological and molecular features of CIMP-high tumors overlap with sporadic MSI cancers; patients with MSI CRCs have a better prognosis [11]; therefore, CIMP statuses are supposed to influence the prognosis of CRC.
Until now, more than 24 papers published focused on the CIMP status and CRC prognosis. Although CIMP-H patients have been reported to be related to poor prognosis of CRC in at least eight studies [5, 6, 12–17], 15 studies reported a null association between CIMP-H and CRC prognosis or even noted a better prognosis [18–32]. One study reported a significant association between CIMP-H and colon cancer specific mortality [33]. Because of the unclear biology cause of CIMP, discrepancy in the methylation markers, and the criteria for CIMP, it still cannot be concluded whether significant association between CIMP-H and CRC prognosis existed. Moreover, no study in China has been published in this aspect. Therefore, we conducted the study to evaluate the association between CIMP status and the prognosis of CRC in Northeast China.
2. Methods
2.1. Study Participants
After obtaining informed written consent from study subjects and approval from Institutional Research Board of Harbin Medical University, we identified CRC patients who underwent surgery at the Cancer Hospital of Harbin Medical University, without preselection and based on pathologic diagnosis alone. Tumor staging was based on the TNM staging system of the American Joint Committee on Cancer [34]. The histological subtypes were classified using the World Health Organization (WHO) criteria [35]. Patients with neuroendocrine carcinoma, malignant melanoma, non-Hodgkin's lymphoma, gastrointestinal stromal tumors, and metastatic colorectal carcinoma were excluded. Patients who had no family history of CRC regardless of the onset age were categorized as sporadic CRC. From June 1, 2004, to January 1, 2008, 453 primary sporadic CRC patients were recruited, and tumor tissue samples from 282 patients were available for the current study.
We followed patients until March 2012 or death. After surgery, the clinical data of patients were collected based on the medical records for analyses, which included age at diagnosis, tumor location, and pathological diagnosis. During follow-up, chemotherapy and radiotherapy protocols were obtained. Meanwhile, we obtained information about disease progression, recurrence, and the date and cause of death (if deceased).
2.2. DNA Extraction
Genomic DNA was successfully extracted from the 282 tumor tissues using the TIAN-amp Genomic DNA kit (Tiangen, Beijing, China).
2.3. Selection of CIMP Markers
The “classic panel” of CIMP included MINT1, MINT2, MINT31, CDKN2A (p16), and MLH1; CIMP was defined as two or more markers methylated [9]. In 2006, a CIMP with new five-gene panel that included CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 was defined as three or more markers methylated [36]. At present, there is no gold standard with respect to gene panels and the number of marker thresholds used to define CIMP [37]. Based on the above two CIMP panels and previous studies [38–40], we used seven genes, which mainly used the classic panel as CIMP markers, which included MINT1, MINT31, p16, MLH1, MGMT, APC, and RUNX3.
2.4. Methylation Sensitive High Resolution Melting (MS-HRM)
Genomic DNA was modified with sodium bisulfite using the EZ Methylation Gold Kit (Zymo Research, Orange, CA). High resolution melting (HRM) was used to assess the methylation status of the seven CIMP-specific promoters (MLH1, MGMT, p16, APC, MINT1, MINT31, and RUNX3). The primers are listed in Table 1. HRM was performed using the following protocol: (1) PCR amplification protocol: denaturation for 10 min at 95°C for 1 cycle, denaturation for 10 s at 95°C, annealing for 30 s at 50°C to 55°C, and extension for 30 s at 72°C for 45 cycles, followed by (2) high resolution melting protocol: 95°C for 1 min, 40°C for 1 min, 74°C for 5 s, and continuous acquisition to 90°C at 25 acquisitions per 1°C (LightCycler 480, Roche, Mannheim, Germany). Each sample was duplicated for two plates. Human methylated and unmethylated DNA sets from Zymo Research were used as 100% methylated and 0% unmethylated controls. The percentages of methylation of 0%, 1%, 5%, 25%, 50%, and 100% were used to draw the standard curve. 5% of methylation was used as cutoff value.
Table 1.
HRM primers and amplicon information.
