Abstract
SSAP method was used to study the genetic diversity of 22 Linum species from sections Linum, Adenolinum, Dasylinum, Stellerolinum, and 46 flax cultivars. All the studied flax varieties were distinguished using SSAP for retrotransposons FL9 and FL11. Thus, the validity of SSAP method was demonstrated for flax marking, identification of accessions in genebank collections, and control during propagation of flax varieties. Polymorphism of Fl1a, Fl1b, and Cassandra insertions were very low in flax varieties, but these retrotransposons were successfully used for the investigation of Linum species. Species clusterization based on SSAP markers was in concordance with their taxonomic division into sections Dasylinum, Stellerolinum, Adenolinum, and Linum. All species of sect. Adenolinum clustered apart from species of sect. Linum. The data confirmed the accuracy of the separation in these sections. Members of section Linum are not as closely related as members of other sections, so taxonomic revision of this section is desirable. L. usitatissimum accessions genetically distant from modern flax cultivars were revealed in our work. These accessions are of utmost interest for flax breeding and introduction of new useful traits into flax cultivars. The chromosome localization of Cassandra retrotransposon in Linum species was determined.
1. Introduction
The genus Linum comprises about 200 species which are distributed throughout the temperate and subtropical regions of the world. The genus is subdivided by Ockendon and Walters into five sections: Linum, Dasylinum (Planch.) Juz., Linastrum (Planchon), Bentham, Syllinum Griseb., and Cathartolinum (Reichenb.) Griseb. [1]. Some taxonomists classified the members of the L. perenne group from section Linum to an independent section Adenolinum (Reichenb.) Juz. [2, 3]. The species L. stelleroides (Planch.), distributed in Far East and China, was classified by Yuzepchuk [2] to a monotype section Stellerolinum Juz. ex Prob. The phylogenetic analyses based on chloroplast (ndhF, trnL-F, and trnK 3′ intron) and nuclear ITS (internal transcribed spacer) DNA sequences revealed that genus Linum was not monophyletic. It contains two major lineages: a yellow-flowered clade (sections Linopsis, Syllinum, and Cathartolinum) and a blue-flowered clade (sections Linum, Dasylinum, and Stellerolinum) [4]. The cultivated flax (L. usitatissimum L.) belongs to sec. Linum from a blue-flowered clade. L. usitatissimum is believed to have originated as a result of domestication of wild species L. angustifolium Huds. approximately 8000 years ago [5–8]. For a long time flax has been cultivating as a dual-purpose crop grown for its fiber and linseed oil.
According to morphological and qualitative traits, cultivated flax was divided into five main types: (1) fiber flax (L. usitatissimum subsp. usitatissimum); (2) oil flax (L. usitatissimum L. subsp. humile Czernom.); (3) dual-purpose flax (L. usitatissimum L. subsp. intermedium Czemom.) that was an intermediate form between the first two ones cultivated for fiber and oil; (4) large seeded flax (L. usitatissimum L. subsp. latifolium Snankev.) which is characterized by a set of specific morphological features and cultivated for oil in the Mediterranean region and North Africa; (5) winter flax (L. usitatissimum L. subsp. bienne Mill. Snankev.) cultivated for fiber and oil in the Caucasus, Turkey, Balkans, and some other south regions of Europe [9, 10]. In addition, collections of flax germplasm maintain accessions of primitive flax forms with dehiscent capsules (L. usitatissimum convar. crepitans [Boenningh.] Kulpa et Danert) [11].
The taxonomy of the genus cannot be considered as finally established one because the phylogenetic linkages between the individual taxa have not been sufficiently investigated. The phylogeny of species of the genus Linum was previously studied by the use of molecular and cytogenetic approaches [4, 12–17], but there are problems that still remain to be solved.
Transposon-based molecular markers are successfully used in phylogenic studies. Transposable elements were shown to influence changing in genomic structure as well as transcriptional regulation occurring during the evolution [18, 19]. The presence of transposons in various species of plants, their high integration activity, conservative sequences, and a large number of copies encouraged the use of transposons in the studies of genetic diversity and profiling of plant varieties [20–22]. Several molecular marker systems based on the information available for the transposable elements sequences were developed for plants [20, 22–27]. SSAP (sequence-specific amplified polymorphism) method was shown to have a number of advantages as compared to other marker systems. SSAP method produces many polymorphic fragments and allows differentiation of most samples using only a single combination of specific primers [23, 28–30]. Different plants were successfully studied by SSAP analysis, but the method has not been applied for the investigation of species of the genus Linum yet. Only recently flax sequences have appeared in databases [31–34], and development of a marker system based on flax transposable elements for the investigation of cultivated and wild species of the genus Linum has become possible.
In this study the SSAP method was used for assessment of genetic diversity. Besides, the possibilities of application of marker-based profiling for identification of L. usitatissimum varieties were analyzed. We studied 46 varieties of L. usitatissimum mainly bred in Russia and a number of varieties which were grown in geographically close or distant regions. We also analyzed different types of cultivated flax (fiber, oilseed, large seeded, winter, and dehiscent flax) together with 21 wild species and subspecies from sections Linum, Adenolinum, Dasylinum, and Stellerolinum to estimate the possibility of using the SSAP method for the investigation of flax domestication history and phylogenic linkages between different taxa of the genus Linum.
2. Materials and Methods
2.1. Plant Materials
For the investigation of genetic diversity of cultivated flax, 46 L. usitatissimum varieties, mainly of Russian origin, were obtained from the All-Russian Research Institute for Flax (VNIIL) (Table 1). For analyzing the reproducibility of the SSAP analysis, flax variety “Stormont cirrus” (IPK genbank, Gatersleben, accession number: LIN 261) was used.
Table 1.
Studied flax varieties.
