Abstract
Methoprene-tolerant (Met) protein is a juvenile hormone (JH) receptor in insects. JH-bound Met forms a complex with the βFtz-F1-interacting steroid receptor coactivator (FISC) and together they regulate JH response genes in mosquitoes. Both proteins contain basic-helix-loop-helix (bHLH) and PAS motifs. Here we demonstrated that FISC is the obligatory partner of Met for binding to JH-response elements (JHREs). Met or FISC alone could not bind a previously characterized JHRE, while formation of the Met-FISC complex was necessary and sufficient to bind to the JHRE. This binding required participation of the DNA-binding domains of both Met and FISC. The optimal DNA sequence recognized by Met and FISC contained a core consensus sequence GCACGTG. While formation of the Met-FISC complex in mosquito cells was induced by JH, heterodimerization and DNA binding of bacterially expressed Met and FISC were JH-independent, implying that additional mosquito proteins were required to modulate formation of the receptor complex.
Keywords: Insects, Hormone receptor, Helix-loop-helix transcription factors, Hormone response element, Protein-protein interaction, Transcription regulation
1. Introduction
Juvenile hormone (JH) is one of the most enigmatic hormones in invertebrate endocrinology. In insects, it is secreted from corpora allata, a pair of endocrine glands connected to the brain (Tobe SS, 1985). JH plays crucial roles in many aspects of insect life, including development, reproduction, diapause, caste differentiation, migratory behavior and longevity (Flatt et al., 2005; Goodman and Cusson, 2012; Nijhout, 1994).
Many functions of JH are mediated by the Methoprene-tolerant (Met) protein, an intracellular receptor of JH. In Drosophila melanogaster, null mutants of Met show resistance to both the toxic and morphogenetic effects of JH and its mimic methoprene (Wilson and Fabian, 1986). RNAi-mediated depletion of Met in the red flour beetle, Tribolium castaneum, causes larvae to pupate prematurely before reaching their final instar (Konopova and Jindra, 2007). In newly emerged Aedes aegypti females, RNAi knockdown of Met stalls the growth of ovarian follicles, similar to the phenotypic effects of JH deprivation (Zou et al., 2013).
The Met protein belongs to the basic-helix-loop-helix Per-Arnt-Sim (bHLH-PAS) family of transcription factors (Ashok et al., 1998). The basic region of the bHLH domain is comprised of 13 amino acids, rich in arginine and lysine residues. The HLH region contains two alpha-helices separated by a loop of variable length. The helices promote formation of homo- or heterodimers, which bring the basic regions of two proteins together to bind to DNA with a hexanucleotide core called E-box (CANNTG) (Sailsbery et al., 2012). The PAS domain functions as a protein dimerization motif and consists of two similar hydrophobic repeats, termed PAS-A and PAS-B, separated by a poorly conserved spacer (Kewley et al., 2004). Recent studies have demonstrated that in vitro synthesized Met binds JH with relatively high affinity through a binding pocket formed by the PAS-B domain (Charles et al., 2011; Miura et al., 2005).
It has been shown in several insect species that Met is essential for the induced expression of JH response genes (Minakuchi et al., 2008; Parthasarathy et al., 2008; Zhu et al., 2010). In newly emerged female Ae. aegypti mosquitoes, the post-eclosion activation of the Krüppel homolog 1 (AaKr-h1) gene and the early-trypsin (AaET) gene requires both AaMet and the βFtz-F1-interacting steroid receptor coactivator (AaFISC). AaFISC also carries the bHLH-PAS domain and has been characterized as a transcriptional coactivator of the ecdysteroid receptor complex (Li et al., 2011; Zhu et al., 2006). AaMet and AaFISC form a heterodimer in the presence of JH. When AaET is upregulated by the elevated JH titer in female adults, AaMet and AaFISC are associated with the AaET promoter in the midgut, indicating that both proteins act directly on the AaET promoter to activate its transcription (Li et al., 2011). In transient transfection assays, AaMet and AaFISC activate the AaET promoter in the presence of JH. A JH response element (JHRE) identified in the AaET promoter, which contains an asymmetric E-box (CACGCG), is sufficient for the JH-induced transactivation by AaMet-AaFISC (Li et al., 2011).
The orthologs of FISC are called Taiman (TAI) in D. melanogaster and Steroid Receptor Coactivator (SRC) in other insect species. JH-induced expression of Kr-h1 in Ae. aegypti, Bombyx mori and T. castaneum all requires the functions of Met and FISC/SRC/TAI (Kayukawa et al., 2012; Kayukawa et al., 2013; Li et al., 2011; Zhang et al., 2011). E-box-like sequences have been identified in the regulatory region of Kr-h1 gene in these three species (Kayukawa et al., 2012; Shin et al., 2012). A more recent study by Raikhel’s lab found similar E-box-like sequences in 68 Ae. aegypti genes, the expression of which is AaMet-dependent in adult female mosquitoes (Zou et al., 2013). Thus, transcriptional activation by recruiting the Met-FISC complex to the E-box-like sequences might be a conserved mechanism in JH action. Besides Met and FISC, expression of AaKr-h1 is also under the control of another bHLH-PAS protein, Cycle (CYC), in newly emerged adult mosquitoes (Shin et al., 2012). CYC dimerizes with Met in a JH-dependent manner and the Met-CYC complex appears to bind independently of FISC to a JHRE containing the CACGCG motif, further complicating the role of FISC in JH signaling (Shin et al., 2012).
Although possessing a putative DNA-binding domain, steroid receptor coactivators do not bind DNA directly. They interact with DNA-binding transcription factors and recruit downstream effectors, including histone acetyltransferases and protein methyltransferases (Xu et al., 2009). While the Met-FISC complex is shown to activate expression of JH response genes, many questions remain unanswered: Are Met and FISC loaded to the target promoters through direct DNA binding or protein-protein interaction? Is FISC the DNA binding partner of Met or a coactivator of the JH receptor? Does Met-FISC recognize other types of JH response elements? Here we report our in vitro DNA-binding assays using bacterially expressed recombinant AaMet and AaFISC proteins. The results indicated that AaMet and AaFISC are required and sufficient for binding to the JHRE identified from AaET. Both AaMet and AaFISC directly bound JHRE through their basic regions located in the bHLH domains. Furthermore, we performed a comprehensive screening of sequences preferably bound by AaMet and AaFISC. A consensus sequence, GCACGTG, was found to bind AaMet and AaFISC with high affinity. Luciferase reporter assay and in vitro DNA-binding assay demonstrated that the consensus sequence was a functional JHRE. This study significantly advances our understanding of the JH-induced gene activation by Met and FISC in molecular details.
2. Material and methods
2.1 Plasmids
pCMA, pCMA-GAD, pCMA-GBD, and UAS×4-188-cc-Luc were from Dr. Lucy Cherbas (Hu et al., 2003). The expression vectors pCMA-AaMet and pCMA-AaFISC have been described previously (Li et al., 2011). To construct 4×JHRE1-luc, a chimeric DNA fragment was inserted into the pGL3 basic plasmid between restriction sites Kpn I and Nco I. The insert consisted of the four tandem repeats of JHRE1 (5′-CCACACGCGAAG-3′) from AaET and the minimal promoter of AaET (−77 to +61). 4×MFBS1-luc was constructed similarly, except that the AaET JHRE1 was replaced with MFBS1 (5′-GCCGCACGTGTC-3′).
2.2 Expression and purification of recombinant proteins
The codon usage of AaMet cDNA was optimized for bacterial expression. A cDNA fragment encoding the amino acid residues 1-597 of AaMet was cloned into expression vector pGEX-6P-1 (GE Healthcare) between restriction sites BamH I and Not I, resulting in an expression plasmid for AaMet, pGEX-6P-1-Metn. Escherichia coli BL21(DE3) strain transformed with the plasmid was cultured in Luria-Bertani (LB) medium at 37°C to approach an OD600 of 0.8. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. The culture was grown at 25°C for three more hours. Bacterial pellets were resuspended in lysis buffer [20 mM sodium phosphate, pH 7.3, 150 mM NaCl, 2 mM DTT, 1 mM PMSF, and 1×Halt protease inhibitor cocktail (Thermo Scientific)]. Cells were lysed using DeBEE high pressure homogenizer (BEE international) and debris was removed by centrifugation at 30,000 × g for 30 minutes. Proteins in the supernatant were affinity-purified using ÄKTA prime and GSTrap FF column (GE Healthcare) at 4°C with binding buffer (20 mM sodium phosphate, pH 7.3, 150 mM NaCl, and 2 mM DTT) and elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione, and 2 mM DTT). Purified GST-AaMet was dialyzed in PBS buffer containing 2 mM DTT and 10% glycerol, and was stored at −80°C until use.
