Abstract
Hypoxia is a common characteristic of many solid tumors. The hypoxic microenvironment stabilizes hypoxia-inducible transcription factor 1α (HIF1A) and 2α (HIF2α/EPAS1) to activate gene transcription, which promotes tumor cell survival. The majority of human genes are alternatively spliced, producing RNA isoforms that code for functionally distinct proteins. Thus, an effective hypoxia response requires increased HIF target gene expression as well as proper RNA splicing of these HIF-dependent transcripts. However, it is unclear if and how hypoxia regulates RNA splicing of HIF targets. This study determined the effects of hypoxia on alternative splicing (AS) of HIF and non-HIF target genes in hepatocellular carcinoma (HCC) cells and characterized the role of HIF in regulating AS of HIF induced genes. The results indicate that hypoxia generally promotes exon inclusion for hypoxia-induced, but reduces exon inclusion for hypoxia reduced genes. Mechanistically, HIF activity, but not hypoxia per se is found to be necessary and sufficient to increase exon inclusion of several HIF targets including pyruvate dehydrogenase kinase 1 (PDK1). PDK1 splicing reporters confirm that transcriptional activation by HIF is sufficient to increase exon inclusion of PDK1 splicing reporter. In contrast, transcriptional activation of a PDK1 minigene by other transcription factors in the absence of endogenous HIF target gene activation fails to alter PDK1 RNA splicing.
Keywords: hypoxia, HIF, alternative splicing, isoform, RNA splicing
Introduction
Hypoxia is a common characteristic of many solid tumors. The hypoxic microenvironment stabilizes hypoxia-inducible transcription factor 1α (HIF1α) and 2α (HIF2α) that are normally degraded under normoxia. The stabilized HIF1α and HIF2α proteins translocate to the nucleus, where they dimerize with aryl hydrocarbon receptor nuclear translocator (ARNT) to form HIF1α/ARNT (HIF1) and HIF2 α/ARNT (HIF2) complexes. HIF1 and HIF2 activate gene transcription to promote tumor progression and metastasis (1-4). Thus, HIF-mediated transcriptional response is very important in cancer biology.
Recently, it was found that 95% of human genes are alternatively spliced and RNA isoforms produced from a single gene code proteins with distinct or opposing functions (5). Thus, it becomes increasing clear that assessing isoforms expression will be more informative than quantifying total transcripts to appreciate the functional consequence of gene regulation.
Exon arrays contain probe sets interrogating every exon of the transcriptome, thereby permitting the distinguishing of different isoforms of a gene as well as measuring gene expression levels. So far, two studies have examined the effect of hypoxia in regulating RNA splicing using exon arrays (6, 7). However, both studies used endothelia cells, a normal cell type. So far, hypoxia-mediated RNA splicing changes have not been measured in cancer cells. Additionally, these previous studies in endothelial cells focused on non-HIF target genes and reported that hypoxia, like other stresses, inhibits RNA splicing of non-HIF target genes by promoting exon skipping and/or intron-inclusion (6, 7). Thus, role of hypoxia in regulating RNA splicing of HIF-induced genes, the most important genes in hypoxia response is still unknown. Furthermore, none of these studies have addressed the molecular mechanisms concerning how hypoxia regulates AS of HIF target genes. In this study, we used exon arrays to assess global effects of hypoxia on RNA splicing of HIF and non-HIF target genes in Hep3B cells, a cancer cell line. Additionally, we studied if HIF or hypoxia per se is important for RNA splicing of several HIF target genes. These findings have important implications for our understanding of how HIFs promote tumorigenesis in response to hypoxia.
Materials and Methods
Cell culture
Hep3B cells were cultured in MEM/EBSS (Hyclone) containing 10% FBS, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 100,000 units/L Penicillin/Streptomycin, 1.5 g/L sodium bicarbonate, and 1X non-essential amino acids (NEAA). HEK293T, RCC4, RCC4T, and UM-SCC-22B cells were grown in high-glucose Dulbecco Modified Eagle Medium (DMEM: Hyclone) with 10% FBS, 2 mmol/L L-glutamine, 100,000 units/L Penicillin/Streptomycin, and 1X NEAA. SK-N-MC cells were grown in RPMI (1640: Hyclone) with 10% FBS, 2 mmol/L L-glutamine, 100,000 units/L Penicillin/Streptomycin, and 1X NEAA. Prior to hypoxia treatment, 25 mM HEPES was added to growth media and cells were incubated under normoxia (21% O2) or hypoxia (1.5% O2) for 12-16 hrs. All parental cell lines were purchased from ATCC. After completing the experiments, the parental (Hep3B, HEK293T, RCC4, UM-SCC-22B, and SKNMC) and modified cell lines (RCC4T) were authenticated by DNA profiling or “fingerprinting” by the University of Colorado DNA Sequencing & Analysis Core.
Exon array analysis of alternative splicing
The genome-wide effect of hypoxia on alternative splicing was determined in Hep3B cells using the Affymetrix GeneChip Human Exon 1.0 ST array. Hep3B cells were cultured, as described above, under normoxic (21%) or hypoxic (1.2%) conditions for 12 hours. RNA was prepared using a Qiagen RNeasy Plus RNA extraction kit. RNA was reverse transcribed and assessed for hypoxic induction of HIF target genes using qRT-PCR. RNAs that passed quality control were submitted to the Gene Expression Core at the University of Colorado Comprehensive Cancer Center for exon array analysis. Three independent replicate samples were analyzed for each treatment. The Core performed target labeling, hybridization, and chip scanning. To assess alternative splicing events, raw CEL files were analyzed using freely available Altanalyze software(http://www.altanalyze.org) (8) with the following settings: Species – Human; Gene and Exon set – both core; gene and exon level normalization FIRMA. The array data can be accessed at GEO (GSE57613). Raw CEL files were alternatively analyzed using easyExon (http://microarray.ym.edu.tw:8080/tools/module/easyexon/index.jsp?mode =home), with default settings and using RMA filtering (9). Additionally, Altanalyze software was used for gene expression analysis. Genes predicted to undergo alternative splicing by Altanalyze and easyExon were then compared to genes that were induced by hypoxia (Altanalyze) using Venny (http://bioinfogp.cnb.csic.es/tools/venny/) to indentify genes that were predicted to undergo alternative splicing and hypoxia induction.
Functional clustering of hypoxia inducible genes and alternatively spliced genes using DAVID Bioinformatics Resources
DAVID Bioinformatics (10) was used to create functionally annotated clusters of 1) genes that were hypoxia induced, 2) genes that were predicted to undergo hypoxia-induced AS, and 3) genes that were hypoxia-induced and exhibiting alternative splicing.
Knockdown of endogenous mRNA using small interfering RNAs (siRNAs)
Control (Qiagen,1027281) or siRNAs specific for human ARNT (Qiagen, equal mix of SI00304220, SI00304234, and SI03020913), HIF1α (Qiagen, SI02664053), HIF2α (Qiagen, SI00380212 ) mRNAs were transfected into Hep3B cells at 40% confluency using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer's protocol. 32hr post-transfection, cells were cultured at 21% or 1.5% O2 for 12-16 hrs and then collected for mRNA or protein purification.