| Number | Variety | Type | Originator | Variety pedigree |
|---|---|---|---|---|
| 1 | Alfa | Fiber | VNIIL | L-1120, Tomskij 4, T-10, Torzhokskij-4, G-4887 |
| 2 | Gorizont | Fiber | VNIIL | L-1120, T-5, T-10, I-2, I-17, Tvertsa. Zarya, Marit Smolenskij, Severyanin, Silva, Natasha, Viera |
| 3 | Lavina | Fiber | Smolenskaja GOSHOS | Sadko × S-108 [X-5 (selected from L-1120) × 1-7] |
| 4 | Vasilyok | Fiber | Belarus | I-7, L-1120, X-5 (selected from L-1120), 221-84-4 |
| 5 | Soyuz | Fiber | Smolenskaja GOSHOS | |
| 6 | Lada | Fiber | VNIIL | Viking, Fibra |
| 7 | Slavnyj 82 | Fiber | VNIIL | from Shokinskij variety, by single plant selection |
| 8 | Krom | Fiber | Pskovskij NIISH | |
| 9 | Tverskoj | Fiber | VNIIL | L-1120, T-4, T-6, T-10, Torzhokskij-4, I-2, G-2782 |
| 10 | Rosinka | Fiber | VNIIL | L-1120, T-5, I-17, Fibra, 1288/12 |
| 11 | Lenok | Fiber | VNIIL | L-1120, M-34 (L-1120 × T-4), T-6, Lazurnyj |
| 12 | A-93 | Fiber | VNIIL | L-1120, T-4, T-6, T-10, Torzhokskij-4 |
| 13 | Tost 3 | Fiber | Sibirskij NIISH | |
| 14 | Mogilevskij-2 | Fiber | Belarus | Stahanovets, L-1120, T-5, T-9 |
| 15 | Belochka | Fiber | Vjatskaja GSHA | L-1120, Tvertsa |
| 16 | Torzhokskij-4 | Fiber | VNIIL | M-34 (L-1120 × T-4) × T-10 (G-360 × G-354) |
| 17 | Tvertsa | Fiber | VNIIL | T-5 × L-1120 |
| 18 | Smolich | Fiber | Smolenskaja GOSHOS | Zarya (X-5 × T-5), mutant A-710, 806/3, severyanin |
| 19 | A-29 | Fiber | VNIIL | L-1120, T-4, T-5, T-10 |
| 20 | Antey | Fiber | Pskovskij NIISH | |
| 21 | Rusich | Fiber | Pskovskij NIISH | Pskovskij 83 (Pryadilshhik, L-1120, T-5, Pobeditel) × Rodnik (T-9, Progress, I-9, L-1120, VNIIL11, Spartak) |
| 22 | Veralin | Fiber | Netherlands | Torzhokskij-4 × Lidiya |
| 23 | Merilin | Fiber | Netherlands | |
| 24 | Smolenskij | Fiber | Smolenskaja GOSHOS | Tvertsa (T-5 × L-1120) × Zarya (X-5—selected from L-1120 × T-5) |
| 25 | Tost-5 | Fiber | Sibirskij NIISH | |
| 26 | S-108 | Fiber | Smolenskaja GOSHOS | X-5 (selected from L-1120) × I-7 |
| 27 | Impuls | Fiber | Smolenskaja GOSHOS | S-108, X-5, I-7, Zarya |
| 28 | Praleska | Fiber | Belarus | |
| 29 | Lider | Fiber | Smolenskaja GOSHOS | Tayga (France selection), mutagen treatment, selection |
| 30 | Voronezhskij 1308/138 | Oil | VNIIMK | |
| 31 | Svetoch | Fiber | VNIIL | Selected from Cherskij kryazh |
| 32 | Ki-5 | Oil | Ukraine | |
| 33 | LM-98 | Oil | VNIIL | |
| 34 | Novotorzhskij | Fiber | VNIIL | L-1120, I-2, I-17, selected from Bogotolskij kryazh, G-2307, G-4523-6-13 |
| 35 | TMP 1919 china 1 | Fiber | china | |
| 36 | k-1147, local form | Oil | Ethiopia | |
| 37 | v-29 | Oil | China | |
| 38 | Aleksim | Fiber | VNIIL | L-1120, T-4, T-5, T-10, Tekstilshhik, M-34, Pryadilshhik, Tvertsa |
| 39 | Ocean k-4497 | Oil | France | |
| 40 | Orshanskij-2 | Fiber | Belarus | I-16 × L-1120 |
| 41 | Κ-6 | Fiber | Pskovskij NIISH | L 1120 × T-5 |
| 42 | Vih oil v-2 | Oil | France | |
| 43 | Donskoj-95 | Oil | Donskaja op.st. | |
| 44 | Diplomat | Fiber | VNIIL | Viking × Fibra (with subsequent selection) |
| 45 | Ford | Fiber | Belarus | |
| 46 | Tomskij-16 | Fiber | Sibirskij NIISH and T | T-9 × G-1077 |
For the investigation of genetic diversity, 47 flax accessions belonging to 22 species from sections Linum, Adenolinum, Dasylinum, and Stellerolinum were used (Table 2). Most of these accessions were obtained from genebank of Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) (Gatersleben, Germany), seed collections of All-Russian Flax Institute (VNIIL) (Torzok, Russian Federation), N. I. Vavilov Research Institute of Plant Industry (VIR) (St. Petersburg, Russian Federation), and Institute of Genetics and Cytology of NAS Belarus (IGC) (Minsk, Belarus). The accessions of L. amurense and L. stelleroides were kindly provided by Dr. L. N. Mironova, Botanic Garden Institute of the Far-Eastern Branch of the Russian Academy of Sciences (BGI) (Vladivostok, Russian Federation). Some accessions were collected in the wild by Dr. A.A. Svetlova, Komarov Botanical Institute RAS (St. Petersburg, Russian Federation), by Dr. N. L. Bolsheva, Engelhardt Institute of Molecular Biology RAS (Moscow, Russian Federation), and by Dr. M. Pavelka, Euroseeds (Novy Jicin, Chech Rep.).
Table 2.
The accessions of studied species of genus Linum.
| Number | Accession name | Source | Genebank number | Origin | Chromosome number (2n) | |
|---|---|---|---|---|---|---|
| Sect. Adenolinum | ||||||
| 1 | L. perenne L. | IPK | LIN 1807 | Russian Federation | 2n = 18 [14] | |
| 2 | L. perenne L. subsp. extraaxillare (Kit.) Nyman | IPK | LIN 1651 | Poland | 2n = 36 [17] | |
| 3 | L. altaicum Ledeb. ex Juz. | IPK | LIN 1632 | Unknown | 2n = 18 [17] | |
| 4 | L. komarovii Juz. | IPK | LIN 1716 | Unknown | 2n = 18 [17] | |
| 5 | L. perenne L. subsp. perenne | IPK | LIN 1521 | Slovakia | 2n = 18 [17] | |
| 6 | L. perenne L. subsp. alpinum (Jacq.) Stoj. and Stef. | IPK | LIN 1905 | Austria | 2n = 18 [17] | |
| 7 | L. perenne L. subsp. anglicum (Mill.) Ockendon | IPK | LIN 1524 | GB | 2n = 36 [17] | |
| 8 | L. perenne L. | VNIIL | K 5500 | Unknown | 2n = 36 [17] | |
| 9 | L. leonii F. W. Schultz | IPK | LIN 1672 | Germany | 2n = 18 [14] | |
| 10 | L. pallescens Bunge | IPK | LIN 1645 | Tajikistan | 2n = 18 [17] | |
| 11 | L. pallescens Bunge | Wild population, collected by N. L. Bolsheva | Russian Federation, Altai | 2n = 18 (present study) | ||
| 12 | L. mesostylum Juz. | IPK | LIN 1774 | Tajikistan | 2n = 18 [17] | |
| 13 | L. mesostylum Juz. | IPK | LIN 1662 | Tajikistan | 2n = 18 [17] | |
| 14 | L. lewisii Pursh | IPK | LIN 1648 | USA | 2n = 18 [17] | |
| 15 | L. lewisii Pursh | IPK | LIN 1550 | USA | 2n = 18 [17] | |
| 16 | L. austriacum L. | IPK | LIN 1608 | Germany | 2n = 18 [17] | |
| 17 | L. austriacum L. | Wild population, collected by A. A. Svetlova | Russian Federation, Rostov | 2n = 18 (present study) | ||
| 18 | L. austriacum L. subsp. euxinum (Juz.) Ockendon | IPK | LIN 1546 | Ukraine | 2n = 18 [17] | |
| 19 | L. austriacum L. | Wild population, collected by A. A. Svetlova | Crimea | 2n = 18 (present study) | ||
| 20 | L. austriacum L. | Wild population, collected by A. A. Svetlova | Ukraine | 2n = 18 (present study) | ||
| 21 | L. amurense Alef. | BGI | outdoors cultivated collection | Russian Federation, Far East, near Pokrovka settlement | 2n= 18 [17] | |
|
| ||||||
| Sect. Dasylinum | ||||||
| 22 | L. hirsutum L. subsp. hirsutum L. | IPK | LIN 1676 | Hungary | 2n = 16 [35] | |