The cDNA region encoding the amino acid residues 1-609 of AaFISC was cloned into expression vector pRSET-A (Invitrogen) between restriction sites BamH I and Kpn I. Recombinant AaFISC was expressed as a His-tag fusion protein under the control of T7 promoter in E. coli BL21(DE3) pLysS strain. Bacterial cells were grown in LB medium at 37°C. When OD600 reached 0.6, IPTG was added to a final concentration of 0.2 mM. Cells were cultured for two more hours at 20°C and were then collected by centrifugation. His6-AaFISC was purified using ÄKTA prime and HisTrap FF column according to the standard protocol provided by GE Healthcare. Buffers used for FISC protein purification were: lysis buffer (20 mM sodium phosphate, pH 7.4, 0.5 mM NaCl, 20 mM imidazole, 2 mM DTT, 1 mM PMSF, and 1×Halt protease inhibitor cocktail); binding buffer (20 mM sodium phosphate, pH 7.4, 0.5 mM NaCl, 20 mM imidazole, and 2 mM DTT); elution buffer (20 mM sodium phosphate, pH 7.4, 0.5 mM NaCl, 0.5 M imidazole, and 2 mM DTT). Dialysis in PBS and protein storage was conducted as described above for GST-AaMet.
2.3 Gel-shift assay
Oligonucleotides used in gel-shift assays were as follows: 5′-CCATCCCACACGCGAAGACGATAAAACCA-3′ (AaET_JHRE1) and 5′-GCCGCACGTGTCGTTGG-3′ (MFBS1). Double-stranded DNA oligonucleotides were end-labeled by T4 Polynucleotide Kinase (New England Biolabs) and [γ-32P] ATP (PerkinElmer), followed by purification with Bio-Spin 6 column (Bio-Rad). For DNA binding, 0.5 μg of purified Met, FISC, or both proteins was added to the binding buffer [20 mM sodium phosphate (pH 7.4), 50 mM NaCl, 1 mM MgCl2, 5 mM DTT, 100 ng/μl BSA, 50 ng/μl poly(dA-dT), and 10 μM JH-III or DMSO carrier]. After 10 minutes incubation at room temperature, 20 fmol of the labeled probe (~20,000 cpm) were added to make a total volume of 20 μl. The reactions were incubated for 20 more minutes followed by electrophoresis at 120V for 50 minutes with a 6% polyacrylamide DNA retardation gel (Invitrogen) in 0.5× TBE buffer. The gel was dried and the 32P-labeled DNA was visualized by autoradiography. Competition experiments were performed by inclusion of a 50-fold molar excess of unlabeled specific or nonspecific competitor DNA in the binding reaction. In super-shift experiments, 3 μg of GST antibody (Santa Cruz), His-tag antibody (Millipore) or mock immunoglobulin G (IgG) were added to the binding reactions 20 minutes after addition of the labeled probe, and the reactions were incubated for an additional 20 minutes before electrophoresis.
2.4 Site-directed mutagenesis
PCR site-directed mutagenesis was carried out as described (Carrigan et al., 2011). Primers containing point mutations were used in PCR to amplify the template plasmids. PCR products were cleaned up with PCR purification kit (Qiagen), followed by Dpn I digestion at 37°C for 1 hour to remove the template plasmids. The DNA was purified again and about 200 ng of the DNA were used to transform E. coli NEB 10-beta competent cells (New England Biolabs) following the manufacturer’s instructions. The mutations were all confirmed by DNA sequencing.
2.5 In vitro selection and amplification of DNA binding site
Screening for DNA-binding site was modified from a method described previously by Swanson et al. (Swanson et al., 1995). A single-stranded DNA library, 5′-CCACCAACAACAACATCAGC-(N)17-CTTCCGATGGATACTGGAGG-3′, was synthesized. It contained all possible 17-bp DNA sequences (417 ≈ 1.7 × 1010 different sequences) flanked by adaptor sequences. To generate double-stranded DNA, the single-stranded DNA library was annealed to a primer complementary to the 3′ adaptor sequence, followed by DNA extension with Taq polymerase at 72°C for 30 minutes. The reaction products were resolved in 2.5% agarose gel and the double-stranded DNA was recovered. Purified DNA was end-labeled with [γ-32P] ATP and T4 Polynucleotide Kinase, followed by purification with Bio-Spin 6 column (Bio-Rad).
Gel-shift assays were conducted by incubating 0.5 μg each of purified AaMet and AaFISC, 1 ng of the labeled DNA in 20 μl binding buffer [20 mM sodium phosphate, pH 7.4, 50 mM NaCl, 1 mM MgCl2, 5 mM DTT, 100 ng/μl BSA, 100 ng of sonicated salmon sperm DNA (GE healthcare), and 10 μM JH-III]. After electrophoresis and autoradiography, shifted band was cut from the gel. The gel slice was placed in 200 μl water and kept in a shaker at 700 rpm at 4°C overnight. Forty microliters of the eluent were used as DNA template for PCR amplification to generate an enriched pool of selected oligonucleotides for the next round of selection. A total of ten rounds of selection were conducted. After the last selection, DNA was cloned into pCR2.1 TOPO TA cloning vectors (Invitrogen) and subjected to sequencing analysis. Consensus motifs were identified by the MEME algorithm (Bailey et al., 2009).
2.6 Luciferase reporter assay
For Ae. aegypti Aag2 cells, 5×105 cells were plated in each well of a 48-well plate. Transfection was carried out according to the manufacturer’s instructions with 2 μl Cellfectin (Invitrogen) and 320 ng of DNA [100 ng of firefly luciferase reporter plasmid, 100 ng of each expression vector for AaMet and AaFISC, and 20 ng of internal control plasmid pRL-CMV (Promega)]. JH-III, farnesol, methoprene and pyriproxyfen were purchased from Sigma Aldrich and dissolved in dimethyl sulfoxide (DMSO). These chemicals were added to the culture medium at 24 hours after transfection. Cells were harvested at 48 hours after transfection and reporter activity was measured using Dual Luciferase Assay kit (Promega).
2.7 Measuring the dissociation constants for the binding of AaMet and AaFISC to JHRE
The apparent equilibrium dissociation constants (Kd) for the binding of Met and FISC to JHREs were measured as described (Riechmann et al., 1996). Gel-shift assays were carried out with a fixed amount of purified Met and FISC proteins (0.5 μg each) and increasing amounts of probes. Probes were used at six concentrations, 1 nM, 2.5 nM, 5 nM, 10 nM, 25 nM, and 50 nM. After gel electrophoresis, the bound and free probe was quantitated with a phosphorimager (Molecular Dynamics). The data were used to calculate the apparent Kd with a Scatchard plot (Scatchard, 1949).
2.8 Protein structure modeling and ligand docking
Homology modeling was carried out as described by Pandini et al (Pandini et al., 2009). The NMR structures of hypoxia-inducible factor 2α (HIF-2α) and the aryl hydrocarbon receptor nuclear translocator (ARNT) PAS-B domains were chosen as the templates for homology modeling. Their coordinate files were obtained from the Protein Data Bank: entries 1P97 for HIF-2α (Erbel et al., 2003) and 1X0O for ARNT (Card et al., 2005). A three-dimensional model of the AaMet PAS-B domain was generated using MODELLER version 9v7 (Sali and Blundell, 1993). Energy minimization was performed to improve overall quality of the generated structures by using the GROMACS 4.0.7 package (Van Der Spoel et al., 2005). Identification and characterization of surface pockets and internal cavities in the modeled PAS-B were carried out by using CASTp with default parameters (Dundas et al., 2006). AutoDockTools (ADT) (Sanner, 1999) and AutoDock 4 (Huey et al., 2007) were used to set up and perform the docking calculations. Images were prepared with Pymol (DeLano Scientific).
2.9 JH-Binding Assays
The [3H]-labeled JH-III (20 Ci/mmol) was from Perkin-Elmer. Dextran-coated charcoal (DCC) assays were performed as described (Charles et al., 2011; Miura et al., 2005). Wild-type and mutants of AaMet were synthesized in vitro using the TNT T7 Coupled Reticulocyte Lysate System (Promega). Non-specific binding was determined in a parallel experiment, where TNT products were incubated with increasing concentrations of [3H] JH-III in the presence of a 100-fold molar excess of unlabeled JH-III. The esterase inhibitor 3-octylthio-1,1,1-trifluoro-2-propanone (OTFP) was added to all the binding assays at a final concentration of 1 μM. Dissociation constants (Kd) were determined using the Scatchard method (Scatchard, 1949).