Viral transduction of Hep3B cells
The GFP-Flag, HIF1αTM-Flag (triple mutation to make HIFα protein active under normoxia, with Flag tag), and HIF2αTM-Flag lentiviral constructs were described (11). The GFP-Flag, HIF1αTM-Flag, or HIF2αTM-Flag plasmid was co-transfected with psPAX2 packaging vector (Addgene), and pMD2.G envelope vector (Addgene) into HEK293T cells. 24 hours after transfection, transfection media was replaced with complete Hep3B media. The following day, viral media was filtered to remove possible 293T cells and viral particles were concentrated using high-speed ultracentrifugation at 26,000 X G for 3 hrs. The viral pellet was re-suspended in 1mL of PBS and added to 6 cm Hep3B cells (30% confluency) for 6 hours. The following day, a second round of concentrated virus was added to the same Hep3B cells. 24 hours after the second viral transduction, Hep3B cells were harvested for RNA and protein analysis.
Plasmid constructs
The CA9P/ADM, PAI1P/ADM, and PAI1PmHRE/ADM constructs were described previously (11). The CA9P/PDK1, PAI1P/PDK1, and PAI1PmHRE/PDK1 constructs were generated by replacing the ADM gene with PDK1 minigene that was PCR-amplified from human genomic DNA and contained exon 3 to exon 5 including introns 3 and 4. The HIF1αTM-Flag, HIF2αTM-Flag, HIF1αDBD-Flag, HIF1αDBD/VP16-Flag, HIF1αDBD/E2F-Flag, and USF2 expression plasmids were described previously (11-13). The G5/PDK1 minigene was generated by replacing the ADM gene with PDK1 minigene in the G5/ADM construct that was described previously (11). The Gal4 DNA binding domain (Gal4), Gal4/HIF1αTAD, Gal4/VP16, and Gal4/E2F1 have been described (11).
PDK1 splicing reporter assay
Typically, HEK293T cells were grown to ~30% confluency in 6-well plates and co-transfected with 200 ng of splicing reporters (CA9P/PDK1, PAI1P/PDK1, PAI1PmHRE/PDK1, or G5/PDK1) and 1.8 µg of His (empty vector control), HIF1αTM, HIF2αTM, USF2, HIF1αDBD, HIF1αDBD/E2F1, or HIF1DBD/VP16 expression constructs or Gal4 expression construct for G5/PDK1. 48 hours after transfection, cells were collected for RNA or protein preparation. Results were the average of at least three experiments.
Protein Analysis
Whole cell lysates were prepared and quantified for protein concentration. Western blot analysis was performed using standard protocols with the following primary antibodies: anti-CA9 (H-120) pAb (SC-25599, Santa Cruz), anti-ANGPTL4 (H-200) pAb (SC-66806, Santa Cruz), anti-Flag mAb (F3165, Sigma), anti-Gal4DBD pAb (SC-577, Santa Cruz), or anti-beta Actin pAb (SC-1616, Santa Cruz).
RNA preparation and reverse transcription PCR or quantitative-PCR (qRT-PCR)
RNA was isolated from cells using the RNeasy Plus mini kit (Qiagen) which removes DNA, then was reverse transcribed using the iSCRIPT Advanced cDNA synthesis kit (Bio-Rad) containing both oligo-dT and random hexamer as primers because using both random hexamers and oligo-dT allows efficiently reverse transcription of the whole mRNA transcript as well as 18 rRNA. Alternative splicing analysis was then validated by RT-PCR using forward and reverse primers flanking alternative splicing events predicted by both Altanalyze and easyExon software. Genes that expressed multiple transcripts were Topo TA cloned and submitted to the Gene Expression Core at the University of Colorado Cancer Center for sequencing. qRT-PCR using iQ Sybr Green supermix (Bio-Rad) in triplicate on the CFX384 Real-Time System (BioRad) were used to quantify the levels of RNA isoforms. All primer sets for qRT-PCR designed to measure mRNA levels were validated for their specificity and amplification efficiency (85%-110%) using melt curve analysis, qRT-PCR product sequencing, and standard dilution analysis. qRT-PCR results were normalized using the PfafflΔΔCT method using18s rRNA and β-Actin as reference genes because their expression are not affected by hypoxia and untreated normoxia samples, GFP lentivirus, or empty vectors (His) as controls. The RNA ratio of splice variants was calculated by dividing the expression level of the FL isoform by the expression level of the ΔE isoform. The expression level for each isoform was determined by the following equation: Expression level = EfficiencytargetdeltaCt target (control-sample)/ EfficiencyreferencedeltaCt reference (control-sample), where the Efficiency = 10−1/slope (slope generated by standard curve analysis). At least three independent experiments were performed to generate the results presented in the Figures. All the primers used for RT-PCR and qRT-PCR were included in Supplemental File 3.
Statistics
One-way analysis of variance was performed unless otherwise stated. Error bars in figures indicate standard deviation. Asterisks indicate statistical significance as follows: *, P< 0.05; **, P< 0.01. Controls for statistical analysis are specified in each figure. All experiments were performed at least three separate times.
Results
Genome-wide exon-array analysis determines that hypoxia alters RNA splicing of HIF and non-HIF target genes in Hep3B cells
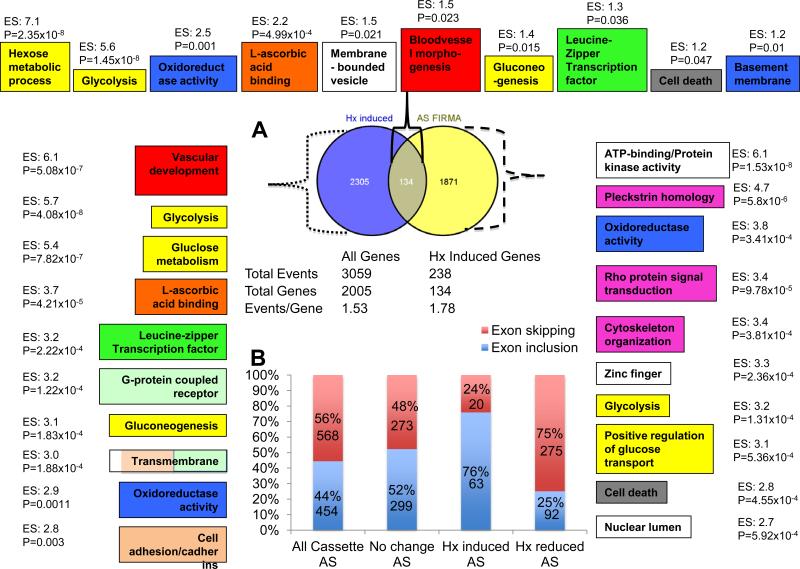
To assess the effects of hypoxia on RNA splicing, Affymetrix GeneChip Human Exon 1.0 ST arrays were used to probe RNAs isolated from normoxic or hypoxic Hep3B cells. Hep3B cells were chosen because of their high expression levels of HIFα proteins and high hypoxic induction of HIF target genes (12-14). Altanalyze (FIRMA) and EasyExon software predicted that hypoxia regulated alternative splicing (AS) of about 2005 (Fig. 1A, yellow and grey) and 3919 genes respectively (Supplemental File 1, AS by Firma or AS by EasyExon). Venn diagrams using Venny indicated that 1012 genes were predicated to undergo AS by both programs while an additional 957 and 2634 genes were predicated to undergo AS by FIRMA and EasyExon respectively. FIRMA was used to further characterize hypoxia mediated splicing events since FIRMA predictions of AS is more stringent than EasyExon. FIRMA analysis indicated that hypoxia produced 3059 AS events in 2005 genes, averaging 1.53 splicing events per gene (Fig. 1A, All Genes). As expected, alternative cassette-exons were the most abundant type of splicing events, making up 51% of all splicing events. Other AS events included alternative 5’ splice-sites (16%), alternative 3’ splice-sites (14%), intron retention (11%), mutually exclusive exon (4%), and others (4%).