| 23 | L. hirsutum subsp. hirsutum L. | IPK | LIN1649 | Romania | 2n = 16 [36] | |
| 24 | L. hirsutum L. subsp. pseudoanatolicum P.H.Davis | Wild population, collected by M. Pavelka | Turkey, Karaman | 2n = 32 (present study) | ||
| 25 | L. hirsutum L. subsp. anatolicum (Boiss.) Hayek | Wild population, collected by M. Pavelka | Turkey, Aksaray | 2n = 32 (present study) | ||
|
| ||||||
| Sect. Linum | ||||||
| 26 | L. marginale A.Cunn. ex Planch | IPK | LIN 1920 | Australia | 2n = 84 (present study) | |
| 27 | L. narbonense L. | IPK | LIN 2002 | Unknown | 2n = 28 [14] | |
| 28 | L. narbonense L. | IPK | LIN1653 | France | 2n = 28 [14] | |
| 29 | L. decumbens Desf. | IPK | LIN 1754 | Italy | 2n = 16 [14] | |
| 30 | L. decumbens Desf. | IPK | LIN1913 | Italy | 2n = 16 [14] | |
| 31 | L. grandiflorum Desf. | IPK | LIN 2000 | Unknown | 2n = 16 [14] | |
| 32 | L. grandiflorum Desf. | IPK | LIN 974 | Unknown | 2n = 16 [14] | |
| 33 | L. angustifolium Huds. | IPK | LIN 1692 | France | 2n = 30 (present study) | |
| 34 | L. angustifolium Huds. | VNIIL | K 5695 | Unknown | 2n = 30 (present study) | |
| 35 | L. angustifolium Huds. | VNIIL | K 3108 | Belgium | 2n = 30 (present study) | |
| 36 | L. angustifolium Huds | IGC | 15 | Belarus | 2n = 30 [14] | |
| 37 | L. angustifolium Huds. | VNIIL | K 4731 | East Germany | 2n = 30 (present study) | |
| 38 |
L. bienne Mill. (syn. L. angustifolium Huds.) |
IGC | 14 | Hungary | 2n = 30 [14] | |
| 39 | L. usitatissimum L. subsp. bienne Mill. Snankev. | VIR | u-303794 | Unknown | winter flax | 2n = 30 (present study) |
| 40 | L. usitatissimum convar.crepitans [Boenningh.] Kulpa et Danert | IGC | 260 | Belarus | dehiscent flax | 2n = 30 (present study) |
| 41 | L. usitatissimum convar.crepitans [Boenningh.] Kulpa et Danert | IPK | LIN 119 | Portugal | dehiscent flax | 2n = 30 (present study) |
| 42 | L. usitatissimum L. variety Giza | IPK | LIN 277 | Egypt | large seeded flax | 2n = 30 (present study) |
| 43 | L. usitatissimum L. G. 12 Ruzokvety | IPK | LIN 633 | Czechoslovakia | dual-purpose flax | 2n = 30 (present study) |
| 44 | L. usitatissimum L.,variety Colchidsky 1 | VIR | u 099845. | Abkhazia | winter flax | 2n = 30 (present study) |
| 45 | L. usitatissimum L. | VNIIL | k 7131 | Morocco | large seeded flax | 2n = 30 (present study) |
|
| ||||||
| Sect. Stellerolinum | ||||||
| 46 | L. stelleroides Planchon | BGI | Outdoors cultivated collection | Russian Federation, Far East, Telyakovsky Inlet | 2n = 20 (present study) | |
| 47 | L. stelleroides Planchon | BGI | Outdoors cultivated collection | Russian Federation, Far East, near Kraskino settlement | 2n = 20 (present study) | |
2.2. Confirmation of Species Determination
Species determination of some accessions of wild Linum species was done during the course of our earlier cytogenetic investigations [14, 17, 37]. To confirm the species determination, the rest of the accessions were planted in the ground, and, additionally, chromosome analysis (determination of chromosome number) using acetocarmine staining according to previously developed approach was performed [14]. Chromosome numbers of all the studied accessions of wild flax are represented in Table 2.
2.3. SSAP Analysis
Genomic polymorphism of different Linum species was studied using SSAP method [23] with modifications described earlier through genomic studies of wheat [38] and strawberry [39]. Total genomic DNA was extracted from young flax leaves according to Edwards et al. [40] with minor modifications. 30 ng of genomic DNA was treated with 10 U of TaqI restriction enzyme (Thermo Scientific, USA) for 3 h at 65°C. Ligation was performed by adding 2.5 U of T4 DNA-ligase (Thermo Scientific, USA), 5 mM ATP, and 50 pmol of double-strand adapter [5′-ACTCGATTCTCAACCCGAAAGTATAGATCCCA; 5′-PO4-CGTGGGATCTATACTT-(C6linker)-NH2] and incubation for 7 h at 37°C. The product was diluted twice with deionized water. The DNA sequence between the LTR (long terminal repeat) region of retrotransposons and the Taq α I restriction site was amplified using the adapter primer (5′-GTTTACTCGATTCTCAACCCGAAAG 3′) and primers to the LTR regions of FL1a, FL1b, FL4, Cassandra, FL10, FL8, FL7, FL12, and FL9 retrotransposons [32]:
-
1826 5′-ACCCCTTGAGCTAACTTTTGGGGTAAG-3′ (FL1a, FL1b)
-
1833 5′-CTTGCTGGAAAGTGTGTGAGAGG-3′ (FL4)
-
1838 5′-TGTTAATCGCGCTCGGGTGGGAGCA-3′ (FL1a, FL1b, Cassandra)
-
1845 5′-AGCCTGAAAGTGTTGGGTTGTCG-3′ (FL11)
-
1846 5′-CTGGCATTTCCATTGTCGTCGATGC-3′ (FL10)
-
1854 5′-GCATCAGCCTGGACCAGTCCTCGTCC-3′ (FL8)
-
1868 5′-CACTTCAAATTTTGGCAGCAGCGGATC-3′ (FL1a, FL1b)
-
1881 5′-TCGAGGTACACCTCGACTCAGG-3′ (FL7)
-
1886 5′-ATTCTCGTCCGCTGCGCCCCTACA-3′ (FL12)
-
1899 5′-TGAGTTGCAGGTCCAGGCATCA-3′ (FL9)
Amplification was performed in two stages. At the first stage, only the primer to LTR of the retrotransposon was used. Amplification was carried out in 25 μL of PCR mix containing 5 μL of ligation mix, 1 U of TrueStart Hot Start Taq DNA polymerase (Thermo Scientific, USA), TrueStart Taq DNA polymerase buffer, 0.5 mM MgCl2, 20 μM dNTP (Thermo Scientific), and 5 pmol of the LTR primer. The program for amplification for the first stage was 95°C for 15 min, 30 cycles (95°C for 30 s, 62°C for 1 min, 72°C for 2 min) and 72°C for 10 min. At the second stage of amplification, the adapter primer 5′-GTTTACTCGATTCTCAACCCGA-3′ and one of the primers to the LTR region of retrotransposons were used. Amplification was carried out in 25 μL of PCR mix containing 12 μL of the first-stage PCR product, 1 U of Taq DNA polymerase (Thermo Scientific, USA), Taq DNA polymerase buffer, 1.5 mM MgCl2, 200 μM dNTP (Thermo Scientific, USA), 25 pmol of LTR primer, and 25 pmol of the adapter primer. The program for amplification for the second stage was 95°C for 15 min, 35 cycles (95°C for 30 s, 62°C for 1 min, 72°C for 2 min), and 72°C for 10 min. The PCR products were separated in 2.5% agarose gel using TВЕ buffer and then stained with ethidium bromide. Ten PCR products were excised from agarose gel and characterized by sequencing on Applied Biosystems 3730 DNA Analyzer to confirm the specificity of SSAP reaction. The Bio-Rad Gel Doc system was used for gel documentation and photography as well as for visual detection of presence or absence of polymorphic fragments in the samples from different accessions. These data were recorded in the form of a binary matrix in which the presence of a fragment was coded as 1 and its absence as 0.
