Abstract
Objective: To elucidate the clinical significance of the methylated status of CpG sites of dapper homolog 1 (DACT1) promoter in the survival prediction in gastric cancer (GC). Methods: The large scale GC patients (n=459) were analyzed for the quantitatively methylated status of CpG sites of DACT1 DNA promoter with the bisulphite sequencing PCR (BSP). With gene sequencing analysis, the methylated statuses of 12 cytosine-phosphate-guanine (CpG) sites in DACT1 promoter were detected to supply detailed information for the precisely prognostic prediction. Associations between molecular, clinicopathologic, and survival data were analyzed. Results: With the MSP detection, different methylated levels of DACT1 promoter were identified in the 25 GC tissues, while none of 25 normal gastric mucosal tissues were found to be methylated. DACT1 promoter methylation was found in 28.32% in 459 patients. GC patients with 4 or more methylated CpG sites of DACT1 promoter was significantly associated with the poorer survival (P=0.19). The methylated statuses of CpG -515, CpG -435, and CpG -430 sites were also identified to provide the elaborate survival discrimination for 459 GC patients, respectively (P=0.049, =0.006, and =0.037). In addition, we demonstrated that the methylated CpG site count had smallest AIC and BIC values than other three methylated status of CpG sites for prediction the survival of 459 GC patients. Conclusions: The methylated CpG site count of DACT1 promoter had the significant applicability for clinical evaluation the prognosis of GC.
Keywords: Stomach, neoplasm, dapper homolog 1, survival, methylation
Introduction
Gastric cancer is highly prevalent in Asia, particularly China, and remains the second leading causes of cancer-related death worldwide [1]. Although diagnostic technique and treatment methods have been improving, the prognosis of gastric cancer (GC) is still dismal [2]. Biomarkers have been hoping to be promising targets for improvement early diagnosis rate and cure rate, despite the low repeatability in the large scale GC patients [3]. It has been fully elucidated that the dysregulation of Wnt signaling leads to cancer development [4,5]. Dishevelled (Dvl) regulates the activation of the Wnt signaling in both the canonical and the noncanonical pathways [5], which can be inhibited by the dapper proteins [6]. Dapper homolog 1 (DACT1), a potential tumor suppressor gene [7], was an important member of dapper protein family. Hypermethylation of the DACT1 DNA promoter was frequently identified to play a regulator of carcinogenesis and prognosis in gastric cancer [7], hepatocellular carcinoma [8], and lung cancer [9]. Authors used to detected methylation of tumour suppressor genes with the qualitative methods [7-9], although the hypermethylation of promoter CpG sites of tumour suppressor genes has been identified in many different cancers [10]. With the qualitative assay of the methylated status of DACT1 promoter, researchers cannot differentiate high-risk methylated CpG sites from low-risk methylated of CpG sites for the accurately prognostic prediction of patients. In this study, we detected the methylated status of CpG sites of DACT1 promoter in the large scale patients with GC (n=459) in order to evaluate the meticulously survival risk of various methylation of CpG sites.
Patients and methods
Data source
After approval from the Tianjin Medical University Cancer Hospital institutional review board, data from the cancer registry of the Tianjin Cancer Institute was obtained. Oral and written inform consents were obtained from the patients who were included in this study. Information which was obtained through participating cancer registry included: age, gender, tumor location, tumor size, depth of tumor invasion (T stage, according to the sixth edition UICC TNM classification for gastric cancer), number of metastatic lymph nodes (N stage, according to the sixth edition UICC TNM classification for gastric cancer), extent of lymph node metastasis, Lauren classification, and follow-up vital status.
Patients and study samples
For DACT1 promoter methylation analysis, we collected 459 fresh GC tissues from gastric cancer patients who underwent curative gastrectomy between April 2003 and December 2007 at the Department of Gastroenterology, Tianjin Medical University Cancer Hospital. In addition, a cohort of 25 normal gastric mucosal epithelial tissues derived from normal people between 2004 and 2007 at the Department of Endoscopic Examination and Treatment, Tianjin Medical University Cancer Hospital. All the tumor and normal gastric mucosal epithelial tissues were histologically verified. All patients were not administered radiation, chemical or biological treatment prior to the potentially curative gastrectomy. Adjuvant chemotherapy or radiotherapy was not routinely administrated in patients routinely. The clinicopathological characteristics of these 459 gastric cancer patients are summarized in Table 1. The patients’ consent was obtained for the use of the tissue samples and records, and the study protocol was approved and permission for use of the clinical data was given by the Institutional Research Ethics Committee of Tianjin Medical University Cancer Hospital.