Figure 1. Hypoxia regulates gene expression and RNA splicing in Hep3B cells.
A) Venn diagrams of genes that are hypoxia induced (blue and grey), genes that are alternatively spliced under hypoxia (yellow and grey), and genes whose expression are induced by hypoxia and alternatively spliced under hypoxia (grey). The left column indicates the top 10 enrichment pathways whose genes are induced at least 1.4 fold by hypoxia. The top row indicates the top 10 enrichment pathways whose genes that are hypoxia-induced and undergo hypoxia-induced alternative splicing. The right column represents the top 10 enrichment pathways whose genes that undergo hypoxia-induced alternative splicing. Numbers next to each box represent the DAVID enrichment score (ES), the higher the more enriched, followed by the Fisher Exact P-Value (P), the smaller the more enriched. P-Values equal to or less than 0.05 are considered strongly enriched. Boxes of the same color, excluding white, represent overlapping or functionally related gene pathways. White boxes are unique pathways for each group. Additional statistics and gene list associated with each group can be found in Supplemental File 2. Below the Venn diagram in Fig. 1A are the splicing events per genes for all alternatively spliced genes (All Genes, left) and hypoxia induced genes (Hx Induced Genes, right). B) Summary of exon skipping or inclusion events in all cassette alternative splicing genes (all cassette AS), genes whose expression are not changed during hypoxia (No Change AS), genes induced by hypoxia (Hx induced AS), and genes reduced by hypoxia (Hx reduced AS).
Additionally, Altanalyze determined that hypoxia induced 2439 genes, for at least 1.4 fold, when compared to normoxic Hep3B controls (Fig. 1A, blue and grey; also see Supplemental File 1, Gene Expression). Venny analysis of hypoxia inducible genes and AS genes indicated that 134 genes were hypoxia induced and alternatively spliced and these 134 genes were called Hx AS genes (Fig.1A, in grey). Interesting, these 134 Hx AS genes had 238 splicing events, averaging 1.78 AS events per gene (Fig. 1A, Hx Induced Genes), suggesting that AS is enriched in hypoxia induced genes, compared to the total population of genes that underwent AS (1.53 AS events per gene). Similar to the total population of AS genes, alternative cassette-exons were the most abundant type of splicing events (62%) in Hx AS genes. Other AS events for these Hx AS genes included alternative 5’ splice-sites (16%), alternative 3’ splice-sites (13%), intron retention (9%), and mutually exclusive exons (0%). However, Hx AS genes exhibited 1.2 fold enrichment in alternative cassette-exon inclusion and a 1.22 fold reduction in intron-retention in comparison with the total population of AS genes.
Since cassette-exon AS is the most abundant splicing event, hypoxia-mediated cassette-exon regulation was further analyzed. In the total population of cassette-exon genes (All Cassette, Fig. 1B), 44% of cassette-exon genes exhibited exon inclusion (454 out of 1022 genes) while 56% genes exhibited exon skipping, suggesting that hypoxia generally promotes exon skipping in Hep3B cells. However, 76% of cassette-exon Hx AS genes underwent exon inclusion (63 out of 83 genes) while 24% of genes exhibited exon skipping, suggesting hypoxia promotes exon inclusion of hypoxia-inducible genes (Fig. 1B, Hx induced AS). In contrast, hypoxia promoted exon skipping of 75% of hypoxia reduced cassette-exon genes (Hx reduced, Fig.1B) whereas the number of exon inclusion to exon skipping events was equal (50%) for genes whose expression was not changed by hypoxia (No Change, Fig. 1B).
Functional clustering of hypoxia-mediated AS genes using DAVID reveals new pathways regulated by hypoxia
To determine if hypoxia-induced AS was enriched in specific cellular pathways, DAVID (10) was used to create functional annotation clusters of genes that were hypoxia induced (Fig. 1A, left column), the entire population of AS genes (Fig. 1A, right column), and the Hx AS genes (Fig. 1A, top row) (see Supplemental File 2 for the gene lists). As expected, hypoxia induced the expression of genes in pathways allowing cancer cells to adapt to a hypoxic microenvironment such as vascular development, glycolysis, oxidoreductase activity, and cell adhesion/cadherins (Fig. 1A, left column). On the other hand, when analyzing all AS genes, pathways involved in oxidoreductase activity, glycolysis, and positive regulation of glucose transport that were enriched in gene expression were also enriched in AS (Fig. 1A, right column and supplemental file 2). However, for the same pathway, the genes involved in AS could be different from the genes enriched in expression analysis, demonstrating the importance of these pathways in the hypoxia response. Importantly, several novel cellular pathways including ATP-binding/protein kinase activity, pleckstrin homology (related to rho protein signal transduction and cytoskeleton organization), rho protein signal transduction, cytoskeleton organization, zinc fingers, cell death, and nuclear lumen genes were also enriched (Fig. 1A, right column and supplemental file 2). Correspondingly, Hx AS genes were largely overlapped with pathways identified in gene expression and/or AS (Fig. 1A, top row).