The genetic distances between varieties were calculated based on the binary matrix of amplified fragments using Dice's formula [41]. The dendrograms were constructed using SplitsTree 4.10 software [42]. Cluster analysis was performed using neighbor-joining method [43] and bootstrap values were determined based on 5000 permutations.
2.4. Preparation of Cassandra Retrotransposon DNA Probe
PCR primers were designed for amplification of Cassandra retrotransposon. We amplified an internal domain (primers IntDom-F AGTGGTATCCGAGCCTCT and IntDom-R CCCATAGGACTCAACGTC) and the LTR with the exception of the 5S rDNA region of this retrotransposon (primers LTR-1-86-F TGTAATGTAACACGTTAGGCA and LTR-1-86-R TTAGTTAGGGACGGATTGTT; LTR-206-279-F AAATAAATCTGTGAGGGATTAGT and LTR-206-279-R ACTTGTAACACCCCGTACT). The amplification was carried out in 20 μL of PCR mixture that contained 1 U of TaqF DNA polymerase (Amplisens, Russia), 1x TaqF buffer, 25 pmol of the forward and reverse primer, 200 μM dNTP (Amplisens, Russia), and 10 ng of genomic DNA. The program for amplification was 95°C for 15 min, 40 cycles (95°C for 10 s, 62°C for 20 s, 72°C for 30 s), and 72°C for 10 min. The amplicons were analyzed in 2% agarose gel and then used as a template for biotin PCR labeling to obtain biotin-labeled probes for FISH. PCR labeling was carried out using Biotin PCR Labeling Core Kit (Jena Bioscience, Germany) according to the manufacture's protocol. Labeled PCR products were precipitated with ethanol.
2.5. FISH with Cassandra Retrotransposon DNA Probe
Chromosome preparation was carried out according to the technique developed earlier for plants having small-sized chromosomes [14]. The hybridization mixture contained 2x SSC, 50% formamide, 10% dextran sulphate, and 2 ng/μL of a biotinylated DNA probe of Cassandra retrotransposon. The probe was hybridized overnight at 31°C. After hybridization the slides were washed twice with 0.1x SSC at 38°C for 10 min, followed by two washes with 2x SSC at 44°C for 5 min and a final 5 min wash in 2x SSC at room temperature. The biotin-labeled DNA probe was detected using a highly sensitive Alexa Fluor 488, Tyramide Signal Amplification system (Invitrogen) according to manufacturer's instructions.
3. Results
3.1. Analysis of SSAP Fingerprints
For analyzing of the reproducibility of the SSAP method, DNA of flax variety “Stormont cirrus” was restricted twice, ligated, and amplified with the primers. The obtained PCR products were visualized in acrylamide and agarose gels (Figure 1). Electrophoretic spectra of PCR products obtained with primers 1826, 1838, 1845, 1868, 1886, and 1899 coincided completely demonstrating high reproducibility of the SSAP results. The fingerprints obtained with primers 1846, 1854, and 1881 varied in individual amplified fragments, so the use of primers 1846, 1854, and 1881 for SSAP analysis of flax varieties will require further optimization of restriction, ligation, or PCR conditions. We selected primers with high reproducibility (1838, 1845, 1868, and 1899) as they yielded PCR products that were easily discernible in an agarose gel. These primers were used for analysis of 46 flax varieties by the SSAP method.
Figure 1.
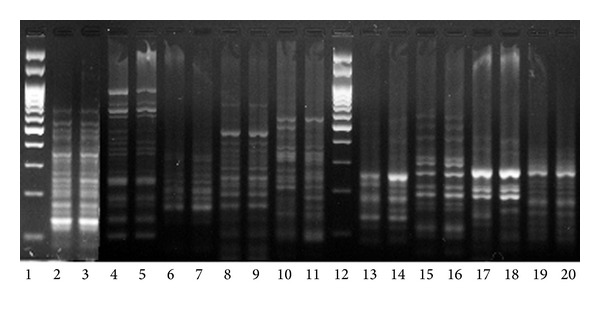
The test of reproducibility of SSAP markers obtained for “Stormont cirrus” flax variety. Lanes 2 and 3, primer 1899; lanes 4 and 5, primer 1826; lanes 6 and 7, primer 1838; lanes 8 and 9, primer 1845; lanes 10 and 11, primer 1846; lanes 13 and 14, primer 1854; lanes 15 and 16, primer 1868; lanes 17 and 18, primer 1881; lanes 19 and 20, primer 1886; lanes 1 and 12, 100 bp DNA ladder.
All the examined varieties produced identical or very similar fingerprints with primers 1838 and 1868 (FL1a, FL1b, and Cassandra). At the same time, the PCR products obtained with primers 1845 and 1899 were unique for different flax varieties. So, primers 1845 and 1899 were chosen for analyzing of genetic diversity of flax varieties. Several of PCR products have been sequenced (Supplementary Material available online at http://dx.doi.org/10.1155/2014/231589), the majority of obtained sequences are significantly similar to the sequences of corresponding retrotransposons.
3.2. Analysis of Genetic Diversity of Flax Varieties
Visual analysis of SSAP fingerprints based on retrotransposons FL11 and FL9 revealed 44 polymorphic retrotransposon insertions (23 fragments for primer 1845 and 21 fragments for primer 1899) in 46 flax varieties. Each of the 46 varieties had their own unique spectrum of retrotransposon insertions. So, we could differentiate all the 46 varieties using only two SSAP primers. In order to analyze genetic diversity of these varieties, we compiled a binary matrix of the presence/absence of polymorphic insertions of the above-mentioned retrotransposons, calculated the genetic distances between the varieties using Dice's formula [41], and constructed a dendrogram by using the neighbor-joining method (Figure 2). The obtained tree branching pattern revealed no distinct clusters among examined varieties.
Figure 2.