3. Results
3.1 Met and FISC bind to JHRE as a complex
To investigate whether Met and FISC proteins are sufficient for binding to JHRE, we carried out gel-shift assays using purified recombinant Ae. aegypti Met and FISC proteins. The bHLH-PAS domain of AaMet (amino acid 1-597) was expressed in E. coli as a fusion protein with an N-terminal GST-tag. The bHLH-PAS domain of AaFISC (amino acid 1-609) was expressed with an N-terminal His6-tag (Fig. 1A). After affinity purification, there was only one major protein band corresponding to the His6-FISC fusion. For GST-Met fusion, we tried many different purification procedures and conditions, and still could not separate two major proteins with sizes of 93 kDa and 60 kDa (Fig. 1B). Mass spectrometry analysis showed that the 93-kDa polypeptide was the expected GST-Met fusion, while the 60-kDa protein was a derivative of GST-Met that lacked the C-terminal portion of the Met PAS domain. The mixture of the 93-kDa and 60-kDa proteins was used as GST-Met in subsequent DNA binding assays.
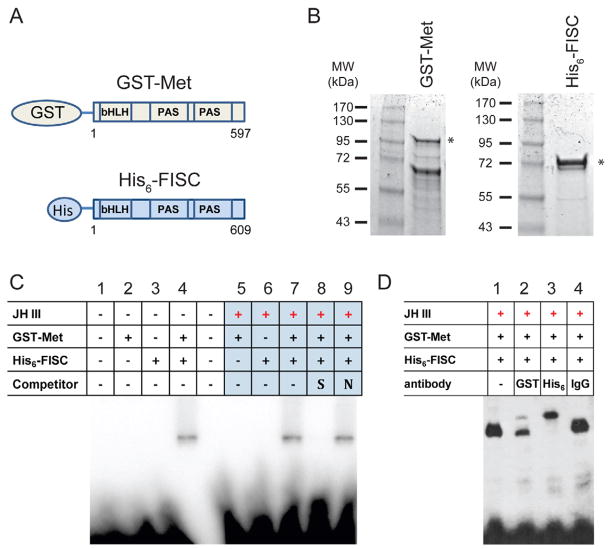
Figure 1.
Met and FISC are sufficient for in vitro binding of JHRE. A) Schematic diagram of E. coli-expressed N-terminal Met fused with GST-tag and FISC with His-tag. B) Coomassie staining of the purified recombinant Met and FISC. The asterisks (*) indicate protein bands with expected sizes. C) Gel-shift assay with the recombinant proteins. Met and FISC, or either protein alone, were incubated with the DNA probe (AaET JHRE1, 5′-CCATCCCACACGCGAAGACGATAAAACCA-3′) in the presence or absence of 10−6 M JH-III for 20 minutes followed by electrophoresis. For competition, 50-fold molar excess of unlabeled specific (S) or non-specific (N) competitor DNA, was mixed with proteins for 20 minutes before addition of the probe. D) Super-shift assay. Antibodies for the GST-tag (GST) or His-tag (His6) were included in the DNA-binding reactions. Non-specific rabbit IgG was used as a control. The experiments were repeated three times with similar results. Representative autoradiographs are shown.
A 29-bp DNA fragment from the AaET promoter, containing a previously characterized JHRE1 (CCACACGCGAAG), was used as a probe in gel-shift assays. The purified GST-Met or His 6-FISC alone was unable to bind the AaET JHRE1 (Fig. 1C, lanes 2, 3, 5 and 6). When GST-Met and His6-FISC together were incubated with the labeled JHRE1, a stable DNA-protein complex was detected in the absence and presence of JH (Fig. 1C, lanes 4 and 7). Formation of the complex in vitro seemed to be JH-independent. The specificity of the Met-FISC-JHRE binding was demonstrated by competition experiments. The binding was abrogated by addition of unlabeled AaET JHRE1 at 50-fold molar excess relative to the probe, but not by unlabeled nonspecific competitor (Fig. 1C, lanes 8 and 9). To verify that both Met and FISC were present in the observed DNA-protein complex, we performed a super-shift experiment. Addition of either GST antibody (Fig. 1D, lane 2) or His-tag antibody (Fig. 1D, lane 3) to the DNA binding reactions resulted in formation of a larger DNA-protein complex, while addition of non-specific IgG did not show similar effect (Fig. 1D, lane 4). The gel-shift experiment thus demonstrated that the purified AaMet and AaFISC proteins bind JHRE as a complex and this in vitro binding does not require other mosquito proteins.
3.2 The basic regions of both Met and FISC proteins are involved in DNA binding
The basic regions of bHLH proteins are usually involved in DNA binding, with the basic residues (arginine and lysine) often forming direct contacts with the major groove of the DNA (Jones, 2004). Seven and five basic residues exist in the basic regions of AaMet and AaFISC, respectively (Fig. 2A and 2B). These residues are highly conserved among Ae. aegypti, D. melanogaster, T. castaneum and B. mori. To test whether the putative DNA-binding domains of AaMet and AaFISC were required for their binding to JHRE, AaMet and AaFISC mutants were created by replacing the individual basic residues in the bHLH domains with glutamine, which is structurally similar to arginine and lysine but has no positive charge on its side chain. The mutants were then tested for their abilities to bind JHRE in vitro and their abilities to activate JH-inducible promoters in transient transfection assays.
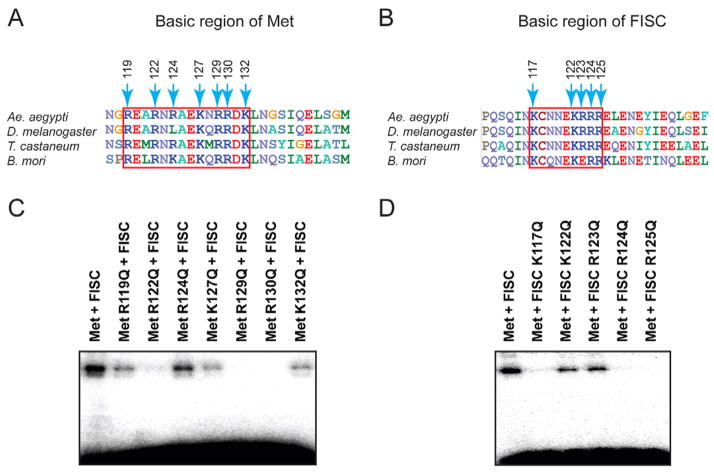
Figure 2.
The basic regions of Met and FISC are both involved in DNA binding. (A and B) Alignments of the first α-helix of the bHLH domain in the orthologs of Met (A) and FISC (B) from several insect species. The basic regions are highlighted in red rectangles. Numbers indicate positions of the basic residues in Ae. aegypti Met and FISC. (C and D) Gel-shift assays with the indicated recombinant Met and FISC that carry mutations in the basic regions. The AaET JHRE1 was used as the radiolabeled probe.
Wild-type and mutant proteins of AaMet and AaFISC were expressed in E. coli and purified using affinity chromatography (Fig. S1). In gel-shift assays, the R122Q, R129Q, R130Q mutations in AaMet completely abolished the binding of Met and FISC to AaET JHRE1, while the R119Q, R124Q, K127Q and K132Q mutations in AaMet diminished the binding to a lesser extent (Fig. 2C). In AaFISC, the K117Q, R124Q and R125Q mutations also eliminated the DNA binding of AaMet-AaFISC (Fig. 2D). The results demonstrated that the binding of AaMet-AaFISC to JHRE requires the DNA-binding domains of both proteins, and implied that each partner perhaps binds to part of the AaET JHRE1.
