Abstract
Processive cytoskeletal motors from the myosin, kinesin, and dynein families walk on actin filaments and microtubules to drive cellular transport and organization in eukaryotic cells. These remarkable molecular machines are able to take hundreds of successive steps at speeds of up to several microns per second, allowing them to effectively move vesicles and organelles throughout the cytoplasm. Here, we focus on single-molecule fluorescence techniques and discuss their wide-ranging applications to the field of cytoskeletal motor research. We cover both traditional fluorescence and sub-diffraction imaging of motors, providing examples of how fluorescence data can be used to measure biophysical parameters of motors such as coordination, stepping mechanism, gating, and processivity. We also outline some remaining challenges in the field and suggest future directions.
Keywords: Molecular motors, single-molecule imaging, sub-diffraction localization, kinesin, dynein, myosin, processivity, intracellular transport, cytoskeletal motors, motility, TIRF, total internal reflection fluorescence microscopy
1. Introduction
A eukaryotic cell depends on a multitude of molecular motors, protein machines that convert chemical energy into mechanical work, to actively maintain the spatial organization and material flux required for the cell’s survival. Molecular motors span several protein superfamilies, exhibiting remarkable diversity in structure and function to fulfill their wide variety of biological roles. The motors of the cytoskeleton are divided into three protein superfamilies. Kinesin and dynein motors bind to and translocate along the microtubule network, whereas myosin motors function on actin (Fig. 1A). These motors share several principal characteristics: they all use adenosine triphosphate (ATP) as the source of chemical energy and perform mechanical work by walking along their respective track. The study of motors through biochemical methods is complicated by the fact that many of the fundamental properties of their motility cannot be readily measured in bulk assays. One such property is the motor’s velocity, which determines how rapidly it can deliver cargo to its destination. Another is processivity, a measurement of how many successive steps a motor can take before dissociating from its track, which is critically important for understanding how teams of motors work together to power long-distance transport while avoiding gridlock and overcrowding. For a more detailed understanding of the motor’s mechanism, it is invaluable to know its stepping pattern - the manner in which the heads move with respect to one another as the motor walks down its track. These properties are all readily amenable to study with single-molecule fluorescence techniques.
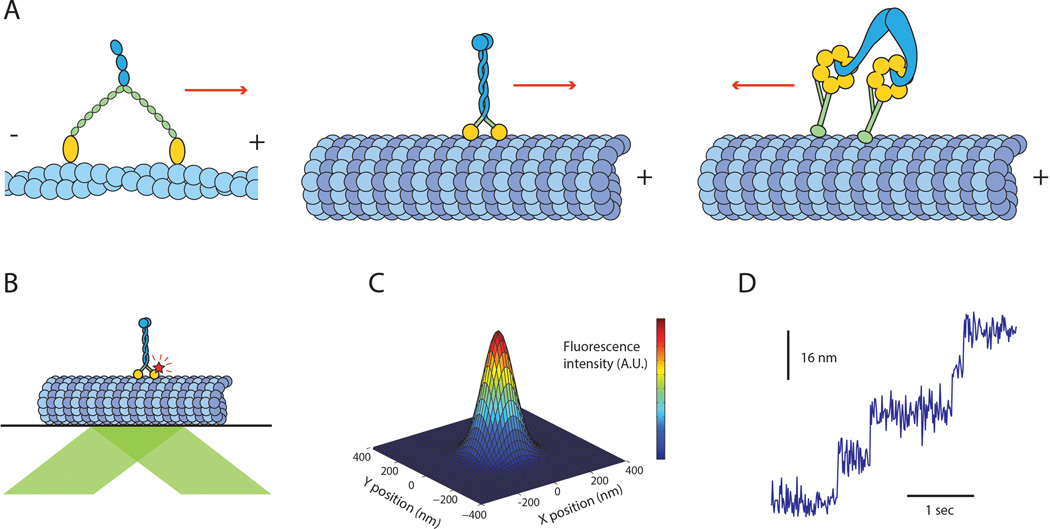
Figure 1.