RT-PCR and qRT-PCR validation of alternative splicing for select HIF target genes identified in exon array analysis
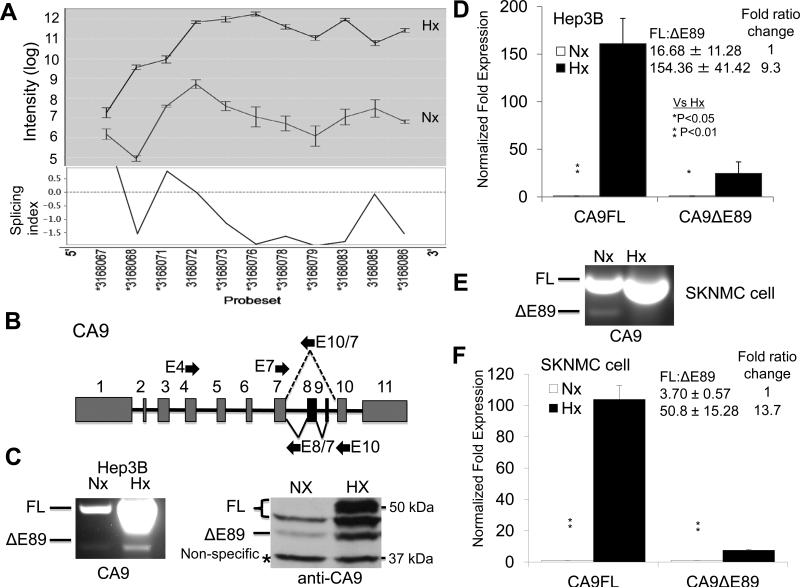
Next, we sought to validate the exon array data using RT-PCR and qRT-PCR. We focused on Hx AS genes since the hypoxia inducible genes are the most important genes in the hypoxia response. First, we selected Hx AS genes as determined by both Altanalyze Firma and easyExon. Next, we used EasyExon to identify regions of potential AS. Then we used RT-PCR to identify the RNA isoforms and qRT-PCR to quantify the levels of these isoforms in normoxic and hypoxic Hep3B cells or other cancer cells. For example, carbonic anhydrase 9 (CA9), a well-known HIF target gene (15, 16), was predicted to undergo AS in exon 2 and in a region spanning exons 6-10 as determined by the splicing index (Fig. 2A). AS was considered significant if the splicing index was greater than +1.5 or less than −1.5 (negative SI indicates exon inclusion by hypoxia) using EasyExon. RTPCR was not able to detect an AS event in exon 2. RT-PCR, using primers that amplify exons 4-10 (Fig. 2B), followed by Topo TA cloning and sequencing confirmed that Hep3B cells expressed two CA9 isoforms, a full-length isoform (FL), and an isoform in which exons 8-9 were skipped (ΔE89) (Fig. 2C left). qRT-PCR determined that hypoxia induced the expression of both isoforms but significantly favored FL in Hep3B cells, thus hypoxia increased the FL/ΔE89 ratio by 9.3 fold (Fig. 2D). Similar CA9 isoform expression patterns and increased CA9 FL/ΔE89 ratio by hypoxia were observed in a neuroblastoma cell-line, SKNMC, suggesting that this splicing event is not cell-type specific (Fig. 2E-F). In addition, western blot of CA9 in Hep3B cells confirmed that both FL and ΔE89 protein isoforms were expressed and FL CA9 protein was preferentially induced by hypoxia (Fig. 2C right). The larger CA9 FL band in hypoxic Hep3B cells was likely the result of post-translation modifications and was also observed previously (17).
Figure 2. Hypoxia regulates AS of the HIF target gene, CA9.
A) EasyExon analysis of differentially expressed probes for the CA9 gene in Hep3B cells. The lines indicate probe intensities for normoxia or hypoxic samples. Differentially expressed probes were considered positive for alternative splicing if the splicing index was greater than 1.5 or less than -1.5 (negative SI indicates exon inclusion is increased by hypoxia). Vertical numbers below the splicing index indicate affymetric probe IDs. B) Diagram of the confirmed CA9 gene isoforms. Grey boxes represent constitutive exons and black boxes represent alternatively spliced exons. Solid arrows indicate primer used for RT-PCR (E4//E10) and qRT-PCR (E7//E10/7 for CA9ΔE89, and E7//E8/7 for CA9 FL). C) RT-PCR and western blot analysis of CA9 mRNA and protein in normoxic and hypoxic Hep3B cells. CA9 FL proteins exhibit two bands with different sizes. D) qRT-PCR analysis of CA9FL and ΔE89 transcripts in normoxic and hypoxic Hep3B cells. E) RT-PCR analysis of CA9 mRNAs in normoxic and hypoxic SKNMC cells. F) qRT-PCR analysis of CA9FL and ΔE89 transcripts in normoxic and hypoxic SKNMC cells. Numbers in the panels D and F here and other qRT-PCR panels represent the ratio of the full-length isoform versus the exon skipping isform ± the standard deviation. Fold of ratio change is also indicated. One-way analysis of variance was performed for this and other studies in this paper unless otherwise stated. *, P= 0.05; **, P =0.01. Controls for statistical analysis are specified in each Figure.
Angiopoietin-like 4 (ANGPTL4) was predicted to undergo AS in exon 4 (Suppl Fig. 1A). RT-PCR, Topo TA cloning and sequencing confirmed that ANGPTL4 expressed a full-length isoform (FL), and an exon 4 skipping isoform (ΔE4) in Hep3B cells (Suppl Fig. 1C left). qRTPCR determined that hypoxia induced the expression of both isoforms but FL was favored; thus hypoxia increased the FL/ΔE4 ratio by 4.9 fold (Suppl Fig. 1D). Additionally, hypoxia increased the ANGPTL4 FL/ΔE4 ratio by 3.5 fold in UM-SCC-22B, a head and neck squamous cell carcinoma cell-line (Suppl Fig. 1E-F). The expression of ANGPTL4 FL and ΔE4 protein isoforms and preferential induction of ANGPTL4 FL protein by hypoxia were also observed in Hep3B cells (Suppl Fig. 1C right).
Expression of multiple isoforms and hypoxia regulating RNA splicing of additional HIF target genes, pyruvate dehydrogenase kinase 1 (PDK1, FL and ΔE4), WNK lysine deficient protein kinase 1 (WNK1, FL and ΔE11-12) and prolyl 4-hydroxylase alpha polypeptide II (P4HA2, FL and ΔE2), were also confirmed by RT-PCR and qRT-PCR (Suppl Fig. 2A-C).
CA9, ANGPTL4, PDK1, WNK1, and P4HA2 exhibited preferential induction of the FL isoform. However, Altanalyze (FIRMA) predicts that hypoxia promotes exon skipping of some HIF target genes including procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2, FL and ΔE14) and enolase 2 (ENO2, FL and ΔE8), which are also confirmed (Suppl Fig. 2D-E). These data demonstrated that hypoxia promotes exon inclusion for most HIF target genes; however, hypoxia promotes exon skipping for some HIF target genes such as PLOD2 and ENO2.
Hypoxia changes isoform ratios of HIF target genes at the transcriptional level
After confirming AS of several HIF target genes, we addressed the molecular mechanism on how hypoxia regulates AS of HIF target genes. Although AS was proposed to explain the observed FL to skipping isoform ratio changes in the above sections, the hypoxia-induced isoform ratio variations could be caused by RNA splicing or other mechanisms. For instance, preferential degradation of exon skipping isoforms under hypoxia might explain the hypoxia-induced isoform ratio changes since exon skipping RNA isoforms may incur premature termination codons and such transcripts are often targeted for nonsense-mediated decay (NMD) (18). In the seven genes analyzed, only RNA of CA9 ΔE89 (E8-9, 172bp), but not ANGPTL4 (E4, 114bp), PDK1 (E4, 138bp), WNK1 (E11-12, 609bp), P4HA2 (E2, 136bp in 5’UTR), PLOD2 (E13, 114bp), and ENO2 (E8, 198bp) had premature stop codon. To test the possibility that preferential instability of CA9 ΔE89 contributes to the increased CA9 FL/ΔE89 ratio under hypoxia, Hep3B cells were placed under hypoxia for 16 hrs to increase the levels of both CA9 FL and ΔE89 transcripts, followed by treatment with actinomycin D to inhibit de novo transcription. Cells were then placed back under normoxia or hypoxia for 0, 2, 4, or 8 hours to allow RNA decay. Using qRT-PCR, both CA9 FL and ΔE89 transcripts were found to be very stable since 90% of the FL and ΔE89 transcripts were detected even after 8 hrs. Moreover, CA9 FL and ΔE89 transcripts exhibited similar stability under normoxia and hypoxia at every time points (data not shown). ANGPTL4 FL and ΔE4 transcripts were far less stable than CA9 transcripts since only 40%, 28%, and 20% of the transcripts were remained after 2, 4, and 8 hrs. However, ANGPTL4 FL and ΔE4 transcripts also exhibited similar stability under normoxia and hypoxia (data not shown). Furthermore, actinomycin D treatment in Hep3B cells blocked hypoxic induction of HIF target genes and blocked splicing change of HIF target genes, indicating that the hypoxia-mediated isoform shift required active transcription. These data supported the idea that transcription regulation, not post-transcriptional regulation is responsible for the hypoxia-induced increased CA9 and ANGPTL4 FL/exon-skipping ratio.