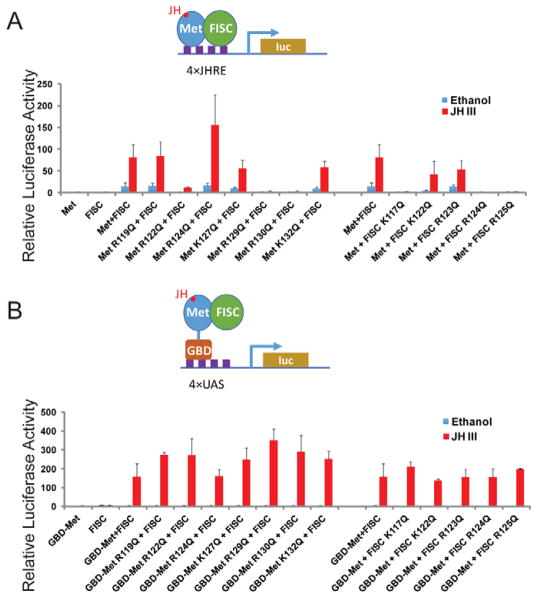
For the cell-based reporter assays, expression vectors for the wild-type or mutant AaMet and AaFISC were transfected into Ae. aegypti Aag2 cells together with a firefly luciferase reporter gene driven by four copies of the AaET JHRE1. The R122Q, R129Q, and R130Q mutations in AaMet all led to a dramatic decrease in the JH-induced expression of the reporter gene (Fig. 3A). Likewise, K117Q, R124Q, and R125Q mutations in AaFISC also displayed similar negative effect (Fig. 3A). There was a good correlation between the reporter assay and the in vitro DNA-binding assay. Mutations in Met and FISC that substantially reduced the JH-induced expression of 4×JHRE1-Luc in the reporter assay, such as MetR122Q, MetR129Q, MetR130Q, FISCK117Q, FISCR124Q, and FISCR125Q, also significantly weakened the binding of AaMet and AaFISC to the AaET JHRE1 (Fig. 2C and 2D). To demonstrate that these mutations affect the DNA binding but not the dimerization or transactivation activity of AaMet and AaFISC, we carried out the reporter assay with some modifications. The wild-type AaMet and its mutants were expressed as fusions to the GAL4 DNA-binding domain (GBD) and the firefly luciferase reporter gene was under the control of four copies of the GAL4-binding sites (UAS). In this system, we expected GBD-AaMet and AaFISC to form a heterodimer in the presence of JH and use the GBD domain to bind UAS of the reporter gene. GBD-AaMet and AaFISC indeed activated expression of the UAS-driven reporter gene when JH was added to the culture medium after transfection (Fig. 3B). None of the mutations of the basic residues in either AaMet or AaFISC showed considerable negative impact on the JH-induced reporter expression (Fig. 3B), indicating that these basic residues are not essential for dimerization and transactivation activity of AaMet and AaFISC. Therefore, the gel-shift assays and these two reporter assays together suggested that the basic regions of both AaMet and AaFISC are required for binding of the AaMet-AaFISC complex to JHRE.
Figure 3.
The mutations in the basic regions primarily affect the DNA-binding properties of Met and FISC. (A) Transfection assay with a JHRE-driven luciferase reporter. Aag2 cells were transfected by the 4×JHRE1-luc reporter construct and expression vectors for the derivatives of Met and FISC. Transfected cells were treated with 10−6 M JH-III or ethanol. (B) Transfection assay with a UAS-driven luciferase reporter. Aag2 cells were transfected by the UAS×4-188-cc-Luc reporter construct and expression vectors for the derivatives of Met and FISC. All the Met derivatives were expressed as fusions to the GAL4 DNA-binding domain.
3.3 Identification of the consensus DNA sequence bound by the JH receptor complex
To identify the DNA sequences bound with high-affinity by the AaMet-AaFISC complex, we screened a random DNA library using multiple cycles of selection and amplification. The synthetic 57-nt DNA library used for selection contained a 17-nt random region flanked by PCR priming sequences. The double-stranded DNA was end-labeled with 32P and incubated with JH-III and the purified GST-Met and His6-FISC in a gel-shift experiment. After electrophoresis and autoradiography, DNA was retrieved from the shifted band and amplified by PCR to get an enriched DNA pool for a second round of selection. After 10 rounds of selection, the enriched DNA was cloned and analyzed by DNA sequencing. Among the 70 sequences that we obtained, 34 were unique sequences. A consensus motif was identified by the MEME motif discovery algorithm. An E-box like sequence, GCACGTG, existed in 67 out of the total 70 sequences (Fig. 4A). For future reference, the consensus sequence was numbered from −4 to +3 as shown in Fig. 4B. The most abundant sequence, GGCCGCACGTGTCGTTG, was named MFBS1 (Met-FISC binding site 1) and used for further study.
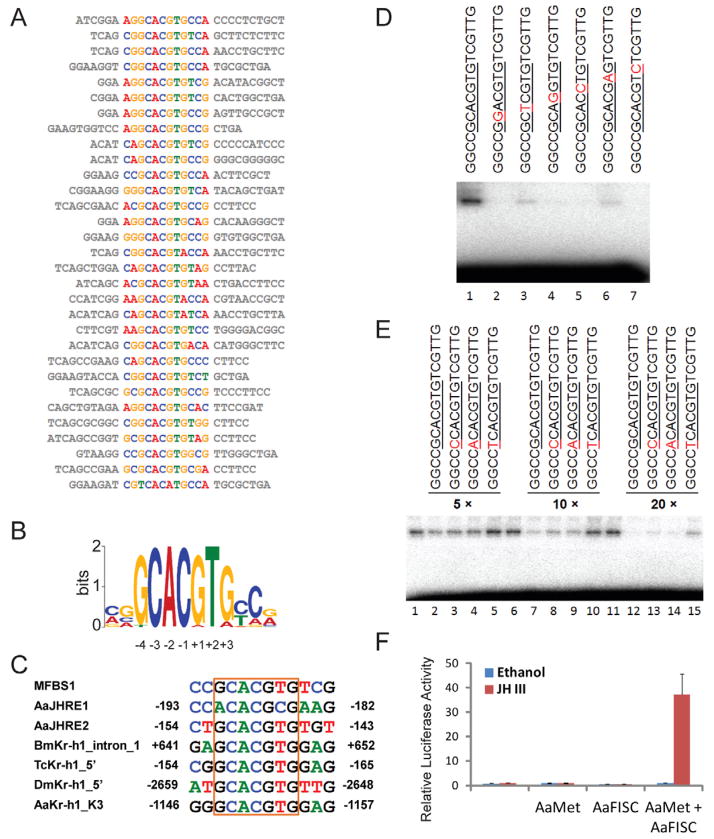
Figure 4.
In vitro selection of DNA sequences bound by Met and FISC. (A) 31 unique sequences after iterative cycles of enrichment and amplification were analyzed by the MEME algorithm. (B) The top-scoring motif. The common motif, GCACGTG, was numbered from −4 to +3. (C) Some previously identified JHREs contain the exact consensus sequence. The AaET JHRE1 was reported by Li et al. (Li et al., 2011). The regulatory regions of Kr-h1 in B. mori (Bm), T. castaneum (Tc), D. melanogaster (Dm) and Ae. aegypti (Aa) were characterized by Dr. Shinoda’s group and Dr. Raikhel’s group (Kayukawa et al., 2012; Shin et al., 2012) (D) Validation of the sequence-specific DNA binding. Gel-shift assays were conducted with purified Met and FISC proteins in the presence of 10−6 M JH-III. MFBS1 and its mutants were labeled individually and used as probes in the experiment. The consensus sequence was underlined and point mutations in the probe sequences were shown in red. (E) Guanosine is preferred at the −4 position in the consensus motif. MFBS1 was labeled as a probe. The indicated oligonucleotides with mutations at the −4 position were used as competitors in the gel-shift assay. The consensus sequence was underlined and point mutations were shown in red. (F) The consensus sequence is a functional JHRE. Aag2 cells were transfected with 4×MFBS1-luc and the expression vectors of AaMet and AaFISC. Transfected cells were treated with either 10−6 M JH-III or ethanol.
The JHRE1 that we have previously identified in AaET shares some similarity with the GCACGTG motif. A sequence downstream of the JHRE1 in the 5′ regulatory region of AaET was found to harbor the exact consensus motif and named AaET_JHRE2. In addition, this motif was discovered in the JH-inducible promoters of the Kr-h1 genes from Ae. aegypti, B. mori, D. melanogaster and T. castaneum (Fig. 4C).
To validate the binding selectivity, gel-shift assay was conducted using the purified recombinant proteins of AaMet and AaFISC. As shown in Fig. 4D, AaMet and AaFISC were able to bind MFBS1 when JH was present. We then introduced point mutations into the consensus motif and used the MFBS1 derivatives as probes in gel-shift assays. Individual point mutation at any position (−3, −2, −1, +1, +2 and +3) within the sequence CACGTG considerably decreased or abolished the DNA binding of AaMet-AaFISC (Fig. 4D).
To examine the role of guanosine at −4 position in the DNA-protein interaction, we carried out a competition experiment. The guanosine at −4 position in MFBS1 was changed into adenine, cytosine or thymine to generate three new MFBS1 derivatives. The 32P-labeled MFBS1 was incubated with the purified AaMet and AaFISC (Fig. 4E). Unlabeled MFBS1 and the three derivatives were added at 5-, 10-, and 20-fold molar excess in the gel-shift assay. The unlabeled MFBS1 was the most effective among the four competitors to inhibit the binding of AaMet-AaFISC to the labeled MFBS1 (Fig. 4E). The in vitro binding assays demonstrated that the GCACGTG motif identified by our selection and amplification approach is a sequence-specific and high affinity binding site of AaMet-AaFISC.