| Gene name | Primer sequences 5′-3′ | GenBank | Number of CpG-sites/length |
|---|---|---|---|
| Accession number | of amplified fragment | ||
| MGMT | Fw GCGTTTCGGATATGTTGGGATAGT Rv AACGACCCAAACACTCACCAAA |
X61657 | 15/110 |
|
| |||
| MLH1 | Fw TTTTTTTAGGAGTGAAGGAGG Rv AACRCCACTACRAAACTAAA |
AY217549 | 13/123 |
|
| |||
| APC | Fw AAGTAGTTGTGTAATTCGTTGGAT Rv CACCTCCATTCTATCTCCAATA |
NT_034772 | 11/149 |
|
| |||
| p16 | Fw GGAGTTTTCGGTTGATTGGTTGGTT Rv AACAACGCCCGCACCTCCTCTA |
AF527803 | 5/69 |
|
| |||
| MINT1 | Fw GGGGTTGAGGTTTTTTGTTAG Rv AATCCCTCTCCCCTCTAAACTT |
AF135501 | 7/137 |
|
| |||
| MINT31 | Fw GGGTGATGGTTTTAGTAAAGTGAG Rv AAAAACACTTCCCCAACATCTAC |
AF135531 | 10/164 |
|
| |||
| RUNX3 | Fw TTTTTAGAGAATGAGGGATTTTTGT Rv CCCTAATCCCTTAAATCTAATACCC |
NC_000001 | 7/115 |
Three levels of CIMP were identified as follows: high-level CIMP (CIMP-H), generally defined as ≥4/7 methylated markers using the seven-marker CIMP panel; low-level CIMP (CIMP-L), generally defined as ≤3/7 methylated markers; CIMP-0, generally defined as 0/7 methylated markers.
2.5. Statistical Analysis
The end point was overall survival, calculated from the first diagnosis of colorectal cancer to the death from any cause or until March 2012. The survival curves were estimated using Kaplan-Meier product-limit method. Cumulative survival probability was calculated at the third, the fifth, and the seventh years, respectively. Proportional hazards-regression models were fitted with computing hazard ratios (HR) and the corresponding 95% confidence intervals (95% CI). All statistical tests were 2 sided; P values <0.05 were considered statistically significant. All the statistical analysis was performed by SAS 9.1 (SAS Institute, Cary, NC, USA).
3. Results
3.1. Characteristics of CRC Patients
The study population consisted of 117 females and 165 males, with a mean age of 58.8 ± 11.2 years (range: 25 to 81 years). The median follow-up time was 53 months (range: 1 to 88 months). During the followup, 100 patients died, and 18 patients were lost to followup. The five-year survival of the 282 CRC patients was 58%.
3.2. CIMP-Specific Promoter Methylation
CIMP-specific promoter methylation was successfully assessed in all cases. Methylation frequencies in the 282 patients were 25.18% for MLH1 (71 cases), 34.75% for MGMT (98 cases), 19.86% for P16 (56 cases), 26.60% for APC (75 cases), 20.92% for MINT1 (59 cases), 48.23% for MINT31 (136 cases), and 19.15% for RUNX3 (54 cases). 12.77% (36/282) of patients were CIMP-0, 74.1% (209/282) of patients were CIMP-L, and 13.12% (37/282) of patients were CIMP-H.
22.22% (8/36) of CIMP-H tumors were poorly differentiated, while 6.41% (15/234) of CIMP-L/0 tumors were poorly differentiated (P = 0.01). In tumor size larger than 5 cm, 60.00% (21/35) were CIMP-H, whereas 36.45% (78/214) were CIMP-L/0 (P = 0.01). CIMP-H tumors were more likely to be in TNM III-IV stages than CIMP-L/0 tumors, 63.89% (23/36) versus 43.80% (106/242) (P = 0.02). No significant difference was observed in other clinicopathological characteristics (gender, age, tumor location, and histological types) among the CIMP phenotypes. Details are shown in Table 2.
Table 2.
Patient characteristics and phenotypic features of colorectal cancers in tissue by CIMP status.
| Variable |
Number (N = 282) |
CIMP-L/0 number (%) (N = 245) |
CIMP-H number (%) (N = 37) |
P |
|---|---|---|---|---|
| Age | 0.77 | |||
| <60 | 146 | 126 (51.43) | 20 (54.05) | |
| ≥60 | 136 | 119 (48.57) | 17 (45.95) | |
| Gender | 0.10 | |||
| Female | 117 | 97 (39.59) | 20 (54.05) | |
| Male | 165 | 148 (60.41) | 17 (45.95) | |
| Location | 0.22 | |||
| Proximal | 54 | 42 (17.87) | 12 (25.53) | |
| Distal | 228 | 193 (82.13) | 35 (74.47) | |
| Differentiation | 0.01§ | |||
| Poorly | 23 | 15 (6.41) | 8 (22.22) | |
| Moderately or well |
247 | 219 (93.59) | 28 (77.78) | |
| Mutinous | 0.91 | |||
| Yes | 59 | 51 (20.82) | 8 (21.62) | |
| No | 223 | 194 (79.18) | 29 (78.38) | |
| Tumor sizes (cm) | 0.01 | |||
| <5 | 150 | 136 (63.55) | 14 (40.00) | |
| ≥5 | 99 | 78 (36.45) | 21 (60.00) | |
| TNM stage | 0.02 | |||
| I-II | 149 | 136 (56.20) | 13 (36.11) | |
| III-IV | 129 | 106 (43.80) | 23 (63.89) |
§Fisher exact test.