HIF activity, not hypoxia per se, is necessary to change AS of HIF target genes
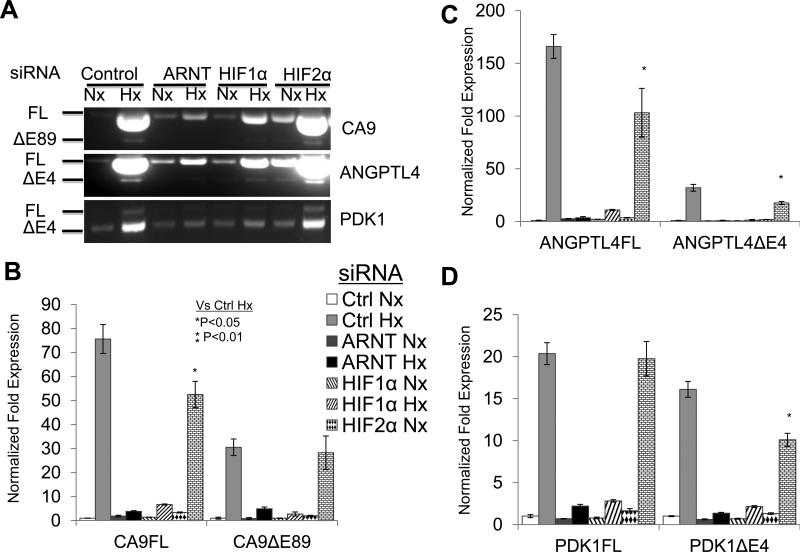
To test whether hypoxic stress or HIF activity is responsible for the splicing changes of HIF target genes, ARNT, HIF1α or HIF2α mRNA levels were reduced by 80% using siRNAs in normoxic or hypoxic Hep3B cells (data not shown). ARNT and HIF1α, but not HIF2α knockdown dramatically reduced the hypoxic induction of CA9, ANGPTL4, and PDK1, consistent with the idea that CA9, ANGPTL4, and PDK1 are primarily regulated by HIF1 in Hep3B cells (Fig. 3A). qRT-PCR confirmed that ARNT and HIF1α knockdown significantly reduced the levels of both FL and exon skipping isoforms of CA9, ANGPTL4, and PDK1 and prevented the splicing changes of these genes (Fig. 3B-D). In contrast, HIF2α knockdown only mildly reduced hypoxic induction of CA9FL (1.44 fold) and PDK1ΔE4 (1.6 fold), similarly reduced hypoxic induction of ANGPTL4FL and ΔE4. Thus, HIF2α knockdown only reduced the CA9FL/ΔE89 ratio by 1.33 fold (Fig. 3B), maintaining the ANGPTL4FL/ΔE4 ratio (Fig. 3C), but enhanced the PDK1FL/ΔE4 ratio 1.5 fold (Fig. 3D). Knockdown of ARNT and HIF1α also inhibited hypoxic induction of the FL and exon skipping isoforms of WNK1, PLOD2, ENO2, and P4HA2 in Hep3B cells and prevented splicing ratio changes for these genes (data not shown). These data suggested that HIF activity, but not hypoxia per se is necessary for increased gene expression as well as hypoxia-mediated splicing changes of these HIF target genes.
Figure 3. HIF activity, but not hypoxia per se is necessary to promote AS of HIF target genes.
A) RT-PCR analysis of CA9, ANGPTL4, and PDK1 RNA transcripts in normoxic or hypoxic Hep3B cells targeted with control (Ctrl), ARNT, HIF1α, or HIF2α siRNAs. B-D) qRTPCR analysis of FL and exon skipping isoforms of CA9 (B), ANGPTL4 (C), and PDK1 (D) in the normoxic or hypoxic Hep3B cells targeted with control or siRNA to ARNT, HIF1α, or HIF2α.
HIF activity is sufficient to regulate AS of HIF target genes
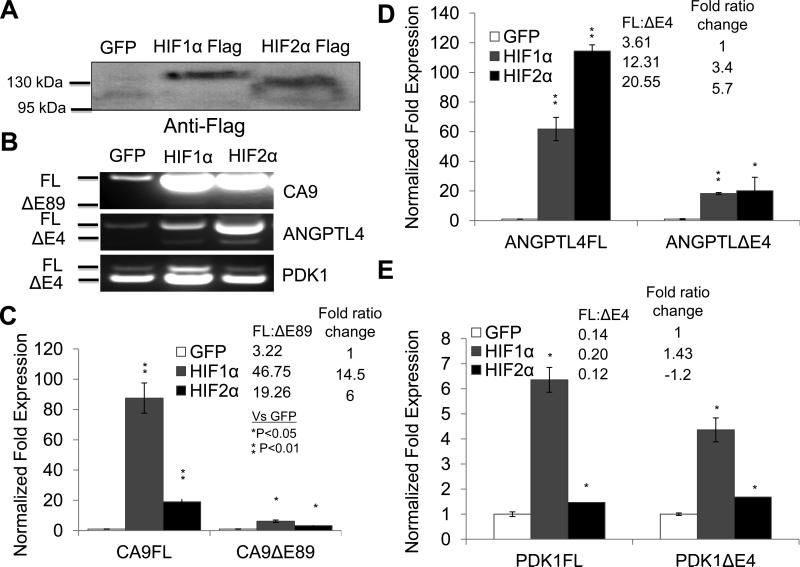
Next, we wanted to determine if HIF activity is sufficient for hypoxia regulated AS of HIF target genes. To test this, normoxic Hep3B cells were transduced with lentiviruses expressing normoxia active, flag-tagged, HIF1α or HIF2α, or GFP as a negative control (Fig. 4A). HIF1α and HIF2α transduction induced the expression of CA9, ANGPTL4, and PDK1 as determined by RTPCR (Fig. 4B). More importantly, qRT-PCR determined that HIF1α and HIF2α increased both FL and exon skipping isoforms of CA9 (Fig. 4C), ANGPTL4 (Fig. 4D), and PDK1 (Fig. 4E). However, FL transcripts of CA9, ANGPTL4, and PDK1 were preferentially induced (Fig. 4C-E). Thus, HIF1 or HIF2 increased the FL/exon skipping ratio for CA9 by 14.5 or 6.0 fold (Fig. 4C), ANGPTL4 by 3.41 or 5.7 fold (Fig. 4D), and PDK1 by 1.43 or -1.2 fold (Fig. 4E).