Table 1.
Patient information
| Gender | |
| Male | 314 (68.41%) |
| Female | 145 (31.59%) |
| Age at surgery | |
| ≤ 60 | 271 (59.04%) |
| > 60 | 188 (40.96%) |
| Tumor size | |
| < 4.0 | 66 (14.38%) |
| ≥ 4.0 | 393 (85.62%) |
| Tumor location | |
| Upper third | 113 (24.62%) |
| Middle third | 117 (25.49%) |
| Lower third | 201 (43.79%) |
| More than 2/3 stomach | 28 (6.1%) |
| Depth of tumor invasion (T stage) | |
| T1 | 6 (1.31%) |
| T2 | 46 (10.02%) |
| T3 | 284 (61.87%) |
| T4 | 123 (26.80%) |
| Number of metastatic lymph nodes (N stage) | |
| N0 | 111 (24.18%) |
| N1 | 163 (35.52%) |
| N2 | 106 (23.09%) |
| N3 | 79 (17.21%) |
| Lauren classification | |
| Intestinal | 122 (26.58%) |
| Diffuse | 319 (69.50%) |
| Mixed | 18 (3.92%) |
| Methylated CpG site count | |
| 3 or less | 434 (94.55%) |
| 4 or more | 25 (5.45%) |
| Methylated status of CpG -515 | |
| Unmethylated | 431 (93.90%) |
| Methylated | 28 (6.10%) |
| Methylated status of CpG -435 | |
| Unmethylated | 432 (94.12%) |
| Methylated | 27 (5.88%) |
| Methylated status of CpG -430 | |
| Unmethylated | 420 (91.50%) |
| Methylated | 39 (8.50%) |
Surgical treatment
Curative resection was defined as a complete lack of grossly visible tumor tissue and metastatic lymph nodes remaining after resection, with pathologically negative resection margins. Primary tumors were resected en bloc with limited or extended lymphadenectomy (D1 or D2-3 according to the Japanese Gastric Cancer Association (JGCA)). Surgical specimens were evaluated as recommended by 6th UICC TNM classification for GC.
DNA extraction and RNA extraction
Genomic DNA was extracted from 459 GC tissues and 25 normal gastric mucosal tissues using QIAamp DNA mini kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. Sodium bisulphite modification of genomic DNA was performed by using the EZ DNA Methylation-GoldTM Kit (Zymo Research, Hornby, Canada). RNA was extracted from 25 of 459 GC tissues and 25 normal gastric mucosal tissues using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Western blotting analysis
25 of 459 GC tissues and 25 normal gastric mucosal tissues were respectively added to 1 mL of 100 mmol/L Tris/HCl (pH 7.5), 100 mmol/L NaCl, 0.5% sodium deoxycholate, 1 mmol/L ethylenediaminetetraacetic acid, 1% Nonidet P40, 0.1% sodium dodecyl sulfate, and protease inhibitor. After blocking, 50 ug sample was incubated for 60 minutes with a rabbit anti- DACT1 (OriGene, TA316654, 1:500 dilution) at room temperature. Gel Imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China) to analyze images and to determine gray values.
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis
The expression of DACT1 mRNA was detected by RT–PCR in 25 of 459 GC tissues and 25 normal gastric mucosal tissues. Total RNA was reverse transcribed to cDNA in a 20 ul volume using Reverse Transcription kit (Invitrogen, Carlsbad, CA). Primers designed and utilized for DACT1 was as follows: Forward sequence: 5’-AGTGAGGACGAGCAGAGCAAT-3’, and Reverse sequence: 5’-AGTTTCAAAGAGCCAGACCGA3’. The GAPDH gene was used as an endogenous control for semi-quantitative DNA-PCR. Primers designed and utilized for GAPDH was as follows: Forward sequence: 5’-GAAGGTGAAGGTCGGAGTC-3’, and Reverse sequence: 5’-GAAGATG GTGATGGGATTTC-3’. The PCR Cycling conditions for all sequences were 35 cycles of denaturation at 95°C for 3 minutes, annealing at 94°C for 30 seconds, and extension at 56°C for 30 seconds followed by a final extension at 72°C for 8 minutes. All PCR product electrophoreses were performed on a 2% agarose gel with ethidium bromide and visualized using the Gel Imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China).