3.3. Survival Analysis
3.3.1. Overall Survival Analysis on Clinical and Pathological Status
In multivariate Cox proportional hazards-regression analysis, differentiation and tumor stage were significantly associated with the prognosis of colorectal cancer. Differentiation and TNM stage were adjusted in the following survival analysis.
3.3.2. Gene-Specific Promoter Methylation and Overall Survival
When analyzing the associations between gene-specific promoter methylation and overall survival of CRC, there was significant association between APC gene promoter methylation and CRC overall survival (HR = 1.61; 95% CI: 1.05–2.46; P = 0.03; Figure 1). There was no significant association between other six-gene promoter methylations and CRC prognoses (Table 3).
Figure 1.
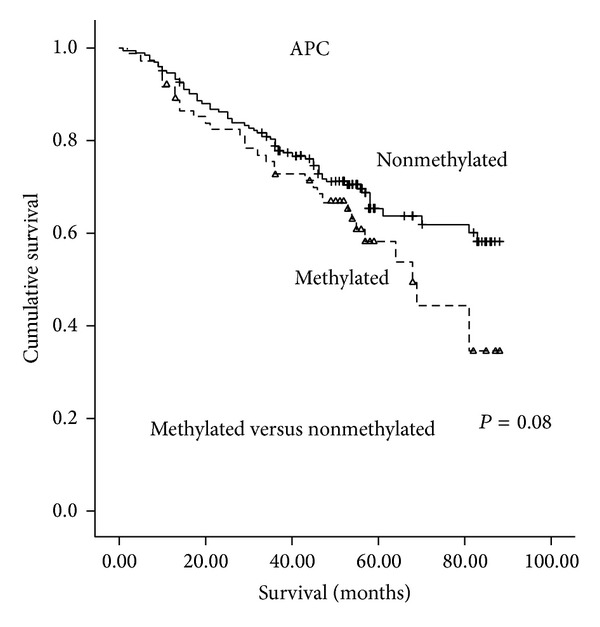
Overall survival according to APC methylation levels in colorectal cancer tissue.
Table 3.
Survival analysis on CRC patients according to methylation of individual methylation marker and CIMP status in tumor tissue.
| Number of patients | Number of deaths | Overall survival (%) | Univariate | Multivariate§ | |||||
|---|---|---|---|---|---|---|---|---|---|
| 3 y | 5 y | 7 y | HR and 95% CI | P | HR and 95% CI | P | |||
| MLH1 | |||||||||
| Unmethylation | 211 | 76 | 71.00 | 56.00 | 48.00 | Ref. | Ref. | ||
| Methylation | 71 | 24 | 69.00 | 64.00 | 64.00 | 0.94 (0.59–1.48) | 0.77 | 1.07 (0.66–1.74) | 0.79 |
| MGMT | |||||||||
| Unmethylation | 184 | 65 | 72.00 | 59.00 | 50.00 | Ref. | Ref. | ||
| Methylation | 98 | 35 | 68.00 | 56.00 | 56.00 | 1.02 (0.68–1.54) | 0.93 | 1.05 (0.69–1.61) | 0.81 |
| p16 | |||||||||
| Unmethylation | 226 | 79 | 72.00 | 60.00 | 53.00 | Ref. | Ref. | ||
| Methylation | 56 | 21 | 67.00 | 47.00 | 47.00 | 1.27 (0.78–2.05) | 0.34 | 1.05 (0.64–1.73) | 0.85 |
| APC | |||||||||
| Unmethylation | 207 | 67 | 72.00 | 63.00 | 59.00 | Ref. | Ref. | ||
| Methylation | 75 | 33 | 67.00 | 44.00 | 34.00 | 1.46 (0.96–2.21) | 0.08 | 1.61 (1.05–2.46) | 0.03 |
| MINT1 | |||||||||
| Unmethylation | 223 | 76 | 73.00 | 59.00 | 51.00 | Ref. | Ref. | ||
| Methylation | 59 | 24 | 62.00 | 53.00 | 53.00 | 1.36 (0.86–2.16) | 0.19 | 1.38 (0.85–2.24) | 0.20 |
| MINT31 | |||||||||
| Unmethylation | 146 | 52 | 72.00 | 56.00 | 50.00 | Ref. | Ref. | ||
| Methylation | 136 | 48 | 69.00 | 59.00 | 54.00 | 1.04 (0.70–1.54) | 0.84 | 1.01 (0.67–1.53) | 0.96 |
| RUNX3 | |||||||||
| Unmethylation | 228 | 83 | 70.00 | 58.00 | 51.00 | Ref. | Ref. | ||
| Methylation | 54 | 17 | 73.00 | 55.00 | 55.00 | 0.90 (0.53–1.52) | 0.69 | 1.14 (0.67–1.95) | 0.63 |
| CIMP status | |||||||||
| CIMP-0 | 36 | 12 | 77.00 | 59.00 | 59.00 | Ref. | Ref. | ||
| CIMP-L | 209 | 70 | 72.00 | 61.00 | 54.00 | 0.99 (0.53–1.82) | 0.96 | 0.95 (0.60–1.52) | 0.84 |
| CIMP-H | 37 | 18 | 58.00 | 33.00 | 33.00 | 1.71 (0.82–3.55) | 0.15 | 3.06 (1.19–7.89) | 0.02 |
| CIMP-H versus CIMP-L | 1.78 (1.07–3.03) | 0.03 | 1.97 (1.11–3.48) | 0.02 | |||||
| CIMP-H versus CIMP-L and CIMP-0 |
1.80 (1.08–3.00) | 0.03 | 2.31 (1.02–5.24) | 0.04 | |||||
§Multivariate analysis, adjusted for age at diagnosis, tumor stage, tumor differentiation, and tumor location.