To test whether MFBS1 actually functions as a JH response element, we performed a reporter assay in Aag2 cells. A reporter plasmid (4×MFBS1-luc) was constructed such that the firefly luciferase expression was controlled by four copies of a shorter version of MFBS1, GCCGCACGTGTC. The reporter gene was readily induced by JH-III when AaMet and AaFISC were over-expressed in Aag2 cells, indicating that the consensus sequence functions as a JHRE that mediates the transcriptional activation by the AaMet-AaFISC complex (Fig. 4F).
3.4 Met and FISC bind the consensus sequence with high affinity
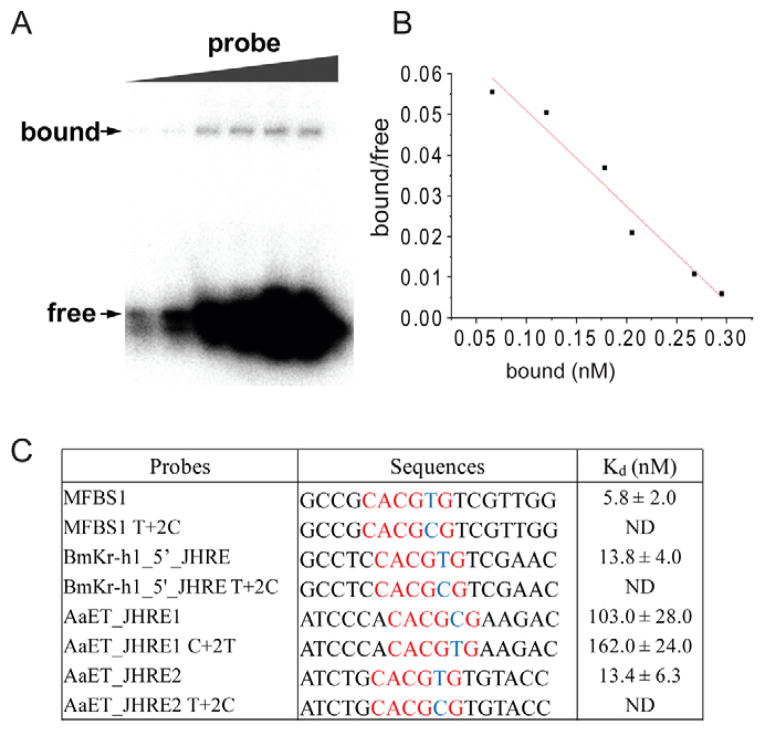
To compare the binding affinity of AaMet-AaFISC to MFBS1 and several naturally occurring E-box-like JHREs, we measured the apparent equilibrium dissociation constants (Kd). A series of gel-shift assays were performed with fixed amounts of the AaMet and AaFISC proteins and increasing amounts of DNA probes. The free and bound DNA probes were quantitated and the data were used to calculate the apparent Kd. as shown in Fig. 5.
Figure 5.
The binding affinities of the AaMet-AaFISC complex to various JHREs. (A) Dissociation constant (Kd) of the binding of Met-FISC to MFBS1 was measured by gel-shift assays. Details were described in the Experimental Procedures. (B) Scatchard analysis of the binding of AaMet-AaFISC to MFBS1. The Kd value was calculated from the slope of the regression line. (C) Comparison of the binding affinities of AaMet-AaFISC to various JHREs. Kd was shown as mean±SD, n=3. ND, Kd was not determined because the DNA binding could not be detected.
The AaMet-AaFISC complex showed the strongest binding to MFBS1, with a Kd of 5.8 nM (Fig. 5C). AaET_JHRE2 and the JHRE identified in the 5′ regulatory region of BmKr-h1 (BmKr-h1_5′_JHRE) each contain a palindromic E-box (CACGTG). Their binding affinities for the JH receptor complex were slightly weaker, with Kd values of 13.4 nM and 13.8 nM for AaET_JHRE2 and BmKr-h1_5′-JHRE, respectively. AaET_JHRE1 harbors a non-canonical E-box (CACGCG) and exhibited a much weaker affinity for the protein complex. Its Kd value was about 17-fold higher than that of MFBS1 (Fig. 5C).
To investigate whether the AaMet-AaFISC complex prefers the DNA sequence of CACGTG over the asymmetric CACGCG, we introduced point mutations to the abovementioned probes at the +2 position. The mutant probes were incubated with AaMet and AaFISC in gel-shift assays and their dissociation constants were measured in the presence of JH-III. When the CACGTG hexameric core was changed into CACGCG in MFBS1, the mutant probe completely lost the ability to bind the AaMet-AaFISC complex (Fig. 5C). Similarly, the thymine-to-cytosine mutation in BmKr-h1_5′_JHRE or AaET_JHRE2 all abolished the binding of AaMet-AaFISC, indicating that AaMet and AaFISC generally prefer a JHRE with a CACGTG core sequence. AaET_JHRE1 shares less similarity with MFBS1 both in the hexameric core and in the flanking sequence. When the CACGCG hexamer in AaET_JHRE1 was replaced with CACGTG, the binding affinity nevertheless remained relatively weak (Fig. 5C). This result suggested that the flanking sequences also contribute significantly to the overall binding affinity of the JHRE.
3.5 Generation of the JH-binding deficient mutants of AaMet
The gel-shift experiment indicated that the purified AaMet and AaFISC bound to AaET_JHRE1 in a JH-independent manner. To examine whether the JH-binding capacity was compromised in the purified AaMet, we measured its dissociation constant. The recombinant AaMet purified from bacteria exhibited a Kd of 160.8 ± 27.6 nM for JH-III, while AaMet produced in rabbit reticulocyte lysates showed a much stronger JH-III binding (Kd=4.4 ± 1.9 nM) (Fig. S2). However, the AaMet protein synthesized in reticulocyte lysates could not be used in gel-shift assays because the unprogrammed reticulocyte lysate contains a strong DNA-binding activity toward the AaET_JHRE1 and MFBS1 (data not shown). To further investigate the effect of hormone binding on the DNA binding of AaMet-AaFISC, we employed homology modeling techniques to generate a three-dimensional model of the AaMet PAS-B domain and constructed several JH-binding deficient mutants of AaMet based on the structural information.
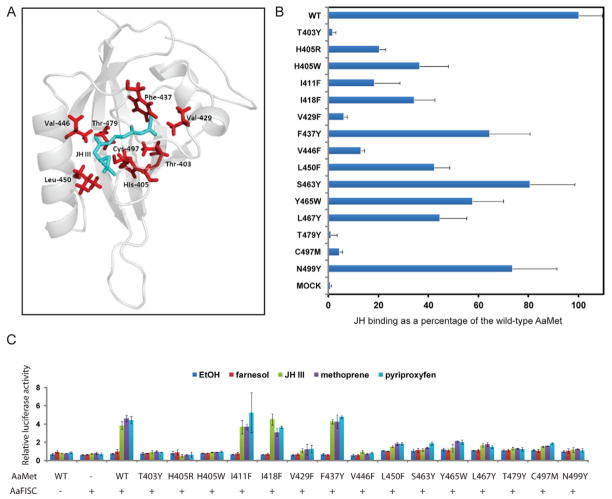
Analysis of structural cavities indicated the presence of a buried cavity between the central anti-parallel β-sheet and several α-helices flanking the sheet (Fig. 6A). Side chains of 22 amino acids, which are conserved in the PAS-B domains of AaMet, DmMet and TcMet, were predicted to be internal to the cavity. Molecular docking calculations were performed to simulate JH-III binding to the PAS-B domain of AaMet. The following 14 residues were predicted to be in close contact with the docked JH-III: Thr403, His405, Ile411, Ile418, Val429, Phe437, Val446, Leu450, Ser463, Tyr465, Leu467, Thr479, Cys497 and Asn499.
Figure 6.