Figure 4. HIF activity is sufficient to promote AS of HIF target genes.
A) Western blot analysis of Flag-tagged proteins in normoxic Hep3B cells transduced with lenti-viruses expressing GFP, or Flag-tagged, normoxia-active HIF1α or HIF2α proteins. B) RT-PCR analysis of CA9, ANGPTL4 and PDK1 transcripts in normoxic Hep3B cells transduced with GFP, or normoxia-active HIF1α or HIF2α proteins. C-E) QRT-PCR analysis of FL and exon-skipping RNA isoforms of CA9 (C), ANGPTL4 (D), and PDK1 (E) in the above described cells.
To further validate these results, the RNA splicing of CA9, ANGPTL4, and PDK1 were examined in RCC4 cells, a renal cell carcinoma cell-line that expresses constitutively active HIF1α and HIF2α proteins due to loss of functional pVHL (15, 19). In addition, the RNA splicing of the above three genes were also assessed in RCC4T cells in which functional pVHL is reintroduced into RCC4 cells and therefore HIF proteins are only active under hypoxia (15, 19). RT-PCR and qRT-PCR analysis determined that hypoxia increased the levels of CA9FL and ΔE89 by 41.8 and 7 fold respectively in RCC4T cells, increasing the FL/ΔE89 ratio by 6.2 fold (Suppl Fig. 3B). In contrast, hypoxia did not induce the levels of CA9FL and ΔE89 isoforms nor did hypoxia enhance the FL/ΔE89 ratio in RCC4 cells (Suppl Fig. 3B). However, the CA9 FL/ΔE89 ratio in RCC4 cells was already high even under normoxia (Suppl Fig. 3B). Similar findings were observed for AS of ANGPTL4; however, hypoxia was still able to enhance the ANGPTL4 FL/ΔE4 ratio in RCC4 (Suppl Fig. 3C). Although, hypoxia was able to increase the expression of the PDK1FL and ΔE4 isoforms and to enhance the PDK1FL/ΔE4 ratio in RCC4T cells, PDK1 FL/ΔE4 ratio was not elevated in normoxic or hypoxic RCC4 cells (Suppl Fig. 3D).
Taken together, these data suggested that HIF activity is necessary and sufficient to regulate alternative splicing of a subset of HIF target genes independent of hypoxia.
PDK1 splicing reporters recapitulate splicing changes observed for the endogenous PDK1 gene when activated by HIF under normoxia
Next, we wanted to see if similar splicing changes could be observed in splicing reporter gene. We selected the PDK1 gene for splicing reporter model as both the FL and ΔE4 isoforms were highly expressed in 293T cells (data not shown). Exons 3-5 including introns 3 and 4 of the PDK1 gene were PCR amplified from human genomic DNA and placed downstream of the CA9 promoter, a HIF1 target gene promoter, or the PAI1 promoter, a HIF2 target gene promoter (Fig. 5A). RTPCR using minigene specific primers determined that the PDK1 minigene expressed both FL and ΔE4 isoforms in 293T and Hep3B cells (Fig. 5B). In addition, the PDK1ΔE4 isoform was the dominantly expressed isoform in both 293T and Hep3B cells (Fig. 5B). Furthermore, PDK FL isoforms were also highly expressed in 293T cells (Fig. 5B). These data demonstrated that the PDK1 splicing reporter recapitulated the expression patterns of the endogenous PDK1 gene in 293T and Hep3B cells (Suppl Fig. 2A and data not shown).
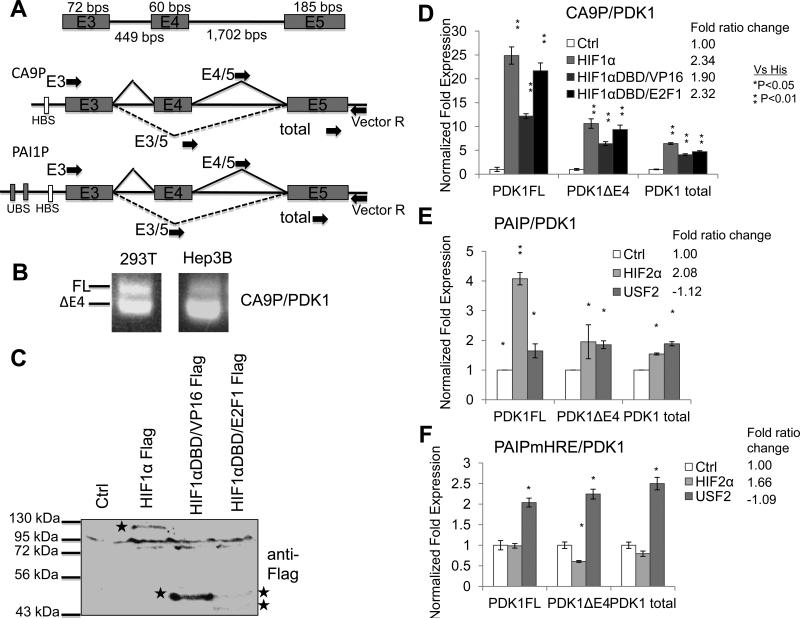
Figure 5. Transcription activation is not sufficient to regulate splicing of a PDK1 minigene.
A) Schematic of a PDK1 splicing reporter driven by CA9 or PAI1 promoter. Arrows represent primers used for RT-PCR (E3//vector R) and qRT-PCR (E3/5, E4/5, or total with Vector R for ΔE4, FL, or total transcript respectively). B) RT-PCR analysis of PDK1 transcripts expressed from CA9P/PDK1 in 293T and Hep3B cells. C) Western blot analysis of Flag-tagged HIF1α, HIF1αDBD/VP16TAD, and HIF1αDBD/E2F1TAD proteins in 293T cells for experiments whose results are presented in Fig. 8D. HIF1αDBD/E2F1TAD construct produces two proteins. D) QRT-PCR analysis of minigene specific PDK1 isoforms in 293T cells co-transfected with CA9P/PDK1 minigene and HIF1α or HIF1αDBD expression constructs. E). qRT-PCR analysis of minigene specific PDK1 isoforms in 293T cells cotransfected with PAI1P/PDK1 minigene and HIF2α or USF2 expression constructs. F). qRTPCR analysis of minigene specific PDK1 isoforms in 293T cells co-transfected with PAI1PmHRE/PDK1 minigene and HIF2α or USF2 expression constructs
To assess AS of the PDK splicing reporter in response to HIF activation, 293T cells were transfected with the PDK1 minigene and normoxia active HIF1α expression plasmids. HIF1α induced the levels of total PDK1 minigene transcripts, FL, and ΔE4 isoform by 3.65, 24.9, and 10.6 fold respectively and enhanced the FL/ΔE4 ratio by 2.34 fold (Fig. 5D). Next, similar experiments were performed using the PAI1P/PDK1 splicing reporter (Fig. 5A). Normoxia active HIF2α induced the expression of PDK1FL and ΔE4 isoforms by 4.22 and 2.36 fold respectively, thus enhancing the FL/ΔE4 ratio 2.08 fold (Fig. 5E). These results indicated that the PDK1 splicing reporters recapitulate splicing changes observed for the endogenous PDK1 gene when activated by HIF under normoxia.