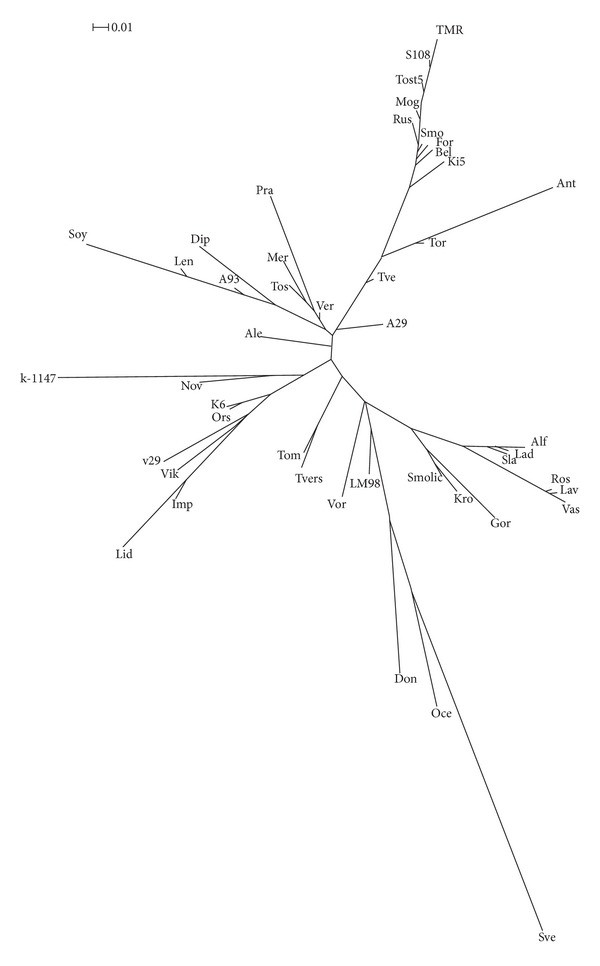
A neighbor-joining dendrogram for SSAP markers. Flax varieties: Alf-Alfa; Gor: Gorizont; Lav: Lavina; Vas: Vasilyok; Soy: Soyuz; Lad: Lada; Sla: Slavnyj 82; Kro: Krom; Tvers: Tverskoj; Ros: Rosinka; Len: Lenok; A93: A-93; Tos: Tost 3; Mog: Mogilevskij 2; Bel: Belochka; Tor: Torzhokskij 4; Tve: Tvertsa; Smolic: Smolich; A29: A-29; Ant: Antey; Rus: Rusich; Ver: Veralin; Mer: Merilin; Smo: Smolenskij; Tost5: Tost 5; S108: S 108; Imp: Impuls; Pra: Praleska; Lid: Lider; Vor: Voronezhskij; Sve: Svetoch; Ki5: Ki-5; Lm98: LM-98; Nov: Novotorzhskij; TMR: TMR-1919; k-1147: k-1147; v29: v-29; Ale: Aleksim; Oce: Ocean k-4497; Ors: Orshanskij 2; K6: K-6; Vic: Viksoil V-2; Don: Donskoj 95; Dip: Diplomat; For: Ford; Tom: Tomskij 16.
3.3. Genomic Diversity of Species of the Genus Linum
For investigation of species of the genus Linum, we chose retrotransposons FL1a, FL1b, and Cassandra which did not show high insertion polymorphism within cultivated flax varieties. Primers 1838 and 1868 were used for SSAP analysis. As the result, 95 bands that originated with primer 1838 and 128 bands with primer 1868 (Figure 3) were scored. All the bands were polymorphic. Based on the SSAP fingerprint similarity, nine groups of closely related species (A-I) were distinguished. Group A included different species of sect. Adenolinum (syn. L. perenne group); group B consisted of L. hirsutum subsp. hirsutum accessions; group C′′ included L. hirsutum subsp. pseudoanatolicum and L. hirsutum subsp. anatolicum. Groups D, E, F, and G comprised species accessions of sect. Linum (L. marginale; L. narbonense, L. decumbens, and L. grandiflorum, resp.). Group H included accessions of L. angustifolium and L. usitatissimum (sect. Linum); and group I consisted of L. stelleroides accessions (sect. Stellerolinum).
Figure 3.
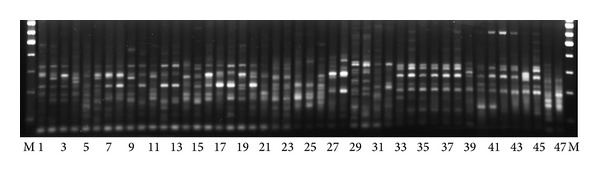
SSAP markers generated using 1868 primer for Linum species: 1: L. perenne LIN 1807; 2: L. perenne subsp. extraaxilare, LIN 1651; 3: L. altaicum, LIN 1632; 4: L. komarovii LIN 1716; 5: L. perenne, LIN 1521; 6: L. perenne susp. alpinum, LIN 1905; 7: L. perenne susp. anglicum, LIN 1524; 8: L. perenne, K 5500; 9: L. leonii, LIN 1672; 10: L. pallescens, LIN 1645; 11: L. pallescens, Altai; 12: L. mesostylum, LIN 1774; 13: L. mesostylum, LIN 1662; 14: L. lewisii, LIN 1648; 15: L. lewisii, LIN 1550; 16: L. austriacum, LIN 1608; 17: L. austriacum, Rostov; 18: L. austriacum subsp. euxinum, LIN 1546; 19: L. austriacum, Crimea; 20: L. austriacum, Ukraine; 21: L. amurense; 22: L. hirsutum, LIN 1676; 23: L. hirsutum, LIN1649; 24: L. hirsutum subsp. pseudoanatolicum; 25: L. hirsutum subsp. anatolicum; 26: L. marginale; 27: L. narbonense, LIN 2002; 28: L. narbonense, LIN1653; 29: L. decumbens, LIN 1754; 30: L. decumbens, LIN1913; 31: L. grandiflorum, LIN 2000; 32: L. grandiflorum, LIN 974; 33: L. angustifolium, LIN 1692; 34: L. angustifolium, K 5695; 35: L. angustifolium, K 3108; 36: L. angustifolium, Belarus; 37: L. angustifolium, K 4731; 38: L. biene (syn. L. angustifolium); 39: winter flax (L. usitatissimum subsp. biene); 40: dehiscent flax (L. usitatissimum convar. crepitans); 260; 41: dehiscent flax (L. usitatissimum convar. crepitans), LIN 119; 42: large seeded flax(L. usitatissimum), LIN 277; 43: dual-purpose flax (L. usitatissimum), LIN 633; 44: winter flax (L. usitatissimum), u 099845; 45: large seeded flax (L. usitatissimum), к 7131; 46: L. stelleroides, Telyakovsky Inlet; 47: L. stelleroides, Kraskino settlement. M—100 bp DNA ladder.
All the groups contained at least one group-specific marker. The results of phylogenetic analysis of Linum species are shown in the dendrogram on Figure 4. As the dendrogram shows nine clearly distinguished groups of species supported by high bootstrap values can be observed.
Figure 4.

Neighbor-joining dendrogram for SSAP markers. Flax species: 1: L. perenne LIN 1807; 2: L. perenne subsp. extraaxilare, LIN 1651; 3: L. altaicum, LIN 1632; 4: L. komarovii LIN 1716; 5: L. perenne, LIN 1521; 6: L. perenne susp. alpinum, LIN 1905; 7: L. perenne susp. anglicum, LIN 1524; 8: L. perenne, K 5500; 9: L. leonii, LIN 1672; 10: L. pallescens, LIN 1645; 11: L. pallescens, Altai; 12: L. mesostylum, LIN 1774; 13: L. mesostylum, LIN 1662; 14: L. lewisii, LIN 1648; 15: L. lewisii, LIN 1550; 16: L. austriacum, LIN 1608; 17: L. austriacum, Rostov; 18: L. austriacum subsp. euxinum, LIN 1546; 19: L. austriacum, Crimea; 20: L. austriacum, Ukraine; 21: L. amurense; 22: L. hirsutum, LIN 1676; 23: L. hirsutum, LIN1649; 24: L. hirsutum subsp. pseudoanatolicum; 25: L. hirsutum subsp. anatolicum; 26: L. marginale; 27: L. narbonense, LIN 2002; 28: L. narbonense, LIN1653; 29: L. decumbens, LIN 1754; 30: L. decumbens, LIN1913; 31: L. grandiflorum, LIN 2000; 32: L. grandiflorum, LIN 974; 33: L. angustifolium, LIN 1692; 34: L. angustifolium, K 5695; 35: L. angustifolium, K 3108; 36: L. angustifolium, Belarus; 37: L. angustifolium, K 4731; 38: L. biene (syn. L. angustifolium); 39: winter flax (L. usitatissimum subsp. biene); 40: dehiscent flax (L. usitatissimum convar. crepitans); 260; 41: dehiscent flax (L. usitatissimum convar. crepitans), LIN 119; 42: large seeded flax (L usitatissimum), LIN 277; 43: dual-purpose flax (L usitatissimum), LIN 633; 44: winter flax (L usitatissimum), u 099845; 45: large seeded flax (L usitatissimum), к 7131; 46: L. stelleroides, Telyakovsky Inlet; 47: L. stelleroides, Kraskino settlement. Bootstrap values that exceeded 70% are shown in italic.