Sodium bisulfite treatment
Sodium bisulphite modification of genomic DNA was performed using the EZ DNA Methylation-GoldTM Kit (Zymo Research, Hornby, Canada).
Methylation-specific PCR (MSP)
25 of 459 GC tissues and 25 normal gastric mucosal tissues were detected the qualitative methylated analysis of DACT1 promoter with the methylation-specific PCR (MSP). DACT1 primers detecting methylated (M) or unmethylated (U) alleles of the DACT1 promoter were: DACT1-MF, 5’-CGGTGTGAGTGGAAATGAGGAGTGGTC-3’ and DACT1-MR, 5’-ACAAAAACCGCGACGAAACGCG-3’ for methylated alleles; DACT1-UF, 5’-TTTG GTGTGAGTGGAAATGAGGAGTGGTT3’ and DACT1-UR, 5’-CCACACAAACAAAAACCACAACAAAACACA-3’ for unmethylated alleles. MSP was performed for 25 cycles using Ampli Taq-Gold (methylation-specific primer, annealing temperature 600°C; unmethylation specific primer, annealing temperature 580°C). MSP primers were first checked for not amplifyling any unbisulfited DNA and the specificity of MSP was further confirmed by direct sequencing of some PCR products. PCR reactions were resolved on a 2% agarose gel.
Bisulphite sequencing (BSP)
All 459 GC tissues and 25 normal gastric mucosal tissues were detected the qualitative methylated analysis of DACT1 promoter with the bisulphite sequencing PCR (BSP). Hot start PCR with the bisulfite-treated DNA was performed with a 195 bp PCR product spanning promoter region -578 bp to -383 bp relative to the transcription start site of DACT1. 12 CpG sites were contained in the promoter region of DACT1. The sequences of PCR primers were as follows: F: 5’-TGTAATATTTTGTTTGGGAAGTGAAAG-3’; R: 5’CTAAAACCCCAACATCCTATTACAATC-3’. The purified PCR products were cloned into the pUC18-T vector (Biodee, Beijing, China), and five clones for each sample were randomly selected and sequenced by Shanghai Sangon Co (Shanghai, China).
Follow-up
After curative surgery, all patients were followed every 3 or 6 months for 2 year at outpatient department, every year from the third to fifth years, and then annually thereafter until the patient died. The median follow-up for the entire cohort was 41 months (range: 1-104). The follow-up of all patients who were included in this study was completed in December 2012. Ultrasonography, CT scans, chest X-ray, and endoscopy were obtained with every visit.
Statistical analysis
The median OS was determined by using the Kaplan-Meier method, and log-rank test was used to determine significance. Factors that were deemed of potential importance on univariate analyses (P < 0.05) were included in the multivariate analyses. Multivariate analysis of OS was performed by means of the Cox proportional hazards model with bootstrapping performance. Hazard ratios (HR) and 95% CI were generated. The Akaike information criterion (AIC), and the Bayesian information criterion (BIC) values within a Cox proportional hazard regression model were calculated for different variables to measure theirs prognostic prediction ability. A smaller AIC or BIC value indicated a better model for predicting outcome [11,12]. With the cut-point survival analysis [13], the optimal cutoff for CpG site conut was identified to be three. Significance was defined as P < 0.05. All statistical analyses were performed with SPSS 18.0 software.
Results
Patient demongraphics
All 459 GC patient clinicopathological characteristics are listed in Table 1. The median OS of all patients was 21 months. Of 459 patients, 61 (13.29%) were alive when the follow-up was over.