Processive cytoskeletal motors and fundamentals of sub-diffraction TIRF imaging. (A) Three classes of processive cytoskeletal motors: myosin V (left) walks towards the plus end of actin filaments, kinesin-1 (center) walks towards the plus end of microtubules, and cytoplasmic dynein (right), walks towards the minus end of microtubules. (B) Schematic depiction of a TIRF motility assay (not to scale). A fluorescently labeled motor (kinesin-1 is shown) walks on a surface-immobilized track. The fluorophore is excited by the evanescent field of a collimated laser beam (green) reflecting off the glass/water interface. (C) The point-spread function (PSF) of a single fluorophore is well approximated by a 2-dimensional Gaussian. By collecting a sufficiently large number of photons per frame, the center of the PSF can be localized with nanometer precision. (D) By localizing a fluorophore attached to a walking motor protein over many consecutive frames and plotting the position of its center as a function of time, one can obtain stepping traces similar to the simulated trace shown here. Such traces can then be processed with a step-finding algorithm and used to extract biophysical parameters such as dwell times, stepping rates, and step sizes.
2. Diffraction-limited single motor imaging
Motors that function in muscle contraction (myosin II) and ciliary beating (inner and outer arm dyneins) work in large groups to generate force on macroscopic scales. While these motors can be studied collectively with filament gliding assays1, individual motors need not be processive and their motility may not be immediately apparent on a single-molecule level. However, it was soon discovered that many other cytoskeletal motors transport cargos in small teams or alone2,3, a function requiring the molecules to be able to take many successive steps without diffusing away from the track. In order to achieve processive motion, a molecular motor must remain tethered to the track throughout its entire mechanochemical cycle, a requirement that potentially explains why the vast majority of processive motors discovered to date possess two or more track binding sites.
The first direct confirmation of motor processivity was achieved by imaging individual kinesin molecules walking along microtubules4, and was soon followed by similar observations on myosin3 and dynein5. Single-motor motility assays are performed under total internal reflection fluorescence (TIRF)6 illumination, in which the evanescent field of a laser beam reflected off the water/glass interface excites fluorescently tagged motors moving along surface-immobilized tracks (Figure 1B). The intensity of the evanescent field falls off exponentially with distance from the coverslip, limiting the depth of the excitation region to a few hundred nanometers and greatly reducing background fluorescence from the bulk solution. Observing the motors directly in real time allows for measurement of a number of fundamental properties. Kinesin-1 was shown to travel on average 600 nm before dissociating from the track, demonstrating that a typical run consisted of ~100 mechanical cycles4 assuming the previously measured 8nm step size7. Repeating the experiment with kinesin constructs lacking their dimerization domain showed that kinesin-1 requires both heads to remain processive. It has furthermore been shown that Unc1048 and myosin VI9 motors transition from diffusional to directional processive motion upon dimerization at high concentrations. The requirement for dimerization for processive motility was also demonstrated in yeast cytoplasmic dynein by designing monomers with chemically inducible dimerization domains 5.
3. Regulation of motors
A cell employs regulatory mechanisms to control the attachment of motors to cargos, to modulate their velocity or force production depending on the specific task they’re performing, or to prevent them from undergoing futile cycles of ATP hydrolysis when not engaged with the track10. Such mechanisms can be grouped into two general categories: autoinhibition and inhibition by small molecules or regulatory proteins. Motility experiments on kinesin-1 mutants with the tail domain either truncated or made less flexible at a prominent hinge showed that both mutants moved 2-3 fold faster than wild-type kinesin and exhibited greatly enhanced processivity. This points towards an autoinhibition mechanism wherein kinesin’s tail acts as a repressor of the motor domain in the absence of bound cargo11. Crystallographic work later showed that this inhibition occurs via a tail-mediated crosslinking of the two motor domains, preventing the separation of the two heads required for neck linker undocking12. Similar autoinhibitory mechanisms appear to be present in kinesin-213, kinesin-314, and myosin V15–17 motors.