3.4. FISH with Cassandra Retrotransposon DNA Probe
The highly sensitive tyramide FISH method was applied for the investigation of abundanceof Cassandra retrotransposons as well as their distribution along chromosomes in three species of sect. Linum (L. usitatissimum, L. grandiflorum, and L. narbonense) and L. amurense (sect. Adenolinum). FISH revealed that Cassandra dispersed along the whole length of chromosomes in karyotypes of four studied species, but its distribution along the chromosomes was nonrandom (Figure 5). In species having small-sized chromosomes (L. usitatissimum, L. grandiflorum, and L. amurense), Cassandra was mainly localized in pericentromeric and subtelomeric chromosome regions. The patterns of Cassandra distribution were chromosome specific and were similar in homologous pairs of chromosomes (Figure 5(e)). In L. narbonense, which possessed large chromosomes, the patterns of Cassandra distribution resembled the patterns observed in karyotypes of the above-mentioned species (having small-sized chromosomes) though they were more regular.
Figure 5.
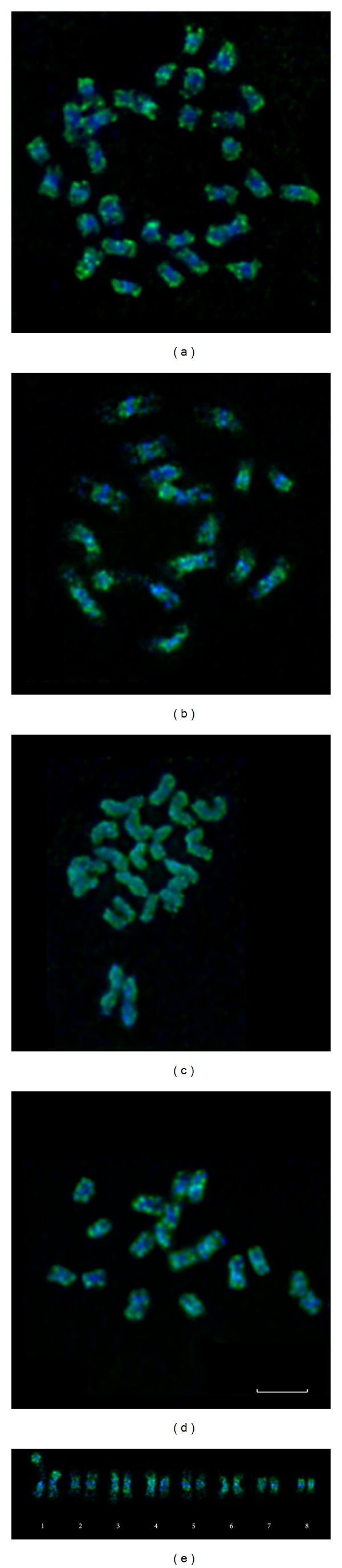
FISH with DNA probe of Cassandra retrotransposons (green). (a) L. usitatissimum (sect. Linum); (b) L. grandiflorum (sect. Linum); (c) L. narbonense (sect. Linum); (d) L. amurense (sect. Adenolinum); (e) a karyotype of L. grandiflorum. Chromosomes were stained with DAPI (blue). Bar—5 μm.
4. Discussion
4.1. Use of SSAP Analysis for Identification of Flax Varieties and Estimation of Genetic Diversity
To estimate genetic polymorphism and to characterize L. usitatissimum varieties, the SSAP method was used, and also high reproducibility of the method was shown. The validity of the SSAP method for molecular genetic studies of flax varieties using FL11 and FL9 retrotransposons was demonstrated. Obtained with primers 1838 and 1868 (retrotransposons FL1a, FL1b, and Cassandra) PCR products were very similar in all the studied varieties. Such low diversity of cultivated flax might be a result of low transposition activity and/or creation of bottleneck effect during flax selection.
In analyzed flax varieties, 44 polymorphic insertions for FL11 (primer 1845) and FL9 (primer 1899) retrotransposons were revealed. Every studied variety possessed a unique set of SSAP markers. Therefore, the SSAP method can be used to mark the genotypes, to identify varieties of L. usitatissimum in genebank collections, to exercise control during of flax variety growth, and to obtain high quality seed material, when the varietal identity is particularly important.
The genetic similarity of 46 flax varieties was characterized by genetic distances calculated based on the SSAP data. The dendrogram (Figure 3) did not contain clearly isolated clusters of varieties. Thus, the studied flax varieties could not be subdivided into distinct groups. Our results were in agreement with earlier obtained data shown that flax accessions examined by IRAP analysis did not form distinct clusters in studies of their origin or the type of commercial use (fiber or oil). These data indicated an overlap in genetic diversity despite of disruptive selection for fiber or seed oil types [32]. In our study, the SSAP method also failed to distinguish fiber or oil seed flax varieties. Since varieties with the best characteristics are commonly used as parents in breeding practice, some valuable flax forms present in the genealogy of most modern varieties. Besides, the lines selected for crossing are usually characterized by low genetic diversity. So, the commercial flax varieties were shown to be less diverse than wild flax species and landraces [32].
Although the examined flax varieties could not be clustered into different groups by SSAP method, it might be used for estimation of their genetic similarity based on polymorphic insertions of retrotransposons. The estimation can be used for choosing the parents in breeding practice and also for creation of core collections which should include genetically diverse accessions.
4.2. Diversity and Phylogeny of Linum Species
In the present study, 20 accessions from sect. Linum, 21 accessions from sect. Adenolinum, 4 accessions from sect. Dasylinum, and 2 accessions from sect. Stellerolinum were analyzed by using SSAP method. All the examined species were clustered into 9 groups mainly according to common taxonomic division of the genus Linum into sections (Figure 4).
4.3. Section Dasylinum
Species from sect. Dasylinum clustered together and formed two related groups B and C. Group B included L. hirsutum subsp. pseudoanatolicum and L. hirsutum subsp. anatolicum and group C included L. hirsutum subsp. hirsutum. Thus, SSAP analysis singled out sect. Dasylinum as a well-supported clade. Our results were in agreement with the AFLP and ITS data as well as chloroplast phylogenies, chromosome studies, and transcriptome analysis of Linum species [4, 13, 37, 44]. It should be mentioned that the subdivision of accessions of sect. Dasylinum into two related clusters correlated with their difference in chromosome numbers and the origin of accessions. Thus, the accessions of L. hirsutum subsp. hirsutum (cluster C) from Europe was characterized by chromosome number of 2n = 16, while accessions from Turkey, L. hirsutum subsp. pseudoanatolicum and L. hirsutum subsp. anatolicum (cluster B) have chromosome number 2n = 32. Chromosome numbers for L. hirsutum subsp. pseudoanatolicum and L. hirsutum subsp. anatolicum were firstly determined in the present study.