Some transcription factors have dual roles in RNA splicing and gene transcription by recruiting splicing factors and transcription co-factors using their transactivation domains (20-22). To determine if the activation domain of HIF1α protein is important for PDK1 pre-mRNA splicing, fusion constructs containing the HIF1α DNA binding domain fused to the transactivation domains from the VP16 or E2F1 transcription factors were used to activate CA9P/PDK1 splicing reporter (Fig. 5C). HIF1αDBD/VP16 induced the expression of PDK1FL (12.2 fold) and PDK1ΔE4 (6.4 fold), and enhanced the FL/ΔE4 ratio by 1.9 fold (Fig. 5D). Similarly, HIF1αDBD/E2F1 induced PDK1FL (21.7 fold) and PDK1ΔE4 (9.4 fold), and increased FL/ΔE4 expression ratio by 2.3 fold (Fig. 5D). These data indicated that HIF transactivation domain is not required for AS of the PDK1 minigene.
In summary, these data indicated that HIF1 or HIF2-mediated activation of the PDK1 minigene is sufficient to increase PDK1 FL/ΔE4 ratio.
Activation of endogenous HIF target genes contributes to increased FL/ΔE4 ratio of PDK1 splicing reporter
As stated above, transcription activation of PDK1 by HIF or HIFDBD hybrid constructs is sufficient to increase the PDK1 minigene FL/ΔE4 ratio. However, HIF and the fusion constructs can activate endogenous HIF target genes; therefore it is possible that activation of endogenous HIF target genes may increase FL/ΔE4 ratio. To rule out or confirm this possibility, we utilized upstream stimulatory factor 2 (USF2) and the PAI1P/PDK1 minigene since USF2 can activate the PAI1 promoter (13), but not endogenous HIF target genes. USF2 induced the expression of the PDK1FL and ΔE4 isoforms by 2.95 and 2.96 fold respectively but was not able to enhance the FL/ΔE4 ratio even though USF2 was able to activate the minigene to a similar extent as HIF2α (USF2=1.97 and HIF2α=1.52 fold induction of total transcripts) (Fig. 5E). To test if activation of endogenous HIF target gene could regulate AS of the PDK1 minigene, HIF-binding site on the PAI1 promoter was mutated to produce the PAI1PmHRE/PDK1 minigene (Fig. 5F). Interestingly, although HIF2α was not able to activate the expression of PAI1PmHRE/PDK1 as assessed by total as well as FL transcripts, HIF2α increased the FL/ΔE4 ratio by reducing the levels of the PDK1 ΔE4 transcript (Fig. 5F).
To further validate that transcription activation of endogenous HIF target genes increases the PDK1 FL/ΔE4 ratio, the PDK1 splicing reporter was placed under the control of a promoter containing 5 copies of the Gal4 DNA binding element (Suppl Fig. 4A, 5xUAS). Fusion constructs containing the Gal4 DNA binding domain fused to the transactivation domains of normoxia active HIF1α, VP16, or E2F1 were used to activate the G5P/PDK1 minigene in the presence or absence of normoxia-active HIF1α and HIF2α that activate endogenous HIF target genes (Suppl Fig. 4B). Interestingly, although Gal4/HIF1αTAD, Gal4/VP16TAD, and Gal4/E2F1TAD increased the PDK1 FL/ΔE4 ratio by 1.27, 2.44, and 1.47 fold, activation of endogenous HIF target genes further increased the PDK1 FL/ΔE4 ratio to 2.04, 3.28, and 3.29 fold. Importantly, co-tranfected HIF1α and HIF2α increased the expression of endogenous HIF target genes including CA9 and LOX (data not shown), but not the expression of G5/PDK1 (Suppl Fig. 4C, same amount of PDK1 total transcripts with or without HIF). These data demonstrated that activation of endogenous HIF target genes contributes to AS of the PDK1 minigene.
Discussion
Most analyses of hypoxia-mediated gene expression changes are conducted at the gene level using traditional microarrays (15, 23-26). However, due to significant functional difference of the proteins encoded by RNA isoforms and the fact that a majority of genes express multiple RNA isoforms (5), gene expression analysis by exon-array or RNA deep sequencing is necessary to fully understand gene expression programs. Our exon-array analysis in normoxic and hypoxic Hep3B cells not only confirms hypoxic induction of previously identified hypoxia-inducible genes but also reveals significant roles of hypoxia in regulating RNA splicing of genes whose transcription are induced by hypoxia or genes whose transcription are not changed or reduced by hypoxia.
Here, we first reported novel hypoxia-regulated genes/pathways that cannot be identified using traditional microarrays since expression levels of these genes are typically not changed by hypoxia or reduced by hypoxia. In addition, we found that hypoxia primarily promotes exon skipping of these non-HIF target genes in Hep3B cells, a result consistent with what is reported in endothelial cells (6, 7). We proposed that increased exon skipping of these non-HIF target genes also have functional consequences. For example, genes involved in ATP binding and protein kinase activity are not hypoxia induced, but exhibit alternative splicing. Increased exon skipping of these genes in hypoxic Hep3B cells, presumably acts to reduce ATP usage to maintain cellular ATP levels. AS of some of these non hypoxia-induced genes are not associated with enriched pathways identified by DAVID Bioinformatics, however, their splicing are also altered and expected to be functionally important. For example, the KRAS 4B isoform (exon 4a is skipped in KRAS 4B, but included in KRAS 4A) is induced in hypoxic Hep3B, thus reducing the KRAS 4A/4B ratio. Interestingly, a decreased KRAS 4A/4B ratio is often observed in human colorectal cancer, but not in normal colon (27, 28). Furthermore, decreased KRAS 4A/4B ratios through reduction of KARS 4A expression or increased KRAS 4B expression promotes DMH-induced colonic tumorigenesis in in vivo mouse models independent of KRAS mutations (29), demonstrating the functional consequence of reduced KRAS 4A/4B ratio in tumor biology.
More importantly, we reported here, for the first time the role of hypoxia in regulating RNA splicing of hypoxia inducible genes. We found that hypoxia promotes exon inclusion for 75% of Hx AS genes. Although no functional differences have been examined for the isoforms of most HIF target genes, FL CA9 protein was reported to be a membrane-associated, tumor-promoting protein by regulating intercellular pH via transporting protons out of the cells during hypoxia. In contrast, the CA9ΔE89 protein is a soluble protein and is not able to transport protons out of the cells (17). Thus, it makes sense that CA9 FL is preferentially induced by hypoxia. Additionally, preferential induction of nerve growth factor receptor TrkA ΔE679 isoform, but not FL leads to nerve growth factor independent receptor activity and neuroblastoma tumor promoting activity (30). Again, specific promotion of Cysteine-Rich protein 61 FL, but not intron-3 retaining isoform (producing no functional protein) promotes tumor angiogenesis (31).