For cytoplasmic dynein, several distinct regulatory proteins were identified. Lis1 impacts dynein motility on a single-molecule level18, effectively anchoring dynein to its track. Interestingly, this mechanism does not prevent futile cycles of ATP hydrolysis, suggesting that dynein may also have an autoinhibitory mechanism yet to be discovered. Lis1-based anchoring potentially configures dynein for low-speed, high-force cellular tasks such as anchoring spindle microtubules during mitosis. Another dynein regulator, She1 diffuses along microtubules until it encounters a walking dynein. She1 binds and pauses the motor, prolonging its attachment to the microtubule19. A small-molecule inhibitor, monastrol, was used to target homotetrameric kinesin Eg5, which slides apart microtubules and contributes to the assembly of the mitotic spindle20. The effect of monastrol in Eg5 motility was tested using single molecule fluorescence assays. Eg5 is a processive plus end-directed motor that occasionally switches into diffusive mode. The addition of the drug monastrol significantly enriches the diffusive state while effectively abolishing directional motion21. Similarly, ciliobrevin D specifically inhibits dynein motors in vivo22. Targeting specific motors may serve as alternatives to tubulin-targeting antimitotic agents used in cancer therapy, with potentially fewer side effects due to their highly specialized roles in mitosis23.
4. Sub-diffraction imaging
Processive motors were found to generally require two head domains to remain motile, which immediately opened the question of how they coordinate their motions to generate successive steps without simultaneously releasing from the track. One may picture the motor walking much like a human would, taking regular alternating steps of equal sizes with its two “feet”. This mechanism, which is referred to as “hand over hand” (HoH), requires a large degree of coordination between the two heads, as each head takes a step in the trailing position and remains firmly attached in the lead24. Another proposed possibility is that the “inchworm” model, wherein the full cycle consists of two nearly simultaneous steps by the heads and results in a translation of the motor without changing the relative orientation of the two heads25. Such a mechanism requires more strict coordination than HoH due to the added timing constraint between the steps of the trailing and leading head. The stochastic stepping model abolishes coordination altogether and allows the heads to move forward independently of their partner. In this case, spontaneous release from the track is prevented by the low probability of simultaneously finding both heads in the unbound state.
The spatiotemporal resolution required to distinguish between these possibilities and investigate the kinetics of stepping resulted in early adoption of sub-diffraction imaging techniques. The diffraction-limited image of a single fluorescent molecule (termed point spread function, or PSF) has a width of approximately λ/(2 N.A.), where λ is the wavelength of light and N.A. is the numerical aperture of the objective lens. Using the highest N.A. (1.49 to 1.65) objectives available, the image of a point-like object emitting visible photons has a width of ~250 nm. This width is an order of magnitude larger than the step size of the motors (8-37 nm). Resolving the stepping pattern requires a significant improvement in resolution, which prompted the use of sub-diffraction fluorophore localization. While the width of a single fluorophore’s image cannot be readily decreased beyond the fundamental limit, its peak position can be determined with high precision by collecting a sufficient number of photons (on the order of 20,000 photons for 1 nm localization accuracy)26 (Figure 1C). With the important limitation that individual molecules must be well-separated on the camera’s detector, a 2-dimensional Gaussian fit can localize their positions to ~1 nm at sub-second frame rates using organic dyes27,28 (Figure 1D).
Because the precision with which a fluorophore can be localized within a frame generally scales as the square root of the number of collected photons27, the photostability of the probe is of paramount importance for the acquisition of high-resolution videos of walking molecules. The most critical parameter is the average total number of photons emitted by the fluorophore before it undergoes photobleaching. Photobleaching generally occurs when a fluorophore in an excited state chemically reacts with a singlet oxygen. This effect can be greatly reduced by removing oxygen from the system, using oxygen scavenging enzymes. Since oxygen enhances fluorescence by acting as an effective triplet state quencher, imaging buffers typically include a separate triplet state quencher to compensate for the absence of oxygen and prevent dye blinking29. As a result, efforts to increase fluorophore lifetimes have centered on triplet state quenchers and enzymatic systems for removal of free oxygen. Recent work showed that while thiol-containing compounds such as β-mercaptoethanol and L-glutathione are efficient triplet state quenchers, they can cause slow blinking of cyanine dyes, a problem that can be overcome by using Trolox as a quencher instead30. To remove free oxygen, one commonly used system is the glucose oxidase/catalase enzyme pair31. More recently, an enzymatic system based on protocatechuate dioxygenase gained popularity because the byproduct of its reaction does not alter the pH of the buffer32. However, despite these improvements, photostability varies greatly between fluorophores33 and the majority of fluorophores are not sufficiently stable for high-resolution tracking. Multi-frame localization with nanometer-scale precision has only been achieved to date using the best small organic probes (such as Cy3, Cy5, TMR and Atto647N)28,34,35 and quantum dots5,36,37.