4.4. Section Stellerolinum
Two accessions of L. stelleroides have rather similar SSAP fingerprints which were differed significantly from all the others Linum species and formed a separated clade. The similar results were obtained by phylogenetic analyses of chloroplast and ITS DNA sequences [4]. Moreover, L. stelleroides was shown to have chromosome number 2n = 20 which was unique for blue-flowered flaxes [45].
4.5. Section Adenolinum
All the members of sect. Adenolinum formed an independent group clustered separately from species of sect. Linum and other sections. Distinct isolation of this species group was also revealed in several molecular and karyological investigations [4, 12–14, 17]. The data were in good agreement with the opinion of Yuzepchuk [2] and Egorova [3] who isolated the group from sect. Linum into an independent section Adenolinum.
SSAP fingerprints of the accessions within sect. Adenolinum were highly polymorphic, but SSAP markers did not allow us to reveal any species subclusters supported by a high bootstrap value. Thus, SSAP analysis used in the present study as well as AFLP and RAPD analyses [13, 17] separated individual accessions but did not identify individual species inside sect. Adenolinum.
4.6. Section Linum
Sect. Linum was subdivided into 5 groups by neighbor-joining clustering. Accessions of L. marginale, L. grandiflorum, L. decumben, and L. narbonense formed four independent single species groups, while the fifth group combined accessions L. angustifolium and L. usitatissimum. Similar results had been obtained earlier by AFLP, RAPD, molecular phylogeny based on chloroplast RbsL sequence, and molecular cytogenetic methods (C/DAPI-banding patterns and localization of rRNA genes on chromosomes) [4, 12–14].
Within a subgroup consisted of L. usitatissimum and L. angustifolium, the accessions of large seeded flax (breeding cultivar), dual-purpose flax, and L. angustifolium were rather similar. Their fingerprints did not differ significantly from fingerprints of studied 46 flax varieties. This data were in a good agreement with the suggestion that L. angustifolium was the progenitor of L. usitatissimum [5, 7].
The accession of large seeded flax landrace, the accessions of winter flax, and the accessions of dehiscent flax differed significantly from the other members of cluster H. Both accessions of dehiscent flax grouped together (supported by a high bootstrap value) and had species-specific SSAP markers.
It should be noted that flax accessions, which are genetically distant from modern flax cultivars, are particular important for flax breeding. The genetic diversity of cultivated flax decreased significantly during the last decades. It might lead to the lack of useful alleles in genomes of modern cultivars [13]. Therefore, introduction of new useful traits from the ancient primitive forms of cultivated flax and wild species could increase the polymorphism of modern flax varieties. SSAP markers allowed us to identify the unique accessions which are important for the investigation of the history of flax domestication.
L. marginale, the last member of sect. Linum, is a wild flax native to Australia. We found that it had the maximal chromosome number (2n = 84) in the genus Linum. The number indicated a high level of ploidy of L. marginale genome. SSAP patterns of the species were significantly different compared with the other species of sect. Linum. Therefore, L. marginale clustered apart from the other species. The obtained results were in contradiction with ITS and chloroplast topologies which clustered the species together with L. bienne and L. usitatissimum [4]. Rogers [46] assumed that Australian and New Zealand species L. marginale Cunn. and L. monogynum Forst. were related to European species L. hologynum Reichenb. (sec. Linum). This assumption based on the fact that diploid chromosome number of L. hologynum (2n = 42) corresponded to haploid chromosome number of L. monogynum and L. marginale. Moreover, all the three species had fused styles and pantoporate pollen grains that were unusual for blue-flowered flaxes. Thus, all the above-mentioned data indicated that the phylogenic lineages of L. marginale need further investigation.
The data obtained in the present study, as well as the results of other molecular phylogenetic and chromosomal investigations, indicated that members of section Linum were not as closely related as members of other sections. Therefore, taxonomic revision of this section is desirable.
4.7. Chromosome Location of Cassandra Retrotransposon
As differences in SSAP fingerprints for several flax species were found, we decided to analyze the distribution of Cassandra retrotransposon along the chromosomes of Linum species. Cassandra is a terminal-repeat retrotransposon in miniature (TRIM) that carries conserved 5S rDNA sequences in its LTRs. Cassandra was found in a number of vascular plants [31]. In our work, we investigated the distribution of this retrotransposon along the chromosomes of Linum species using tyramide FISH. We revealed that Cassandra localized in pericentromeric and subtelomeric regions of chromosomes that was typical for transposable elements [47]. A more uniform distribution of Cassandra retrotransposon was found in L. narbonense in comparison with L. usitatissimum, L. grandiflorum, and L. amurense. It was probably due to a higher content of transposable elements correlated with a larger size of its chromosomes (therefore its genome).
5. Conclusions
The availability of LTR sequences of flax retrotransposons and high polymorphism of SSAP markers offer a promising potential for SSAP analysis of genus Linum. Applications of SSAP analysis, for example, evolutionary and phylogenetic studies, assessment of genetic diversity, accession identification, and search for exotic genepools of cultivated flax, could be applied to L. usitatissimum and other Linum species. SSAP analysis was shown to be very useful for characterization of flax varieties and identification of accession belonging to different species or sections and provided new information about of phylogenetic relationships within the genus Linum.
Supplementary Material
Sequences of SSAP PCR products.
Acknowledgments
The work was financially supported by the Russian Foundation for Basic Research (Grants 12-04-01469-а, 13-04-01770-a, and 14-08-01167); by Fundamental Research Program of the Russian Academy of Sciences “Dynamics of Plant, Animal and Human Genofonds”; and by the Ministry of Education and Science of the Russian Federation under state Contract 14.621.21.0001. Part of this work was performed at the EIMB RAS “Genome” center.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Nataliya V. Melnikova, Anna V. Kudryavtseva, and Alexander V. Zelenin contributed equally to this work.