Protein and mRNA expression of dact1 in 25 gc tissues and 25 normal gastric mucosal tissues
DACT1 protein expression was detected in 25 of 459 GC tissues and 25 normal gastric mucosal tissues by Western blot, simultaneously (Figure 1A). We found there were significant differences of DACT1 protein expression in 25 GC tissues. The mean value of relative protein expression of DACT1 in 25 GC tissues was 0.645±0.137 (range, 0.140-1.428), while the mean value of relative protein expression of DACT1 in 25 normal gastric mucosal tissues was 1.381±0.137 (range, 0.758-1.901). The mean value of relative protein expression of DACT1 in 25 GC tissues was much lower than that in 25 normal gastric mucosal tissues (P=0.024).
Figure 1.
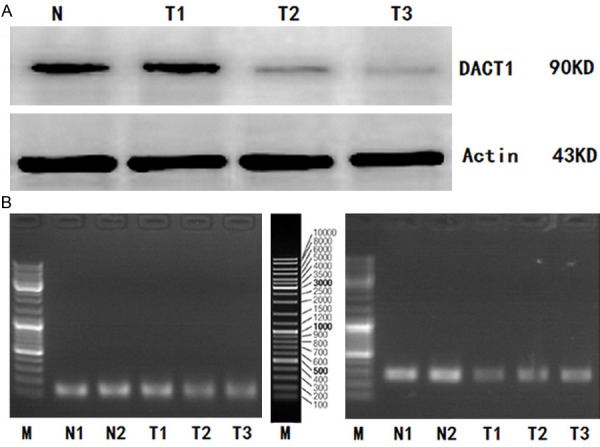
A. Western Blot analysis for DACT1 protein expression in GC tissues and in normal gastric mucosal tissues; B. DACT1 mRNA expression (RT-PCR) in GC tissues and in normal gastric mucosal tissues. (Representation: T, GC tissues; N, normal gastric mucosal tissues).
Similarly, DACT1 mRNA expression was also detected in 25 of 459 GC tissues and 25 normal gastric mucosal tissues by RT-PCR (Figure 1B). We also found there were significant differences of DACT1 mRNA expression in 25 GC tissues. The mean value of relative mRNA expression of DACT1 in 25 GC tissues was 0.712±0.233 (range, 0.073-0.664), while the mean value of relative mRNA expression of DACT1 in 25 normal gastric mucosal tissues was 1.482±0.521 (range, 1.136-1.772). The mean value of relative mRNA expression of DACT1 in 25GC tissues was lower than that in 25 normal gastric mucosal tissues (P=0.017).
Methylation detection of DACT1 promoter
We detected the different levels of DACT1 promoter methylation (including methylation, non-methylation, and partial methylation) in 25 of 459 GC tissues with the MSP analysis, while no DACT1 promoter methylation was found in 25 normal gastric mucosal tissues (Figure 2).
Figure 2.
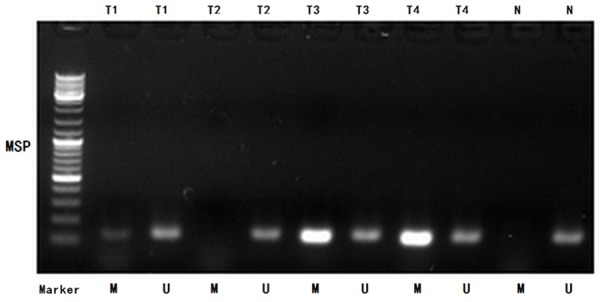
MSP detection of DACT1 promoter methylation in different GC tissues and normal gastric mucosal tissues. (Representation: T, GC tissues; N, normal gastric mucosal tissues; M, methylated; U, unmethylated).
Subsequently, we adopted the BSP analyzed the methylated status of all CpG sites of DACT1 promoter to obtain the precisely quantitative methylated degree of DACT1 promoter in all 459 GC patients. Methylated CpG site count of 459 GC patients ranged between 0 and 12. Of the 459 patients included in the study, 130 patients (28.32%) presented one or more methylated CpG sites and 329 patients (71.68%) presented no methylated CpG site. Although patients without methylated CpG site had longer median OS than those with one or more methylated CpG sites (21 vs 20 months), there is not statistical difference between two groups of patients (P=0.995). According to the result of cut-point analysis for the methylated CpG site count, 25 patients (5.45%) presented with four or more methylated CpG sites and 434 patients (94.55%) presented with three or less methylated CpG sites. No methylated CpG site was found in the normal gastric mucosal epithelial tissues. The methylation sequencing pictures and CpG site charts were shown in Figure 3.