5. Motor stepping studied with sub-diffraction imaging
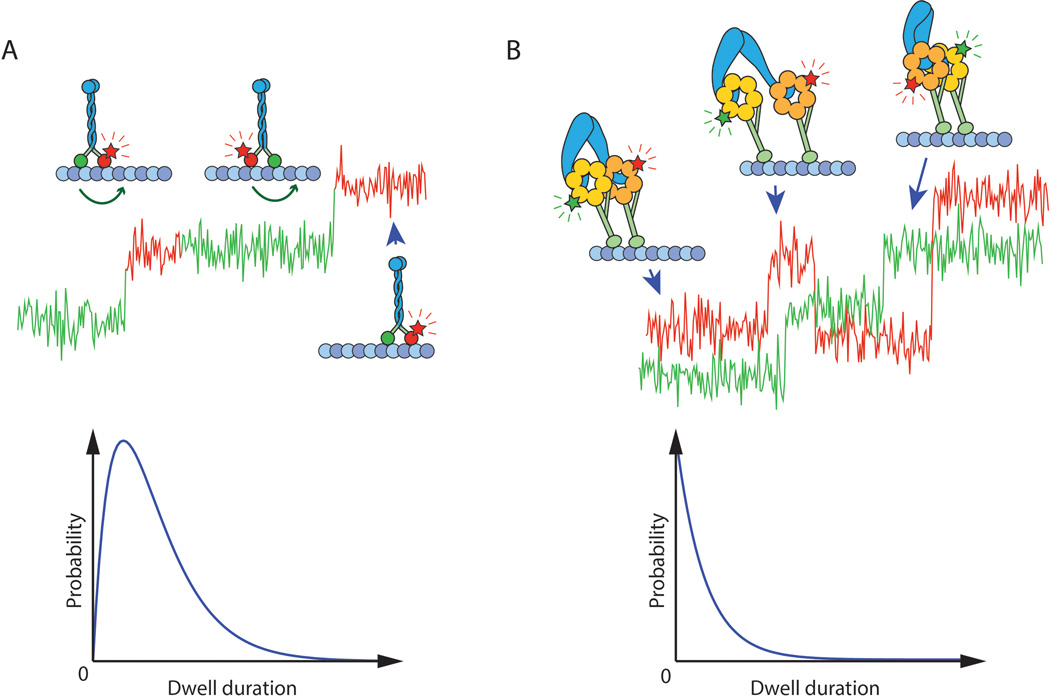
Single-molecule tracking methods provided sufficient spatiotemporal accuracy to detect individual steps taken by cytoskeletal motors. Using optical traps, it had previously been shown that myosin V’s cargo binding domain moves on average in 37 nm steps38,39, equivalent to the half helical pitch of the actin. To determine whether these steps resulted from an inchworm or HoH mechanism, myosin V motors were fluorescently labeled on one head and tracked with 1 nm precision as they walked on surface-immobilized actin filaments28. Had myosin been an inchworm motor, one would expect to see the heads step in 37 nm increments just like the cargo-binding domain. However, in practice each molecule exhibited a distinctly bimodal step size distribution, moving in 74nm steps alternating with invisible “0 nm” steps28. The presence of the “0 nm” steps is revealed by the characteristic shape of the dwell time histogram, which changes from a single exponential decay into a convolution of two exponentials as the step of a single head consists of two consecutive events (Figure 2A). Later, kinesin-1 and myosin VI were demonstrated to be HoH steppers in a similar manner35,40,41, while more recent work suggested that the two modes of stepping are not necessarily mutually exclusive as myosin VI motors are able to switch between HoH and inchworm-like steps36.
Figure 2.