3.3.3. CIMP Status and Overall Survival
Compared to CIMP-0, CIMP-H was significantly associated with worse prognosis of CRC (HR = 3.06; 95% CI: 1.19–7.89; P = 0.02). CIMP-H was also significantly associated with poor prognosis of CRC compared with CIMP-L (HR = 1.97; 95% CI: 1.11–3.48; P = 0.02). CIMP-H was also significantly associated with worse prognosis of CRC comparing to the combine of CIMP-L and CIMP-0 (CIMP-L/0) (HR = 2.31; 95% CI: 1.02–5.24; P = 0.04) (Table 3, Figure 2).
Figure 2.
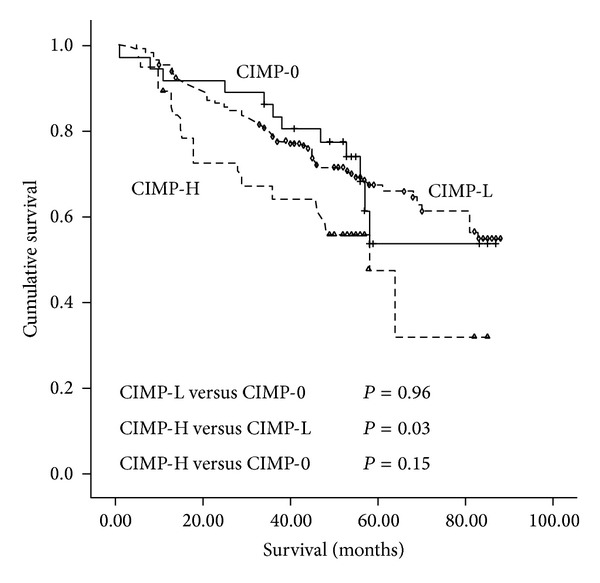
Survival curves in three molecular subgroups according to status of CpG island methylator phenotype (CIMP) in colorectal cancer tissue.
When analyses stratified according to tumor stage, CIMP-H CRC patients demonstrated a marginally worse prognosis than CIMP-0 CRC patients in stages III to IV group (HRadj = 1.67; 95% CI: 1.00–2.81; P = 0.05). However, when comparing to CIMP-L, CIMP-H was not significantly associated with CRC prognosis in stages III-IV group (P = 0.20). When comparing to CIMP-L/0, CIMP-H was also not significantly associated with CRC prognosis in stages III-IV group (P = 0.07). In stages I-II group, there were no significant differences between the CIMP-H CRC patients and the CIMP-L, CIMP-0, and CIMP-L/0 CRC patients in survival, respectively. Details are shown in Table 4.
Table 4.
Survival analysis on CRC patients according to CIMP status in tumor tissue in different tumor stages.
| CIMP status | Stages I-II (n = 149) | Stages III-IV (n = 129) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate§ | Univariate | Multivariate§ | |||||
| HR and 95% CI | P | HR and 95% CI | P | HR and 95% CI | P | HR and 95% CI | P | |
| CIMP-H versus CIMP-L/0 | 0.69 (0.17–2.91) | 0.62 | 0.52 (0.12–2.22) | 0.38 | 1.81 (1.02–3.24) | 0.04 | 1.75 (0.95–3.23) | 0.07 |
| CIMP-H versus CIMP-L | 0.74 (0.17–3.14) | 0.68 | 0.57 (0.13–2.48) | 0.46 | 1.73 (0.96–3.11) | 0.07 | 1.50 (0.80–2.81) | 0.20 |
| CIMP-L versus CIMP-0 | 0.66 (0.27–1.60) | 0.35 | 0.57 (0.23–1.41) | 0.22 | 1.42 (0.60–3.34) | 0.43 | 2.06 (0.77–5.53) | 0.15 |
| CIMP-H versus CIMP-0 | 0.71 (0.32–1.58) | 0.40 | 0.56 (0.21–1.49) | 0.24 | 1.55 (0.96–2.49) | 0.07 | 1.67 (1.00–2.81) | 0.05 |
§Multivariate analysis, adjusted for age at diagnosis, tumor stage, tumor differentiation, and tumor location.