While isoform ratio changes of several HIF target genes have been reported under hypoxia (17, 30-34), none of these studies have addressed the molecular mechanisms concerning how hypoxia regulates isoform ratio changes of HIF target genes. We demonstrated in this study that the increased FL/exon-skipping ratio of several HIF target genes including CA9 and ANGPTL4 is the result of alternative splicing, not preferential degradation of exon-skipping isoforms. Moreover, we determined that HIF activity, but not hypoxic stress per se is necessary and sufficient to regulate AS of CA9, PDK1, and ANGPTL4 pre-mRNAs. Furthermore, we found that activation of endogenous HIF target genes contributes to AS of PDK1 minigenes.
Our studies also indicated that there is a opposing effect on HIF target genes versus non-HIF target genes for RNA splicing in which hypoxia primarily promotes exon inclusion of HIF target genes, but inhibits exon inclusion of genes repressed under hypoxia. This opposing effect is similarly observed for gene transcription and protein translation in which, transcription and protein translation of non-HIF target genes are generally reduced while HIF target genes are transcriptionally induced and protein translation of genes involved in hypoxia response such as HIF1A, HIF2A, and VEGF are maintained due to internal-ribosome-entry-sites (IRES). Our data indicated that exon inclusion of HIF target genes is due to HIF activity, but not hypoxic stress. However, we do not know if exon skipping of hypoxia-repressed genes is dependent on HIF activity or hypoxic stress. This is a very interesting question that should be assessed in future studies.
In summary, this study suggests that hypoxia regulates alternative splicing of hypoxia induced genes, hypoxia reduced genes, and genes whose transcription is not changed by hypoxia. Additionally, HIF activity, not hypoxic stress is found to be necessary and sufficient to regulate AS of a subset of hypoxia inducible genes. Moreover, activation of endogenous HIF target genes contributes to AS of some HIF target genes. These findings significantly increase our understanding of how cells regulate gene transcription as well as RNA splicing to adapt to a hypoxic microenvironment. In the future, it will be interesting to examine the role of hypoxia in regulating RNA splicing in primary tumors.
Supplementary Material
IMPLICATIONS.
This study demonstrates a novel function of HIF in regulating RNA splicing of HIF target genes.
Acknowledgments
This work was supported by grants from the National Cancer Institute (RO1CA134687, Hu) and Cancer League of Colorado (Hu). Johnny Sena was supported by “Research Supplemental to Promote Diversity” (RO1CA134687-S3 and RO1CA134687-S4).
Financial supports
National Cancer Institute (RO1CA134687, Hu) and Cancer League of Colorado (Hu). Johnny Sena was supported by “Research Supplemental to Promote Diversity” (RO1CA134687-S3 and RO1CA134687-S4)
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Finger EC, Giaccia AJ. Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer metastasis reviews. 2010;29:285–93. doi: 10.1007/s10555-010-9224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harbor symposia on quantitative biology. 2011;76:335–45. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PloS one. 2012;7:e42697. doi: 10.1371/journal.pone.0042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hang X, Li P, Li Z, Qu W, Yu Y, Li H, et al. Transcription and splicing regulation in human umbilical vein endothelial cells under hypoxic stress conditions by exon array. BMC genomics. 2009;10:126. doi: 10.1186/1471-2164-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic acids research. 38:W755–62. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang TY, Li YY, Jen CH, Yang TP, Lin CH, Hsu MT, et al. easyExon--a Java-based GUI tool for processing and visualization of Affymetrix exon array data. BMC bioinformatics. 2008;9:432. doi: 10.1186/1471-2105-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sena JA, Wang L, Pawlus MR, Hu CJ. HIFs Enhance the Transcriptional Activation and Splicing of Adrenomedullin. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-13-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Molecular biology of the cell. 2007;18:4528–42. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pawlus MR, Wang L, Ware K, Hu CJ. Upstream stimulatory factor 2 and hypoxia-inducible factor 2alpha (HIF2alpha) cooperatively activate HIF2 target genes during hypoxia. Molecular and cellular biology. 2012;32:4595–610. doi: 10.1128/MCB.00724-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. Faseb J. 2004;18:1462–4. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 15.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Molecular and cellular biology. 2003;23:9361–74. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2013 doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barathova M, Takacova M, Holotnakova T, Gibadulinova A, Ohradanova A, Zatovicova M, et al. Alternative splicing variant of the hypoxia marker carbonic anhydrase IX expressed independently of hypoxia and tumour phenotype. British journal of cancer. 2008;98:129–36. doi: 10.1038/sj.bjc.6604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Current opinion in cell biology. 2005;17:309–15. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 20.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, et al. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2270–4. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Molecular and cellular biology. 2004;24:442–53. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Molecular cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Hu CJ, Iyer S, Sataur A, Covello KL, Chodosh LA, Simon MC. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Molecular and cellular biology. 2006;26:3514–26. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer research. 2005;65:3299–306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 25.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer research. 2003;63:6130–4. [PubMed] [Google Scholar]
- 26.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. The Journal of biological chemistry. 2006;281:15215–26. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 27.Plowman SJ, Berry RL, Bader SA, Luo F, Arends MJ, Harrison DJ, et al. K-ras 4A and 4B are co-expressed widely in human tissues, and their ratio is altered in sporadic colorectal cancer. J Exp Clin Cancer Res. 2006;25:259–67. [PubMed] [Google Scholar]
- 28.Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. The Journal of pathology. 2009;219:435–45. doi: 10.1002/path.2625. [DOI] [PubMed] [Google Scholar]
- 29.Luo F, Ye H, Hamoudi R, Dong G, Zhang W, Patek CE, et al. K-ras exon 4A has a tumour suppressor effect on carcinogen-induced murine colonic adenoma formation. The Journal of pathology. 2010;220:542–50. doi: 10.1002/path.2672. [DOI] [PubMed] [Google Scholar]
- 30.Tacconelli A, Farina AR, Cappabianca L, Desantis G, Tessitore A, Vetuschi A, et al. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer cell. 2004;6:347–60. doi: 10.1016/j.ccr.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Hirschfeld M, zur Hausen A, Bettendorf H, Jager M, Stickeler E. Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer research. 2009;69:2082–90. doi: 10.1158/0008-5472.CAN-08-1997. [DOI] [PubMed] [Google Scholar]
- 32.Gang H, Hai Y, Dhingra R, Gordon JW, Yurkova N, Aviv Y, et al. A Novel Hypoxia-Inducible Spliced Variant of Mitochondrial Death Gene Bnip3 Promotes Survival of Ventricular Myocytes. Circulation research. doi: 10.1161/CIRCRESAHA.110.238709. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi Y, Kakeya T, Nakajima O, Sakai A, Ikeda K, Yamaguchi N, et al. Hypoxia induces expression of a GPI-anchorless splice variant of the prion protein. The FEBS journal. 2008;275:2965–76. doi: 10.1111/j.1742-4658.2008.06452.x. [DOI] [PubMed] [Google Scholar]
- 34.Elias AP, Dias S. Microenvironment Changes (in pH) Affect VEGF Alternative Splicing. Cancer Microenviron. 2008;1:131–9. doi: 10.1007/s12307-008-0013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.