Extracting information from high-resolution stepping traces. (A) If a single head of a dimeric motor protein is labeled with a fluorophore, the degree of coordination between the two heads can be measured by analyzing the dwell time distribution. If the heads take strictly alternating steps, as is the case in kinesin-1 (shown here), every other step will be taken by an unlabeled head and thus invisible to the observer (the simulated stepping trace is colored according to which head is in the leading position). The dwell time distribution of such a stepper will follow the functional form of a convolution of two identical exponentials. (B) Cytoplasmic dynein is distinct from kinesin-1 and myosin V in that its two heads are nearly uncoordinated. If the heads are labeled with fluorophores of different color and imaged simultaneously, one will expect to see traces similar to the simulated data shown here. The heads do not strictly alternate, take occasional backward steps, and swap leading and trailing positions at random times. At limiting ATP concentrations, the dwell time distribution for a single head of such an uncoordinated walker will follow a single exponential decay because the head does not need to wait for its partner to take a step before it can step again.
The finding that a motor such as kinesin walks in strictly alternating steps necessitates some form of “gating” mechanism between the two heads that would prevent the leading head to release from the track until the trailing head completes its step. Several theories have been proposed for motor protein gating. The tubulin binding gate model postulates that the stepping head cannot bind to the next site on the track until its partner head binds a new ATP molecule42. Tension gating proposes that mechanical strain generated between the two heads plays a role in keeping the two heads out of phase. In this model, rear-head gated scenario suggests that the trailing head is mechanically pulled off the track by strain produced in the front head. The front head-gated scenario suggests that ATP binding to the front head is suppressed by rearward strain from the trailing head43. The importance of mechanical strain between the two heads was demonstrated by inserting flexible linkers of varying length between the heads and the dimerization domain. When examined in single-molecule fluorescence, these extended constructs exhibited reduced velocity and a highly variable stepping pattern, suggesting that the loss of intramolecular strain led to a substantial decrease in gating efficiency44. A later optical trapping study established that kinesin spends the majority of its time in a one head-bound state and the intramolecular strain makes the off-pathway two-head bound states more unfavorable, rather than triggering release in a two head-bound state45.
The stepping pattern of cytoplasmic dynein labeled with a single fluorophore was somewhat inconclusive about the way dynein walks along microtubules. Unlike kinesin, which takes regular 8 nm steps in a HoH fashion, dynein displays a large variability in step size, as well as frequent movement in sideways and backward directions5. To visualize how dynein’s heads move with respect to one another, two groups simultaneously pursued the goal of labeling dynein’s two heads with fluorophores of different color and obtaining simultaneous stepping traces for them. An important challenge in dual-color super-resolution tracking is overlaying the images from both channels with sub-pixel accuracy. This can be achieved, for example, by scanning a fiducial marker (such as a broad-spectrum fluorescent bead) in small increments across the field of view, localizing it with a 2D Gaussian fit in each channel, and using the resulting position pairs to generate a map between the two channels46. Provided that the fiducial markers fluoresce at the same wavelengths as the actual fluorophores used in the experiment, mapping can correct for optical aberrations, CCD pixel-to-pixel variation, and physical differences between the two imaging paths, resulting in an overall mapping accuracy of up to 1 nm46–48. In the two-color dynein experiments, while Qiu et al. used organic dyes and DeWitt et al. opted for the larger but more photostable quantum dots, both reached the same conclusion that cytoplasmic dynein displays a large variability in stepping pattern compared to myosin and kinesin34,47. Both the size and timing of a dynein head’s step are only weakly affected by the position of the partner head (Figure 2B). Dynein’s case clearly illustrates that gating is not necessary for processive motility and that long-range movement can be achieved by two mechanically linked heads, provided each spends the majority of its time bound to the track and only releases briefly to take the next step.
6. Imaging teams of motors
Intracellular cargoes such as lipid droplets, organelles, and endoplasmic reticulum vesicles are commonly transported by small teams of motors rather than individual proteins. Time-lapse observation of mitochondrial transport in axons and dendrites of cultured neurons exhibited diverse motility, ranging from slow unidirectional movement to rapid switching between bursts of fast retrograde and anterograde runs49. Similar bidirectional behavior was observed using a variety of techniques for phagosomes50, endosomes51, lipid droplets52, neurofilaments53, intraflagellar transport (IFT) trains54, and other cargos. The cell may control the overall distribution of cargos in the cytoplasm by affecting either motor recruitment to cargos or the engagement of cargo-bound motors with the track55. Two commonly proposed and not mutually exclusive models for how bidirectional transport may be achieved and regulated are stochastic tug-of-war between competing teams of opposite-polarity motors bound to the same cargo56 and regulated directional switching52. Such switching can be carried out through different pathways, such as inactivation/unbinding of motors of one polarity or specific inhibition of a particular class of motors.