Figure 3.
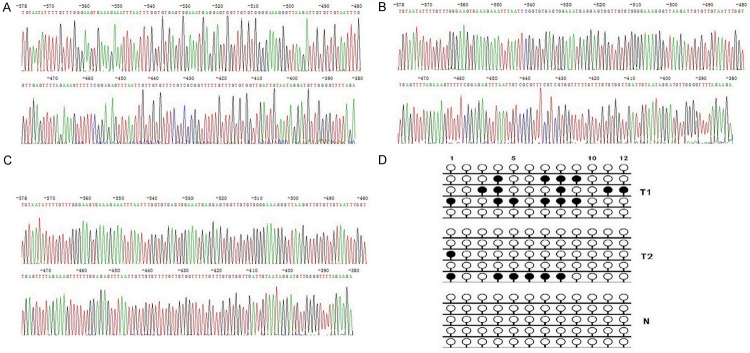
(A) Bisulphite sequencing figure of DACT1 in GC tissue 1; (B) Bisulphite sequencing figure of DACT1 in GC tissue 2; (C) Bisulphite sequencing figure of DACT1 in normal gastric mucosal tissue; and (D) Bisulfite sequencing results in GC tissues and in normal gastric mucosal tissue.
Survival analysis
With the univariate survival analysis, four clinicopathological characteristics were found to have statistically significant associations with OS of 459 GC patients. They were as follows: T stage (P < 0.01), N stage (P < 0.01), tumor size (P=0.033), and tumor location (P=0.048) (Table 2). We also demonstrated that the methylated status of three CpG sites (CpG -515, CpG -435, and CpG -430) of DACT1 promoter had significant association with the survival of 459 GC patients (P=0.049, =0.006, and =0.037), respectively. In addition, we found that the methylated CpG site count (P=0.019) was significantly associated with the OS of patients with the Kaplain-Meier curves discrimination (Table 2, Figure 4).
Table 2.
Survival analysis of 459 GC patients
| Variables | Median OS (mo) | X2 value | Univariate P value | HR value | Multivariate P value |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 21 | 0.544 | 0.461 | ||
| Female | 20 | ||||
| Age at surgery (years) | |||||
| ≤ 60 | 20 | 0.128 | 0.720 | ||
| > 60 | 23 | ||||
| Tumor location | |||||
| Upper third | 21 | 7.891 | 0.048 | 1.000 (0.898-1.115) | 0.989 |
| Middle third | 18 | ||||
| Lower third | 24 | ||||
| More than 2/3 stomach | 16 | ||||
| Tumor size (cm) | |||||
| < 4.0 | 26 | 4.551 | 0.033 | 0.945 (0.701-1.274) | 0.733 |
| ≥ 4.0 | 20 | ||||
| Lauren classification | |||||
| Intestinal | 26 | 5.774 | 0.056 | ||
| Diffuse | 20 | ||||
| Mixed | 17 | ||||
| Depth of tumor invasion (T stage) | |||||
| T1 | 70 | 44.871 | < 0.001 | 1.559 (1.309-1.855) | 0.001 |
| T2 | 27 | ||||
| T3 | 24 | ||||
| T4 | 12 | ||||
| Number of metastatic lymph nodes (N stage) | |||||
| N0 | 37 | 98.573 | < 0.001 | 1.552 (1.399-1.720) | 0.001 |
| N1 | 23 | ||||
| N2 | 17 | ||||
| N3 | 10 | ||||
| Methylated CpG site count | |||||
| 3 or less | 21 | 5.484 | 0.019 | 0.968 (0.293-3.198) | 0.958 |
| 4 or more | 12 | ||||
| Methylated status of CpG -515 | |||||
| Unmethylated | 21 | 3.866 | 0.049 | 1.257 (0.735-2.147) | 0.389 |
| Methylated | 14 | ||||
| Methylated status of CpG -435 | |||||
| Unmethylated | 22 | 7.465 | 0.006 | 1.554 (0.849-2.843) | 0.266 |
| Methylated | 12 | ||||
| Methylated status of CpG -430 | |||||
| Unmethylated | 21 | 4.362 | 0.037 | 0.839 (0.302-2.335) | 0.764 |
| Methylated | 15 | ||||
Figure 4.