Generation of JH-binding deficient mutants of AaMet by structure-guided mutagenesis. (A) Cartoon representation of the modeled AaMet PAS-B domain with the lowest-energy docked conformation of JH-III (shown as green sticks). (B) Effect of the mutations within the PAS-B domain of AaMet on the binding of [3H]-labeled JH-III. The binding was normalized against the AaMet inputs measured in Figure S3. (C) The JH-induced transactivation activity decreased substantially in the JH-binding deficient mutants of AaMet. Drosophila L57 cells were transfected by the 4×JHRE1-luc reporter plasmid and the expression vectors for AaFISC and the indicated AaMet derivatives. The mutations in AaMet did not markedly affect the expression of AaMet and AaFISC in L57 cells (Fig. S4). Transfected cells were treated with 1 μM of farnesol, JH-III, methoprene or pyriproxyfen. Methoprene and pyriproxyfen are two JH agonists. Farnesol is a biosynthetic intermediate for JH-III. Ethanol was used as control. The mean and standard deviation from triplicate samples were indicated.
To validate this model, we performed site-directed mutagenesis of these selected residues and tested the effects of mutations on JH binding. The bHLH-PAS domains of the wild-type and mutant AaMet were synthesized in vitro in coupled transcription/translation reactions (Fig. S3). JH-III binding of AaMet was nearly abolished by the T403Y, V429F, T479Y and C497M mutations. H405R, I411F and V446F each reduced the JH-III binding to 10%–20% of that of the wild-type AaMet (Fig. 6B). The hormone binding assay demonstrated that these residues are crucial for the high-affinity binding of AaMet to JH-III. The corresponding residues of T403, I411, V429, V446, T479 and C497 in T. castaneum have been demonstrated previously as essential for the binding of TcMet to JH-III (Charles et al., 2011), suggesting that the structure signatures of the JH-binding pocket in Met are highly conserved in insects.
Furthermore, we carried out a transient transfection experiment to examine how these mutations in AaMet affect the transactivation activity of the AaMet-AaFISC complex. The JH-induced expression of the 4×JHRE1-luc reporter gene was substantially dampened when the 6 AaMet mutants (T403Y, H405R, V429F, V446F, T479Y and C497M) showing considerably reduced JH-binding affinities were used in lieu of the wild-type AaMet, consistent with the predicted roles of those residues in binding of JH (Fig. 6C). The result also implied that these mutations affected the binding of AaMet to JH analogs, such as methoprene and pyriproxyfen. The S463Y, Y465W and N499Y mutants, while retained 60–80% of the JH-binding capacity of the wild-type AaMet, failed to activate the JH-induced expression of the reporter gene, suggesting that these residues play important roles in the protein interaction between Met and FISC or other transcriptional cofactors.
3.6 JH-independent interaction between the bacterially expressed Met and FISC
The T403Y, V429F and T479Y AaMet mutants were then compared with the wild-type AaMet in the in vitro DNA-binding assays. In the presence of JH and AaFISC, the dissociation constants of the three mutants for MFBS1 were 6.1–7.3 nM, similar to 5.8 nM for the wild-type AaMet (Table 1). In the absence of JH, the wild-type AaMet and AaFISC showed a slightly higher Kd value of 13.8 nM. Therefore, the results confirmed that the purified recombinant AaMet and AaFISC bound to JHRE in a JH-independent manner.
Table 1.
Dissociation constants of AaMet-AaFISC for MFBS1
| Proteins and Hormone | Kd (nM) | p-value (comparing with “AaMet + AaFISC + JH- III”) |
|---|---|---|
| AaMet + AaFISC + Ethanol | 13.8 ± 1.2 | p<0.01 |
| AaMet + AaFISC + JH-III | 5.8 ± 2.0 | |
| AaMetT403Y + AaFISC + JH-III | 6.1 ± 2.0 | p>0.05 |
| AaMetV446F + AaFISC + JH-III | 7.3 ± 3.2 | p>0.05 |
| AaMetT479Y + AaFISC + JH-III | 8.1 ± 3.1 | p>0.05 |
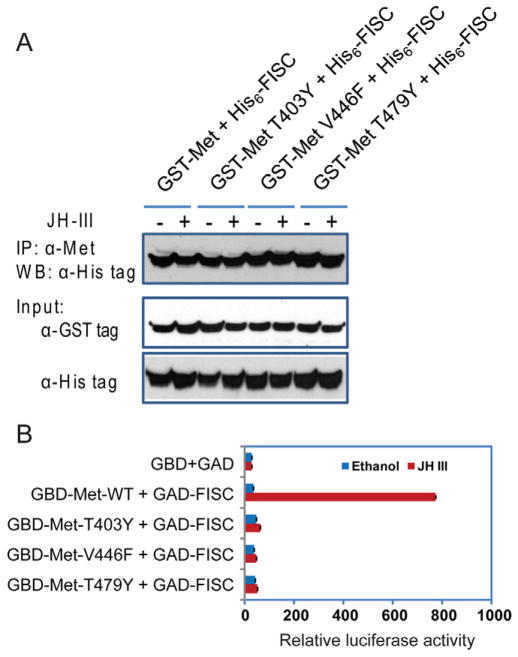
The purified AaMet derivatives were also compared for their protein-protein interaction with AaFISC in an in vitro binding assay. The GST-AaMet fusion protein was readily co-immunoprecipitated with His6-AaFISC regardless of the presence of JH (Fig. 7A). The T403Y, V429F or T479Y mutations showed no negative effect on the AaMet-AaFISC interaction when JH was present (Fig. 7A), indicating that the interaction between the bacterially expressed AaMet and AaFISC does not require the JH binding of AaMet.
Figure 7.
The in vitro AaMet-AaFISC dimerization does not require JH. (A) Co-immunoprecipitation of the purified GST-Met and His6-FISC. The proteins purified from bacteria were incubated in the absence or presence of 10 μM JH-III. The protein complexes were precipitated with the antibody against AaMet. FISC in the pellets was detected in Western blot with antibody against the His-tag. (B) Interaction of AaMet and AaFISC in Aag2 cells. The cultured cells were transfected with the UAS×4-188-cc-Luc reporter plasmid and expression vectors for the indicated GAD and GBD fusion proteins. The transfected cells were treated with 10−6 M JH-III or ethanol.
In parallel, we performed a two-hybrid assay to test the protein interaction in Aag2 cells. The bHLH-PAS domain of AaMet was fused to the GAL4 DNA-binding domain and the bHLH-PAS domain of AaFISC was expressed as a fusion to the activation domain of GAL4. Activation of the UAS-reporter gene by the two fusion proteins was enhanced more than 20-fold when the transfected cells were treated with JH-III. The T403Y, V429F or T479Y mutations each eliminated the JH-induced expression of the reporter gene, confirming that the AaMet-AaFISC interaction in Aag2 cells is JH-dependent (Fig. 7B).
4. Discussion
4.1 Met and FISC are sufficient to bind JHRE
Previous studies have suggested that Met and FISC regulate gene expression via recognition of short DNA sequences (JHRE) in the target genes (Kayukawa et al., 2012; Li et al., 2011). FISC and its Drosophila ortholog TAI have been characterized as steroid receptor coactivators of the p160 family. They function as a coactivator of the ecdysone receptor (EcR), enhancing gene expression induced by ecdysteroids (Bai et al., 2000; Zhu et al., 2006). It is widely speculated that FISC also acts as a coactivator of Met and that Met binds JHRE either alone or with a protein other than FISC.
To test this hypothesis, we first attempted to use cellular protein extracts or the rabbit reticulocyte lysate system for the in vitro DNA-binding assays. High levels of background binding were a huge problem because the AaET JHRE1 contains an E-box-like sequence which is recognized by many bHLH transcription factors. In this study, we used purified Met and FISC proteins in gel-shift assays to test their DNA-binding properties. We demonstrated that Met or FISC alone was unable to bind the probe, but the two proteins together were able and sufficient to bind with specificity to JHRE1. To elucidate the molecular function of FISC in JH signaling, we explored whether the conserved basic helix-loop-helix region in FISC was a functional DNA-binding domain. Mutations in the basic region of FISC led to a dramatic decrease in the binding of Met and FISC to JHRE, consistent with the studies of other bHLH transcription factors showing that the basic regions contact DNA and determine DNA sequence specificity (Shimizu et al., 1997). Taken together, we conclude that FISC functions as an obligatory DNA-binding partner of Met in mediating gene regulation by JH.