An important challenge for understanding the mechanism of cargo transport is the difficulty of measuring the number of motors of each polarity that are attached to the cargo and engaged with the track at any given point in time. The total number of motors attached to a cargo can be measured by several methods, such as fluorescence bleaching counting assays or quantitative blotting56. However, not all of the cargo-bound motors may be active at a time. The number of actively engaged motors pulling the cargo was estimated to be relatively low (1–5 motors of each polarity) by in vivo and in vitro optical trapping assays, with an important underlying assumption that motor stall forces are additive at low copy numbers51,56–59. High-resolution imaging of reconstituted neuronal transport vesicles revealed that even at such small motor numbers vesicles moved bidirectionally and exhibited rapid direction switching similarly to their in vivo counterparts, just as predicted by the mechanical tug-of-war model56. On the other hand, IFT trains in Chlamydomonas reinhardtii were found to move in a clearly coordinated manner, with motors of only one polarity active at a time60, illustrating that regulation of transport in vivo is in no way limited to tug-of-war. An artificial DNA origami scaffold helps overcome the limitation of the motor number per cargo variability, by assembling well-defined groups of motors in vitro61. The presence of mechanical tug-of-war between multiple dyneins and kinesins were demonstrated by changing the relative numbers of the opposing motors on a scaffold. Cargoes with 2.5 times more kinesins than dyneins still moved in the retrograde direction despite dynein’s lower stall force, suggesting that parameters other than stall force (such as tenacity of microtubule attachment) may be more relevant for a motor’s tug-of-war performance.
7. Conclusion
The relatively non-invasive nature of fluorescence imaging, together with the high resolution tracking ability, enables direct observation of actively translocating motors under physiological conditions. Trajectories of single motors are used to measure parameters such as processivity, velocity, stepping pattern, interhead coordination, and regulation, which are critical for understanding how motors work alone or in teams. Even though much has been learned about how cytoskeletal motors operate, many more questions remain unanswered. Only a handful of motors have been studied in detail, and the evolutionary diversity of the myosin, kinesin, and dynein families suggests that novel properties and peculiarities will be revealed as new family members are isolated and subjected to scrutiny. Technical advances in the field, perhaps smaller and more photostable fluorescent probes or improved image analysis algorithms, will enable more detailed mechanistic studies and help resolve small-scale motions that lie below the current detection limit. As the individual stepping mechanisms of isolated motors become increasingly well understood, the field’s focus will likely continue to shift towards interactions between motors and proteins that modulate their behavior, such as other motors or dedicated regulatory proteins. The ultimate goal of this field, a comprehensive understanding of how powered intracellular transport is organized and regulated, will require a large concerted effort spanning several length scales in both living cells and artificial reconstituted systems.