A total of 48 (46.2%) of 104 stage III and 38 (21%) of 115 stage II patients received chemotherapy after curative resection of tumor. In CIMP-H patients, 5-FU based adjuvant chemotherapy did not significantly improve overall survival of CRC (HR = 0.71; 95% CI: 0.20–2.54; P = 0.60). Patients also did not benefit from 5-FU based adjuvant chemotherapy in CIMP-L/0 group (HR = 1.09; 95% CI: 0.65–1.82; P = 0.75).
4. Discussion
Epigenetic aberrations are thought to be an important mechanism in human carcinogenesis [41]. One of the epigenetic regulations influencing gene expression is DNA methylation, a postreplicative DNA modification that occurs in genome regions rich in cytosine and guanosine (CG) dinucleotides that are called CpG islands. Modification of bases by addition of a methyl group can physically inhibit binding of transcription factors and also permit recruitment of the methyl-CpG-binding domain proteins to promote regions, which can repress transcription initiation [42]. A subset of colon cancers exhibits widespread promoter methylation, referred to as the CpG island methylator phenotype (CIMP) [43–46]. CRC tumors characterized by CIMP are thought to arise via the serrated neoplasia pathway [47]. An early event in CIMP tumors appears to be a mutation in the BRAF protooncogene, which inhibits normal apoptosis of colonic epithelial cells [48]. CRCs with high-level CIMP present distinct clinicopathological and molecular profiles [9, 10]. Clinically, there is evidence to suggest that CIMP-H patients had a shorter cancer-specific survival compared with CIMP-0 patients in CRC [14]. However, another study indicated that CIMP-H appears to be an independent predictor of a low colon cancer-specific mortality [33].
The CpG island methylation phenotype (CIMP), characterized by the extensive hypermethylation of multiple gene CpG islands, was originally described in 1999 by Toyota et al. [49], who defined a subgroup of cancers with a high rate of methylation. Initially, CIMP-positive group was defined as methylation at three or more of seven MINT markers [50]. Subsequently, the so-called “classic panel,” which includes MINT1, MINT2, MINT31, CDKN2A (p16), and MLH1, was described by Issa in 2004; CIMP-H was defined by high level of methylation at two or more markers [9]. In 2006, Weisenberger et al. [36] introduced a new five-gene panel, which includes the genes CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1; CIMP-H was defined by high level of methylation at three or more markers.
At present, many different combinations of gene panel/marker thresholds/laboratory methods were used to define CIMP. Moreover, there is also no standard protocol for choosing primers and/or location of methylation in the markers [37]. The location of core regions and the density of methylation required for gene silencing can vary in every gene; therefore, the classical dogma of promoter CpG island methylation and gene silencing may not be sufficient to interpret data on DNA methylation, gene expression, and clinicopathological associations [51]. Therefore, we used seven genes which mainly used the classic panel as CIMP markers, which included MINT1, MINT31, p16, MLH1, MGMT, APC, and RUNX3. A high-throughput platform MS-HRM protocol [52] was used to screen for methylation changes. 0.1% difference can be detected by the high sensitive MS-HRM, and 0.1–10% [53–57] have been used as a cutoff for the scoring criteria of gene methylation. Therefore, 5% of methylation was used as cutoff value. Furthermore, there is debate whether CIMP should be distinguished as two categories (“CIMP-positive” and “CIMP-negative”) [36, 49] or three categories (either “CIMP-H, CIMP-L, or CIMP-0”) [58]. Nosho et al. [59], using a large population-based sample, demonstrated that CIMP-H was independently associated with MSI-high and BRAF mutation. Moreover, researchers discovered that when KRAS mutation was found in CIMP CRCs, it is associated with CIMP-L. Accumulating evidence suggests that CIMP-L represents a distinct phenotype in CRC [12, 60, 61]. Thus, we used CIMP classification as three categories (either “CIMP-H, CIMP-L, or CIMP-0”).
In our study, we observed significant associations between APC promoter methylation and CRC overall survival. CIMP-H was significantly associated with poor CRC prognosis, when comparing to CIMP-L, CIMP-0, and CIMP-L/0, respectively. CIMP-H was a marginally worse prognosis in stages III+IV group, when comparing to CIMP-0 CRC patients.
As to gene-specific promoter methylation, APC promoter methylation increased the hazard risk of CRC by 1.61-fold in our study. Hypermethylation of APC may provide an important mechanism for impairing APC function and could be involved in the progression of human CRC [62, 63]. However, the APC promoter methylation demonstrates significantly lower hazards for CRC death (HR = 0.43; 95% CI: 0.19–0.96; P = 0.04) in a Taiwanese study [64]. The different methylation detection method may explain the discrepancy. Methylation-specific PCR (MSP) was used in the Taiwanese study, which has a higher false positive rate than MS-HRM [65]. However, there was no significant association between other individual gene methylations and overall survivals of CRC in our study.