Some bHLH-PAS family proteins can function as either transcription factors or transcription cofactors, depending on the circumstances. The aryl hydrocarbon receptor nuclear translocator protein (ARNT) is such an example. ARNT and the aryl hydrocarbon receptor (AHR) are both bHLH-PAS domain proteins. Upon binding of environmental pollutants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin, AHR translocates from the cytoplasm to the nucleus where it binds ARNT and activates expression of proteins involved in xenobiotic metabolism. Both AHR and ARNT bind directly to the xenobiotic response element, T(C/T)GCGTG. AHR binds the 5′-T(C/T)GC half sites with high affinity while ARNT prefers the GTG-3′ half-sites (Swanson et al., 1995). ARNT can also serve as a coactivator of estrogen receptor (ER)-dependent transcription. It has been shown that ARNT is recruited to ER target gene promoters via physical interaction with ER (Brunnberg et al., 2003). Here we show that FISC also functions in similar ways. Aside from it being a transcription coactivator of the ecdysone receptor, FISC is the DNA-binding partner of the JH receptor and directly binds to the JH-inducible promoters. Many studies have shown that JH can exert its functions by modulating the ecdysteroid signaling pathway (Dubrovsky, 2005; Jindra et al., 2013). It will be intriguing to study whether FISC is involved in the crosstalk between the two pathways and whether EcR and Met compete for the binding of FISC when both JH and 20E are present.
4.2 DNA sequence heterogeneity in JHREs
The DNA binding sites of Met and FISC have been characterized so far in only a few JH response genes (Kayukawa et al., 2012; Li et al., 2011). It was not clear whether the E-box-like sequence we had identified represents a typical binding site of Met-FISC. In vitro unbiased selection from random DNA library indicates that E-box-like sequences are indeed the DNA-binding sites of Met and FISC. Although the optimal binding site contains a core sequence of CACGTG, the flanking sequences also contribute significantly to the DNA binding affinity. For example, the AaET JHRE1 and JHRE2 differ in core sequence and flanking sequence (Fig. 5). After making a point mutation at the +2 position, the JHRE1 mutant (AaET_JHRE1_C+2T) and JHRE2 both harbor identical core sequence (CACGTG). However, the dissociation constants of Met-FISC for AaET_JHRE1_C+2T and JHRE2 are 162.0 nM and 13.4 nM, respectively.
The upstream regulatory regions of Kr-h1 from several insect species each contain several copies of the E-box-like sequences. Because of the DNA sequence heterogeneity, individual JHRE may exhibit a distinct binding affinity to the orthologs of Met and FISC. Copy number and spacing of the multiple JHREs may define the expression pattern of the JH target genes, activating different sets of genes precisely at the proper concentration of JH.
4.3 DNA binding of the homodimer and heterodimer of Met
Met can form a homodimer in the absence of JH (Godlewski et al., 2006). Met monomer or homodimer was not able to bind to AaET JHRE1 in our gel-shift assay (Fig. 1). We tried to use the amplification and selection protocol to identify the binding sites of Met. The experiment was performed without adding JH to the incubation. We could not enrich any specific DNA sequence after 10 rounds selection. This result does not support the hypothesis that the Met homodimer can bind to specific target genes.
In newly emerged adult female mosquitoes, RNAi-mediated silencing of Met, FISC or CYC all leads to considerable down-regulation of AaKr-h1 (Li et al., 2011; Shin et al., 2012). So far there is no solid evidence indicating that Met, FISC or CYC actually bind to the AaKr-h1 promoter in vivo and directly regulate the JH-dependent expression of AaKr-h1. Four E-box-like elements were found in the regulatory region of AaKr-h1. K1, K2 and K4 contain the CACGCG motif, while K3 harbors the CACGTG motif. When nuclear extracts from adult female mosquitoes at 2 days post eclosion were incubated with the four DNA elements, only K1 was able to form a stable DNA-protein complex. Met and CYC, but not FISC, were detected in the DNA-protein complex, suggesting that the in vitro binding of the Met-CYC complex to K1 does not require FISC (Shin et al., 2012). Our previous gel-shift experiment showed that JHRE1 from AaET was bound by Met and FISC in the nuclear extracts from female adults at 30 hours post eclosion (Li et al., 2011). Additional in vitro DNA binding experiments need to be conducted carefully under the same conditions using the same mosquito nuclear extracts to explicitly determine whether the Met-FISC and Met-CYC complexes preferentially bind to distinct JHREs. K1 and AaET_JHRE1 share the CACGCG motif. If selective binding of distinct protein complexes to K1 and AaET_JHRE1 is confirmed, this would suggest that sequences flanking the E-Box-like elements influence DNA binding specificity of bHLH-PAS transcription factors. It will be also interesting to examine whether Met, FISC and CYC are all associated in vivo with the AaKr-h1 promoter at the same time and whether Met and CYC are sufficient to bind to JHREs.
Not all JH target genes require the function of FISC for their JH-induced expression in newly emerged mosquitoes (Li et al., 2011). Met may interact with other proteins to mediate JH signaling. A 9-mer Met-binding motif, CACG(C)/TG(A)/G(T)/AG, has been identified from the mosquito genes that are regulated by Met in previtellogenic stage (Zou et al., 2013). It is similar to the GCACGTG motif that we report in this study. In light of the Met-FISC and Met-CYC interactions, identity of the transcriptional factors that recognize this 9-mer Met-binding motif may need to be further investigated.
4.4 Effect of JH binding on the Met-FISC interaction
In the absence of ligands, the aryl hydrocarbon receptor is associated with several chaperone proteins, including the 90-kDa heat shock protein (Hsp90). The chaperones assist AHR to achieve a mature ligand-binding conformation (Coumailleau et al., 1995; Pongratz et al., 1992). The Hsp90 homolog in bacteria is unable to interact with AHR, and the bacterially expressed AHR fails to bind its specific ligand (Coumailleau et al., 1995). In our experiment, the bacterially expressed AaMet displayed a much weaker binding affinity to JH-III, compared with the AaMet produced in rabbit reticulocyte lysates. We are currently examining whether Hsp90 and other chaperones are associated with AaMet in vivo and play a role in the JH binding of AaMet.
In our in vitro assays, the concentration of JH-III was 10 μM, well above the equilibrium dissociation constant between the purified AaMet and JH-III (Kd=160.8 ± 27.6 nM). The protein interaction between AaMet and AaFISC and the binding of the complex to JHRE were all JH-independent. This conclusion was substantiated by further comparing the wild-type AaMet with the JH-binding deficient AaMet mutants in the in vitro assays. In contrast, two-hybrid assay indicated that the AaMet-AaFISC interaction in Ae. aegypti cells was considerably enhanced when JH was present. Similar to our observation with the JH receptor, ligand binding also does not affect the DNA-binding properties of AHR and ARNT that are expressed and purified from E. coli (Kikuchi et al., 2003). In the absence of a ligand, AHR is associated with Hsp90, co-chaperone protein p23, and several other proteins in vivo. These proteins serve to stabilize the ligand-binding conformation of the receptor and inhibit constitutive dimerization with ARNT. After ligand binding, these proteins are released from AHR (Beischlag et al., 2008). Studies have demonstrated that in vitro dissociation of the chaperone proteins from the unliganded AHR results in formation of the AHR-ARNT heterodimer in the absence of ligand (Kazlauskas et al., 1999; Pongratz et al., 1992). The interaction between Met and Hsp90 has been reported in Helicoverpa armigera cells (Liu et al., 2013). It remains to be tested whether Hsp90 has a similar inhibitory function in preventing the constitutive interaction between Met and FISC.
5. Conclusions
We demonstrated using in vitro approaches that a steroid receptor coactivator, FISC, acts as a DNA-bound transcription factor in mediating JH responses. Binding to JHREs requires intact DNA-binding domains of both FISC and the JH receptor Met. These two proteins are sufficient to bind to a consensus motif GCACGTG and activate the transcription of various JH target genes. This study elucidates a key step in JH signaling.
Supplementary Material
Purification of the derivatives of AaMet and AaFISC that carry point mutations in the basic regions. Coomassie staining of the partially purified recombinant Met, FISC, and their mutants. The arrows point to the protein bands with expected sizes.
Binding of [3H]-JH-III to recombinant AaMet. The binding of JH-III to the purified AaMet was analyzed by the DCC assay method. Non-specific binding was determined by including a 100-fold excess of unlabeled JH-III in the binding reactions. Saturation curve and Scatchard plot analysis of specific binding data are shown. Data are the average of three independent experiments. The calculated Kd for the bacterially expressed AaMet is 160.8 ± 27.6 nM. Binding of JH-III to the AaMet protein synthesized in rabbit reticulocyte lysates was measured in the same way and the Kd is 4.4 ± 1.9 nM.
Synthesis of the bHLH-PAS domain (aa. 1-597) of AaMet with mutations in PAS-B. The AaMet products were expressed as His-tag fusions by in vitro coupled transcription and translation reactions. AaMet was quantitatively measured with an antibody against the His-tag in western blot analysis. The arrow points to the band of the expected size.