Acknowledgements
We are grateful to F. Cleary for critical reading of this manuscript. This work was supported by NIH (GM094522 (AY)), NSF CAREER Award (MCB-1055017 (AY)) and NSF Graduate Research Fellowship (DGE 1106400 (VB)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rock RS, Rief M, Mehta aD, Spudich Ja. In vitro assays of processive myosin motors. Methods. 2000;22:373–381. doi: 10.1006/meth.2000.1089. [DOI] [PubMed] [Google Scholar]
- 2.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 4.Vale Ronald D, Funatsu Takashi, Pierce Daniel W, Romberg Laura, Harada Yoshie, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelrod D, Thompson NL, Burghardt TP. Total internal reflection fluorescent microscopy. J. Microsc. 1983;129:19–28. doi: 10.1111/j.1365-2818.1983.tb04158.x. [DOI] [PubMed] [Google Scholar]
- 7.Svoboda Karel, Schmidt Christoph F, Schnapp Bruce J, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 8.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 9.Park H, et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Barlan K, Rossow MJ, Gelfand VI. The journey of the organelle: teamwork and regulation in intracellular transport. Curr. Opin. Cell Biol. 2013;25:483–488. doi: 10.1016/j.ceb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 12.Kaan HYK, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond JW, et al. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, et al. Regulated conformation of myosin V. J. Biol. Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem. Biophys. Res. Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 Acts as a “Clutch” between the ATPase and Microtubule-Binding Domains of the Dynein Motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markus SM, Kalutkiewicz Ka, Lee W-L. She1-Mediated Inhibition of Dynein Motility along Astral Microtubules Promotes Polarized Spindle Movements. Curr. Biol. 2012:1–10. doi: 10.1016/j.cub.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapitein LC, Peterman EJG, Kwok BH. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 21.Kwok BH, et al. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 22.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergnes G, Brejc K, Belmont L. Mitotic Kinesins: Prospects for Antimitotic Drug Discovery. Curr. Top. Med. Chem. 2005;5:127–145. doi: 10.2174/1568026053507697. [DOI] [PubMed] [Google Scholar]
- 24.Kuo SC, Gelles J, Steuer E, Sheetz MP. A model for kinesin movement from nanometer-level movements of kinesin and cytoplasmic dynein and force measurements. J. Cell Sci. Suppl. 1991;14:135–138. doi: 10.1242/jcs.1991.supplement_14.27. [DOI] [PubMed] [Google Scholar]
- 25.Hua W, Chung J, Gelles J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 26.Bobroff N. Position measurement with a resolution and noise-limited instrument. Rev. Sci. Instrum. 1986;57:1152. [Google Scholar]
- 27.Thompson R. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yildiz A, et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 29.Levitus M, Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011;44:123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 30.Rasnik I, Mckinney SA, Ha T. Nonblinking and long- lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 31.Englander SW, Calhoun DB, E J. Biochemistry without oxygen. Anal Biochem. 1987;161:300–306. doi: 10.1016/0003-2697(87)90454-4. [DOI] [PubMed] [Google Scholar]
- 32.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 34.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 2012 doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa S, et al. Switch between Large Hand-Over-Hand and Small Inchworm-like Steps in Myosin VI. Cell. 2010;142:879–888. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 37.Warshaw DM, et al. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys. J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock RS, et al. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali MY, et al. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 2002;9:464–467. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 40.Yildiz A, et al. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 41.Okten Z, Churchman LS, Rock RS, Spudich Ja. Myosin VI walks hand-over-hand along actin. Nat. Struct. Mol. Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 42.Alonso MC, et al. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeWitt Ma, Chang AY, Combs Pa, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertsinidis A, Zhang Y, Chu S. Subnanometre single-molecule localization, registration and distance measurements. Nature. 2010;466:647–651. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- 49.Ligon La, Steward O. Movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J. Comp. Neurol. 2000;427:340–350. doi: 10.1002/1096-9861(20001120)427:3<340::aid-cne2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 50.Blocker A, et al. Molecular requirements for bi-directional movement of phagosomes along microtubules. J. Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. USA. 2009;106:19381–19386. doi: 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunwar A, et al. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. PNAS. 2011;108:18960–18965. doi: 10.1073/pnas.1107841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchida A, Alami NH, Brown A. Tight Functional Coupling of Kinesin-1A and Dynein Motors in the Bidirectional Transport of Neurofilaments. Mol. Biol. Cell. 2009;20:4997–5006. doi: 10.1091/mbc.E09-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engel BD, et al. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 2009;93:157–177. doi: 10.1016/S0091-679X(08)93009-0. (Elsevier, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross SP, Vershinin M, Shubeita GT. Cargo transport: two motors are sometimes better than one. Curr. Biol. 2007;17:R478–R486. doi: 10.1016/j.cub.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 56.Hendricks AG, et al. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shubeita GT, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gross SP. Hither and yon: a review of bi-directional microtubule-based transport. Phys. Biol. 2004;1:R1–R11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- 59.Sims Pa, Xie XS. Probing dynein and kinesin stepping with mechanical manipulation in a living cell. Chemphyschem. 2009;10:1511–1516. doi: 10.1002/cphc.200900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih SM, et al. Intraflagellar transport drives flagellar surface motility. Elife. 2013;2:e00744. doi: 10.7554/eLife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derr ND, et al. Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold. Science. 2012:80. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]