Our study investigated that CIMP-H could increase the hazard risk of CRC by 1.97-fold, 3.06-fold, and 2.31-fold, when comparing to CIMP-L, CIMP-0, and CIMP-L/0, respectively. Other eight studies also observed a significant association between CIMP-H and worse CRC prognosis (the HR varied from 1.45 to 5.5) [5, 6, 12–17]. However, one study reported an inverse association between CIMP-H and colon cancer-specific mortality [33]; 15 studies reported a null association between CIMP-H and CRC prognosis [18–32]. However, no clear biological cause for CIMP has been determined, so comparing results across studies is a challenge [37]. The discrepancy might be due to the differences in patient cohorts, as well as the following variation. First, the differences in methylation markers and criteria for CIMP may result in inconsistence. Among the published papers, five papers [12, 18, 20, 28, 66] selected the classical panel of CIMP, where ≥2/5 or 4/5 was defined as CIMP-H; six papers [5, 16, 22, 25, 66, 67] selected the new panel of CIMP, where ≥3/5 was defined as CIMP-H. Two papers [13, 32] selected the classical panel and other genes, where ≥3/n was defined as CIMP-H; seven papers [14, 15, 17, 19, 26, 30, 33] selected the new panel and other genes, where ≥5/8 or 6/8 was defined as CIMP-H. Two papers [5, 6] selected methylation markers according to their own criteria, where ≥1/4 or 2/3 was defined as CIMP-H. Meanwhile the variance in the primers and/or location of methylation in the markers may also affect the result. For example, the CIMP marker panel used in Lee et al. study [20] and Barault et al. study [12] was a classic five-marker panel (MLH1, MINT1, MINT2, MINT31, and p16); different primers of MLH1, MINT1, and MINT2 may explain differences of the prognosis of CIMP+ tumors between two studies. In addition, variance in the laboratory method and criteria of methylation could be a reason for the inconsistency of these results of the studies. MSP was used in 12 papers [5, 6, 12, 16, 18, 20, 21, 23, 25, 28, 32, 66]; methylation was assessed by the positive bands in the agarose electrophoresis. MethyLight was used in 10 papers [5, 14, 17, 19, 22, 27, 29, 30, 33, 67]; the percentage of methylated reference (PMR) was used in classification; if the PMR was greater than 4 or 10, it was considered methylated. Bisulfite pyrosequencing was used in 3 papers [13, 24, 68]; each marker was classified as methylated when the mean percentage was higher than 5% or 10%. Moreover, the variable inclusion of other potential confounders in multivariate analysis models may affect the results. Because of the different methylation markers and criteria for CIMP, we could not systematically analyze the association between CIMP status and CRC prognosis upon meta-analysis to obtain a stable result.
When analyses stratified by tumor stage, CIMP-H increased the hazard risk of CRC by 1.67-fold, when comparing to CIMP-0 in stages III-IV group (P = 0.05). However, when comparing to CIMP-L, CIMP-0, and CIMP-L/0, respectively, CIMP-H was not associated with worse prognosis of CRC in stages III-IV group. In stages I-II group, there were no significant associations between CIMP and CRC prognosis. One study in Korea yielded a consistent significant association between CIMP-H and worse disease free survival of stage III proximal CRC (P = 0.015) [13]. The other study in Germany also demonstrated a significant association between CIMP-H and worse disease free survival of stage II/III rectal cancer (HR = 5.5; 95% CI: 2.1–13.9) [16]. Another study in the USA found that CIMP-H cancers experienced a nonsignificantly low colon cancer-specific mortality in stage III and stage IV colon cancers, respectively (HR = 0.52 (95% CI: 0.17–1.59) for stage III; HR = 0.47 (95% CI: 0.18–1.21) for stage IV) [33]. Stages III and IV are strong determinants for CRC prognosis; CIMP-H was more frequent in stages III and IV CRC than in stages I and II CRC, which may explain the significant association between CIMP and CRC prognosis in stages III and IV CRC. The variant CIMP panels as well as the different cancer sites and different cancer stages may explain the discrepancy.
CIMP-H has been proposed to be having potential application for 5-FU based chemotherapy treatment response prediction [6, 24]. Therefore, we conducted the stratified analyses by 5-FU based chemotherapy. However, we failed to observe any significant association between CIMP-H and the prognosis of stages II+III CRC treated with 5-FU based chemotherapy. A large-scale study also failed to demonstrate a significant association between CIMP-positive and the overall survival of stages II+III CRC treated with adjuvant chemotherapy [32], while CIMP-H was reported to be an independent predictor of survival benefit from 5-FU adjuvant chemotherapy in stage III CRC in another study in Australia [6]. Different CIMP panels and different stages of CRC may explain the discrepancy.
In conclusion, CIMP-H may be a predictor of poor prognosis of CRC, especially for stages III+IV CRC. APC gene promoter methylation indicated a poor prognosis of CRC.