Expression of AaMet proteins in L57 cells. In the experiment described in Fig. 6C, aliquots of cell lysates were separated by SDS-PAGE, and immunoblot analyses were performed with antibodies for AaMet and AaFISC (Li et al., 2011; Zhu et al., 2006).
The p160 coactivator FISC is a DNA-binding partner of the juvenile hormone receptor Met.
Binding to juvenile hormone response elements requires intact DNA-binding domains of Met and FISC.
Met and FISC are sufficient to bind to a consensus motif GCACGTG.
This study reveals mechanistic details in a key step in signal transduction of juvenile hormone.
Acknowledgments
We thank Dr. Bruce Hammock for 3-octylthio-1,1,1-trifluoro-2-propanone (OTFP), Dr. Richard Helm for his help with mass spectrometry, and Ms. Anne Brown for helping with molecular docking. This work was supported by NIH grant R01 AI099250 to JZ. This work was also supported by a National Science Foundation S-STEM grant (DUE-0850198) to JDW.
Abbreviations
- 20E
20-hydroxyecdysone
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- bHLH-PAS
basic helix-loop-helix Per-ARNT-Sim
- CYC
Cycle
- DCC
Dextran-coated charcoal
- EcR
ecdysone receptor
- ER
estrogen receptor
- FISC
βFTZ-F1-interacting steroid receptor coactivator
- GAD
GAL4 activation domain
- GBD
GAL4 DNA-binding domain
- HIF-2α
hypoxia-inducible factor 2α
- JH
juvenile hormone
- JHRE
juvenile hormone response element
- Kd
dissociation constants
- Kr-h1
Krüppel homolog 1
- Met
Methoprene-tolerant
- MFBS1
Met-FISC binding site 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashok M, Turner C, Wilson TG. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc Natl Acad Sci US A. 1998;95:2761–2766. doi: 10.1073/pnas.95.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Critical reviews in eukaryotic gene expression. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnberg S, Pettersson K, Rydin E, Matthews J, Hanberg A, Pongratz I. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc Natl Acad Sci U S A. 2003;100:6517–6522. doi: 10.1073/pnas.1136688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. Journal of molecular biology. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Carrigan P, Ballar P, Tuzmen S. Site-directed mutagenesis. In: DiStefano JK, editor. Disease Gene Identification. Humana Press; 2011. pp. 107–124. [DOI] [PubMed] [Google Scholar]
- Charles JP, Iwema T, Epa VC, Takaki K, Rynes J, Jindra M. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc Natl Acad Sci U S A. 2011;108:21128–21133. doi: 10.1073/pnas.1116123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumailleau P, Poellinger L, Gustafsson JA, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB. Hormonal cross talk in insect development. Trends in endocrinology and metabolism: TEM. 2005;16:6–11. doi: 10.1016/j.tem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Wang S, Wilson TG. Interaction of bHLH-PAS proteins involved in juvenile hormone reception in Drosophila. Biochem Biophys Res Commun. 2006;342:1305–1311. doi: 10.1016/j.bbrc.2006.02.097. [DOI] [PubMed] [Google Scholar]
- Goodman W, Cusson M. The Juvenile Hormones. In: Gilbert L, editor. Insect Endocrinology. Elsevier; Amsterdam, The Netherlands: 2012. pp. 310–365. [Google Scholar]
- Hu X, Cherbas L, Cherbas P. Transcription activation by the ecdysone receptor (EcR/USP): identification of activation functions. Mol Endocrinol. 2003;17:716–731. doi: 10.1210/me.2002-0287. [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. Journal of computational chemistry. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annual review of entomology. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome biology. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukawa T, Minakuchi C, Namiki T, Togawa T, Yoshiyama M, Kamimura M, Mita K, Imanishi S, Kiuchi M, Ishikawa Y, Shinoda T. Transcriptional regulation of juvenile hormone-mediated induction of Kruppel homolog 1, a repressor of insect metamorphosis. Proc Natl Acad Sci U S A. 2012;109:11729–11734. doi: 10.1073/pnas.1204951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukawa T, Tateishi K, Shinoda T. Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Scientific reports. 2013;3:1570. doi: 10.1038/srep01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J Biol Chem. 1999;274:13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Ohsawa S, Mimura J, Ema M, Takasaki C, Sogawa K, Fujii-Kuriyama Y. Heterodimers of bHLH-PAS protein fragments derived from AhR, AhRR, and Arnt prepared by co-expression in Escherichia coli: characterization of their DNA binding activity and preparation of a DNA complex. Journal of biochemistry. 2003;134:83–90. doi: 10.1093/jb/mvg115. [DOI] [PubMed] [Google Scholar]
- Konopova B, Jindra M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc Natl Acad Sci U S A. 2007;104:10488–10493. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mead EA, Zhu J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc Natl Acad Sci U S A. 2011;108:638–643. doi: 10.1073/pnas.1013914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhang FX, Cai MJ, Zhao WL, Li XR, Wang JX, Zhao XF. The hormone-dependent function of Hsp90 in the crosstalk between 20-hydroxyecdysone and juvenile hormone signaling pathways in insects is determined by differential phosphorylation and protein interactions. Biochimica et biophysica acta. 2013;1830:5184–5192. doi: 10.1016/j.bbagen.2013.06.037. [DOI] [PubMed] [Google Scholar]
- Minakuchi C, Zhou X, Riddiford LM. Kruppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech Dev. 2008;125:91–105. doi: 10.1016/j.mod.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Oda M, Makita S, Chinzei Y. Characterization of the Drosophila Methoprene -tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 2005;272:1169–1178. doi: 10.1111/j.1742-4658.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Insect Hormones. Princeton University Press; 1994. [Google Scholar]
- Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry. 2009;48:5972–5983. doi: 10.1021/bi900259z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Tan A, Palli SR. bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech Dev. 2008;125:601–616. doi: 10.1016/j.mod.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267:13728–13734. [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Research. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailsbery JK, Atchley WR, Dean RA. Phylogenetic analysis and classification of the fungal bHLH domain. Molecular Biology and Evolution. 2012;29:1301–1318. doi: 10.1093/molbev/msr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sanner MF. Python: a programming language for software integration and development. Journal of molecular graphics & modelling. 1999;17:57–61. [PubMed] [Google Scholar]
- Scatchard G. The attractions of proteins for small molecules and ions. Annals of the New York Academy of Sciences. 1949;51:660–672. [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. The EMBO journal. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Zou Z, Saha TT, Raikhel AS. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc Natl Acad Sci U S A. 2012;109:16576–16581. doi: 10.1073/pnas.1214209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Tobe SS, SB Structure and regulation of the corpus allatum. Advances in Insect Physiology. 1985:305–432. [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wilson TG, Fabian J. A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev Biol. 1986;118:190–201. doi: 10.1016/0012-1606(86)90087-4. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nature reviews Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Sheng Z, Sui Y, Palli SR. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J Biol Chem. 2011;286:8437–8447. doi: 10.1074/jbc.M110.191684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Busche JM, Zhang X. Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem Mol Biol. 2010;40:23–29. doi: 10.1016/j.ibmb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen L, Sun G, Raikhel AS. The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Molecular and cellular biology. 2006;26:9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Saha TT, Roy S, Shin SW, Backman TW, Girke T, White KP, Raikhel AS. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci U S A. 2013;110:E2173–2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of the derivatives of AaMet and AaFISC that carry point mutations in the basic regions. Coomassie staining of the partially purified recombinant Met, FISC, and their mutants. The arrows point to the protein bands with expected sizes.
Binding of [3H]-JH-III to recombinant AaMet. The binding of JH-III to the purified AaMet was analyzed by the DCC assay method. Non-specific binding was determined by including a 100-fold excess of unlabeled JH-III in the binding reactions. Saturation curve and Scatchard plot analysis of specific binding data are shown. Data are the average of three independent experiments. The calculated Kd for the bacterially expressed AaMet is 160.8 ± 27.6 nM. Binding of JH-III to the AaMet protein synthesized in rabbit reticulocyte lysates was measured in the same way and the Kd is 4.4 ± 1.9 nM.
Synthesis of the bHLH-PAS domain (aa. 1-597) of AaMet with mutations in PAS-B. The AaMet products were expressed as His-tag fusions by in vitro coupled transcription and translation reactions. AaMet was quantitatively measured with an antibody against the His-tag in western blot analysis. The arrow points to the band of the expected size.
Expression of AaMet proteins in L57 cells. In the experiment described in Fig. 6C, aliquots of cell lysates were separated by SDS-PAGE, and immunoblot analyses were performed with antibodies for AaMet and AaFISC (Li et al., 2011; Zhu et al., 2006).