References
- 1.Ockendon DJ, Walters SM. Linaceae. Cambridge, Mass, USA: Cambridge University Press; 1968. [Google Scholar]
- 2.Yuzepchuk SA. Genus Linum—Linaceae Dumort. In: Shishkin BK, editor. Flora SSSR (Flora of the Soviet Union) Vol. 14. Leningrad, Russia: Izdatel’stvo Akademii Nauk; 1949. pp. 84–146. [Google Scholar]
- 3.Egorova TV. Genus Linum, Linaceae. St Petersburg Publishing; 1996. [Google Scholar]
- 4.McDill J, Repplinger M, Simpson BB, Kadereit JW. The phylogeny of Linum and Linaceae subfamily Linoideae, with implications for their systematics, biogeography, and evolution of heterostyly. Systematic Botany. 2009;34(2):386–405. [Google Scholar]
- 5.Helbaek H. Domestication of food plants in the old world. Science. 1959;130(3372):365–372. doi: 10.1126/science.130.3372.365. [DOI] [PubMed] [Google Scholar]
- 6.van Zeist W, Bakker-Heeres JAH. Evidence for linseed cultivation before 6000 bc. Journal of Archaeological Science. 1975;2(3):215–219. [Google Scholar]
- 7.Diederichsen A, Hammer K. Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.) Genetic Resources and Crop Evolution. 1995;42(3):263–272. [Google Scholar]
- 8.Zohary D, Hopf M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe, and the Nile Valley. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 9.Sisov IA. Flax. Leningrad, Russia: Selhosgiz; 1955. [Google Scholar]
- 10.Cernomorskaja NM, Stankevic AK. K voprosu o vnutryvidovoj klassifikacii lna obyknovennogo (Linum usitatissimum L.). To the problem of intraspecific classification of common flax (Linum usitatissimum L.) Selekcija I Genetika Tehnicheskih Kultur. 1987;113:53–63. [Google Scholar]
- 11.Diederichsen A. Ex situ collections of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genetic Resources and Crop Evolution. 2007;54(3):661–678. [Google Scholar]
- 12.Fu Y, Peterson G, Diederichsen A, Richards KW. RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genetic Resources and Crop Evolution. 2002;49(3):253–259. [Google Scholar]
- 13.Vromans J. Molecular genetic studies in flax (Linum usitatissimum L.) [Ph.D. thesis] Wageningen, The Netherlands: Wageningen University; 2006. [Google Scholar]
- 14.Muravenko OV, Yurkevich OY, Bolsheva NL, et al. Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica. 2009;135(2):245–255. doi: 10.1007/s10709-008-9273-7. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Allaby RG. Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genetic Resources and Crop Evolution. 2010;57(5):667–677. [Google Scholar]
- 16.Soto-Cerda BJ, Saavedra HU, Navarro CN, Ortega PM. Characterization of novel genic SSR markers in Linum usitatissimum (L.) and their transferability across eleven Linum species. Electronic Journal of Biotechnology. 2011;14(2) [Google Scholar]
- 17.Yurkevich OY, Naumenko-Svetlova AA, Bolsheva NL, et al. Investigation of genome polymorphism and seed coat anatomy of species of section Adenolinum from the genus Linum. Genetic Resources and Crop Evolution. 2013;60(2):661–676. [Google Scholar]
- 18.Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology. 2000;42(1):251–269. [PubMed] [Google Scholar]
- 19.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nature Reviews Genetics. 2002;3(5):329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 21.Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 22.Kalendar R, Flavell AJ, Ellis THN, Sjakste T, Moisy C, Schulman AH. Analysis of plant diversity with retrotransposon-based molecular markers. Heredity. 2011;106(4):520–530. doi: 10.1038/hdy.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waugh R, McLean K, Flavell AJ, et al. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Molecular and General Genetics. 1997;253(6):687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- 24.Ellis THN, Poyser SJ, Knox MR, Vershinin AV, Ambrose MJ. Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Molecular and General Genetics. 1998;260(1):9–19. doi: 10.1007/pl00008630. [DOI] [PubMed] [Google Scholar]
- 25.Queen RA, Gribbon BM, James C, Jack P, Flavell AJ. Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Molecular Genetics and Genomics. 2004;271(1):91–97. doi: 10.1007/s00438-003-0960-x. [DOI] [PubMed] [Google Scholar]
- 26.Melnikova NV, Konovalov FA, Kudryavtsev AM. Long terminal repeat retrotransposon Jeli provides multiple genetic markers for common wheat (Triticum aestivum) Plant Genetic Resources: Characterisation and Utilisation. 2011;9(2):163–165. [Google Scholar]
- 27.Kim H, Terakami S, Nishitani C, et al. Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breeding Science. 2012;62(1):53–62. doi: 10.1270/jsbbs.62.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce SR, Knox M, Ellis THN, Flavell AJ, Kumar A. Pea Ty1-copia group retrotransposons: transpositional activity and use as markers to study genetic diversity in Pisum. Molecular and General Genetics. 2000;263(6):898–907. doi: 10.1007/s004380000257. [DOI] [PubMed] [Google Scholar]
- 29.Vershinin AV, Allnutt TR, Knox MR, Ambrose MJ, Ellis THN. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Molecular Biology and Evolution. 2003;20(12):2067–2075. doi: 10.1093/molbev/msg220. [DOI] [PubMed] [Google Scholar]
- 30.Sanz AM, Gonzalez SG, Syed NH, Suso MJ, Saldaña CC, Flavell AJ. Genetic diversity analysis in Vicia species using retrotransposon-based SSAP markers. Molecular Genetics and Genomics. 2007;278(4):433–441. doi: 10.1007/s00438-007-0261-x. [DOI] [PubMed] [Google Scholar]
- 31.Kalendar R, Tanskanen J, Chang W, et al. Cassandra retrotransposons carry independently transcribed 5S RNA. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(15):5833–5838. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smýkal P, Bačová-Kerteszováč N, Kalendar R, Corander J, Schulman AH, Pavelek M. Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theoretical and Applied Genetics. 2011;122(7):1385–1397. doi: 10.1007/s00122-011-1539-2. [DOI] [PubMed] [Google Scholar]
- 33.Ragupathy R, Rathinavelu R, Cloutier S. Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genomics. 2011;12, article 217 doi: 10.1186/1471-2164-12-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.González LG, Deyholos MK. Identification, characterization and distribution of transposable elements in the flax (Linum usitatissimum L.) genome. BMC Genomics. 2012;13(1, article 644) doi: 10.1186/1471-2164-13-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosova IV, Semenova OY, Samatadze TE, et al. Investigation of karyotype structure and mapping of ribosomal genes on chromosomes of wild linum species by FISH. Biologicheskie Membrany. 2005;22(3):244–248. [Google Scholar]
- 36.Bolsheva NL, Semenova OY, Muravenko OV, Nosova IV, Popov KV, Zelenin AV. Localization of telomere sequences in chromosomes of two flax species. Biologicheskie Membrany. 2005;22(3):227–231. [Google Scholar]
- 37.Muravenko OV, Bol'sheva NL, Iurkevich OI, et al. Karyogenomics of species of the genus Linum L. Genetika. 2010;46(10):1339–1342. [PubMed] [Google Scholar]
- 38.Konovalov FA, Goncharov NP, Goryunova S, Shaturova A, Proshlyakova T, Kudryavtsev A. Molecular markers based on LTR retrotransposons BARE-1 and Jeli uncover different strata of evolutionary relationships in diploid wheats. Molecular Genetics and Genomics. 2010;283(6):551–563. doi: 10.1007/s00438-010-0539-2. [DOI] [PubMed] [Google Scholar]
- 39.Melnikova NV, Kudryavtseva AV, Speranskaya AS, et al. The FaRE1 LTR-retrotransposon based SSAP markers reveal genetic polymorphism of strawberry (Fragaria x ananassa) cultivars. Journal of Agricultural Science. 2012;4(11):111–118. [Google Scholar]
- 40.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research. 1991;19(6):p. 1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 42.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Sveinsson S, McDill J, Wong GK, et al. Phylogenetic pinpointing of a paleopolyploidy event within the flax genus (Linum) using transcriptomics. Annals of Botany. 2014;113(5):753–761. doi: 10.1093/aob/mct306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokolovskaya AP, Probatova NS. Chromosome numbers in the vascular plants from the Primorye territory, Kamchatka, region, Amur valley and Sakhalin. Botanicheskii Zhurnal SSSR. 1985;70(4):997–999. [Google Scholar]
- 46.Rogers CM. A further note on the relationships of the European Linum hologynum and the Australian species of Linum (Linaceae) Plant Systematics and Evolution. 1984;147(3-4):327–328. [Google Scholar]
- 47.Heslop-Harrison JSP, Schwarzacher T. Organisation of the plant genome in chromosomes. Plant Journal. 2011;66(1):18–33. doi: 10.1111/j.1365-313X.2011.04544.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of SSAP PCR products.