Acknowledgments
This study was supported by the Grants from National Natural Science Foundation of China (NSFC 30972538) and the Graduate Foundation and supported by the Scientific Research of Heilongjiang Province (YJSCX2009-224HLJ) and Harbin Medical University (HCXB2009009).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Yashuang Zhao designed the study and revised the paper. Xia Li, Yibaina Wang, Xiaoping Yao, and Zuoming Zhang participated in the experiment. Xia Li and Fulan Hu participated in the data analysis and cowrote the paper. Guizhi Sun participated in the collection of colorectal cancer samples. All authors read and approved the final paper.
References
- 1.Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Modern Pathology. 2003;16(4):376–388. doi: 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- 2. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 3.Coleman MP, Forman D, Bryant H, et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): an analysis of population-based cancer registry data. The Lancet. 2011;377(9760):127–138. doi: 10.1016/S0140-6736(10)62231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman MP, Quaresma M, Berrino F, et al. Cancer survival in five continents: a worldwide population-based study (CONCORD) The Lancet Oncology. 2008;9(8):730–756. doi: 10.1016/S1470-2045(08)70179-7. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, Catalano PJ, Benson AB, III, O'Dwyer P, Hamilton SR, Issa JJ. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil-based chemotherapy. Clinical Cancer Research. 2007;13(20):6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clinical Cancer Research. 2003;9(8):2898–2903. [PubMed] [Google Scholar]
- 7.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. Journal of Molecular Diagnostics. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issa J. CpG island methylator phenotype in cancer. Nature Reviews Cancer. 2004;4(12):988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 10.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 11.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. Journal of Clinical Oncology. 2005;23(3):609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 12.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Research. 2008;68(20):8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 13.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117(9):1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlin AM, Palmqvist R, Henriksson ML, et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clinical Cancer Research. 2010;16(6):1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 15.Rhee Y, Kim MJ, Bae JM, et al. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on l1 methylation level. Annals of Surgical Oncology. 2012;19(11):3441–3448. doi: 10.1245/s10434-012-2410-7. [DOI] [PubMed] [Google Scholar]
- 16.Jo P, Jung K, Grade M, et al. CpG island methylator phenotype infers a poor disease-free survival in locally advanced rectal cancer. Surgery. 2012;151(4):564–570. doi: 10.1016/j.surg.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Bae JM, Kim JH, Cho NY, Kim T, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. British Journal of Cancer. 2013;109(4):1004–1012. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samowitz WS, Curtin K, Wolff RK, Tripp SR, Caan BJ, Slattery ML. Microsatellite instability and survival in rectal cancer. Cancer Causes and Control. 2009;20(9):1763–1768. doi: 10.1007/s10552-009-9410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Archiv. 2009;455(6):485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Cho N, Choi M, Yoo EJ, Kim J, Kang GH. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathology International. 2008;58(2):104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferracin M, Gafà R, Miotto E, et al. The methylator phenotype in microsatellite stable colorectal cancers is characterized by a distinct gene expression profile. Journal of Pathology. 2008;214(5):594–602. doi: 10.1002/path.2318. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez JA, Krumroy L, Plummer S, et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. British Journal of Surgery. 2009;96(10):1196–1204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- 23.Kakar S, Deng G, Sahai V, et al. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Archives of Pathology and Laboratory Medicine. 2008;132(6):958–964. doi: 10.5858/2008-132-958-CCCIMP. [DOI] [PubMed] [Google Scholar]
- 24.Jover R, Nguyen T, Prezcarbonell L, et al. 5-fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140(4):1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donada M, Bonin S, Barbazza R, Pettirosso D, Stanta G. Management of stage II colon cancer—the use of molecular biomarkers for adjuvant therapy decision. BMC Gastroenterology. 2013;13, article 36 doi: 10.1186/1471-230X-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S, Lee H, Bae JM, et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. International Journal of Cancer. 2013;132(9):2209–2216. doi: 10.1002/ijc.27888. [DOI] [PubMed] [Google Scholar]
- 27.Min B, Bae JM, Lee EJ, et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer. 2011;11, article 344 doi: 10.1186/1471-2407-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Research. 2005;65(14):6063–6070. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 29.Kohonen-Corish MR, Tseung J, Chan C. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. International Journal of Cancer. 2014;134(12):2820–2828. doi: 10.1002/ijc.28619. [DOI] [PubMed] [Google Scholar]
- 30.Kim JH, Rhee YY, Bae JM, Cho NY, Kang GH. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. The American Journal of Surgical Pathology. 2013;37(10):1532–1541. doi: 10.1097/PAS.0b013e31829ab1c1. [DOI] [PubMed] [Google Scholar]
- 31.Samadder NJ, Vierkant RA, Tillmans LS, et al. Associations between colorectal cancer molecular markers and pathways with clinicopathologic features in older women. Gastroenterology. 2013;145(2):348–356. doi: 10.1053/j.gastro.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. Journal of Clinical Oncology. 2003;21(20):3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 33.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80(9):1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Hamilton SR, Aaltonen LA. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Digestive System. Lyon, France: IARC Press; 2000. [Google Scholar]
- 36.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genetics. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 37.Hughes LAE, Khalid-de Bakker CAJ, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochimica et Biophysica Acta—Reviews on Cancer. 2012;1825(1):77–85. doi: 10.1016/j.bbcan.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Archiv. 2007;450(5):529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 40.Hinoue T, Weisenberger DJ, Lange CPE, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Research. 2012;22(2):271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers. 2013;5(2):676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YS, Deng G. Epigenetic changes (aberrant DNA methylation) in colorectal neoplasia. Gut Liver. 2007;1(1):1–11. doi: 10.5009/gnl.2007.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colussi D, Brandi G, Bazzoli F. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International Journal of Molecular Sciences. 2013;14(8):16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathology Research International. 2011;2011:8 pages. doi: 10.4061/2011/902674.902674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JH, Kang GH. Molecular and prognostic heterogeneity of microsatellite-unstable colorectal cancer. World Journal of Gastroenterology. 2014;20(15):4230–4243. doi: 10.3748/wjg.v20.i15.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annual Review of Pathology. 2009;4:343–364. doi: 10.1146/annurev.pathol.4.110807.092317. [DOI] [PubMed] [Google Scholar]
- 48.Snover DC. Update on the serrated pathway to colorectal carcinoma. Human Pathology. 2011;42(1):1–10. doi: 10.1016/j.humpath.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JJ. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 51.van Vlodrop IJH, Niessen HEC, Derks S, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clinical Cancer Research. 2011;17(13):4225–4231. doi: 10.1158/1078-0432.CCR-10-3394. [DOI] [PubMed] [Google Scholar]
- 52.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nature Protocols. 2008;3(12):1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 53.Meng W, Huebner A, Shabsigh A, Chakravarti A, Lautenschlaeger T. Combined RASSF1A and RASSF2A promoter methylation analysis as diagnostic biomarker for bladder cancer. Molecular Biology International. 2012;2012:8 pages. doi: 10.1155/2012/701814.701814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balic M, Pichler M, Strutz J, et al. High quality assessment of DNA methylation in archival tissues from colorectal cancer patients using quantitative high-resolution melting analysis. Journal of Molecular Diagnostics. 2009;11(2):102–108. doi: 10.2353/jmoldx.2009.080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morandi L, Franceschi E, de Biase D, et al. Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer. 2010;10, article 48 doi: 10.1186/1471-2407-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avraham A, Uhlmann R, Shperber A, et al. Serum DNA methylation for monitoring response to neoadjuvant chemotherapy in breast cancer patients. International Journal of Cancer. 2012;131(7):E1166–E1172. doi: 10.1002/ijc.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive melting analysis after real time-methylation specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Research. 2008;36(7, article e42) doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. Journal of Molecular Diagnostics. 2006;8(5):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3(11) doi: 10.1371/journal.pone.0003698.e3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogino S, Kawasaki T, Kirkner GJ, Suemoto Y, Meyerhardt JA, Fuchs CS. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56(11):1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawasaki T, Ohnishi M, Nosho K, et al. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Modern Pathology. 2008;21(3):245–255. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- 62.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Research. 2000;60(16):4366–4371. [PubMed] [Google Scholar]
- 63.Hiltunen MO, Alhonen L, Koistinaho J, et al. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. International Journal of Cancer. 1997;70(6):644–648. doi: 10.1002/(sici)1097-0215(19970317)70:6<644::aid-ijc3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 64.Chen S, Chiu S, Wu C, et al. The association of methylation in the promoter of APC and MGMT and the prognosis of Taiwanese CRC patients. Genetic Testing and Molecular Biomarkers. 2009;13(1):67–71. doi: 10.1089/gtmb.2008.0045. [DOI] [PubMed] [Google Scholar]
- 65.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee S, Cho N, Yoo EJ, Kim JH, Kang GH. CpG island methylator phenotype in colorectal cancers: comparison of the new and classic CPG island methylator phenotype marker panels. Archives of Pathology and Laboratory Medicine. 2008;132(10):1657–1665. doi: 10.5858/2008-132-1657-CIMPIC. [DOI] [PubMed] [Google Scholar]
- 67.English DR, Young JP, Simpson JA, et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiology Biomarkers and Prevention. 2008;17(7):1774–1780. doi: 10.1158/1055-9965.EPI-08-0091. [DOI] [PubMed] [Google Scholar]
- 68.Kim JC, Choi JS, Roh SA, Cho DH, Kim TW, Kim YS. Promoter methylation of specific genes is associated with the phenotype and progression of colorectal adenocarcinomas. Annals of Surgical Oncology. 2010;17(7):1767–1776. doi: 10.1245/s10434-009-0901-y. [DOI] [PubMed] [Google Scholar]