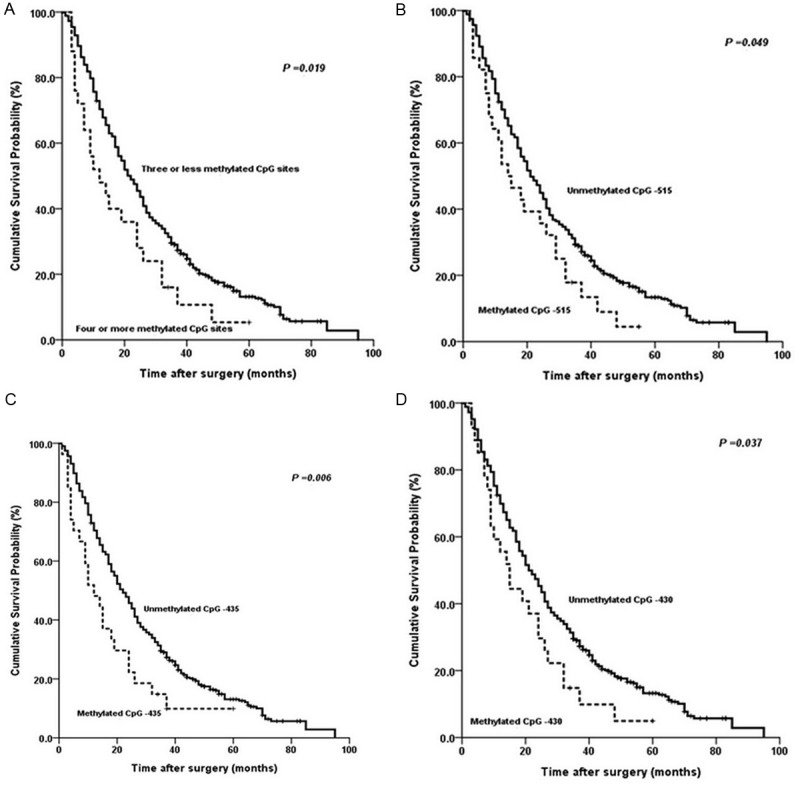
Kaplan-Meier survival curves comparing months of survival in gastric cancer patients are shown for (A) methylated CpG site count of DACT1 promoter; (B) methylated status of CpG-515; (C) methylated status of CpG-435; and (D) methylated status of CpG-430.
All above eight factors were included in a multivariate Cox proportional hazards model with bootstrapping performance to adjust for the effects of covariates. With the multivariate analysis, the independent predictors with the OS of all 459 GC patients were identified as N stage (HR=1.552, P < 0.01) and T stage (HR=1.559, P < 0.001) rather than anyone of the above-mentioned four CpG site variables (Table 2).
Lastly, we demonstrated that the methylated CpG site count of DACT1 promoter had the smaller AIC and BIC values than anyone of the methylated status of three CpG sites (CpG -515, CpG -435, and CpG -430) calculated within the Cox proportional hazard regression model, representing the optimal prognostic predictor of GC (Table 3).
Table 3.
AIC and BIC values test for survival prediction
| Variables | AIC Value | BIC Value |
|---|---|---|
| Methylated CpG site count | 29.718 | 46.234 |
| Methylated status of CpG -515 | 30.045 | 46.561 |
| Methylated status of CpG -435 | 29.841 | 46.357 |
| Methylated status of CpG -430 | 29.792 | 46.368 |
Discussion
Dact1, a negative regulator of both canonical and noncanonical Wnt signaling, not only regulates embryo development but may have an important role in tumorigenesis [14]. Many researchers reported that DACT1 was found to be down-regulated by promoter methylation in many malignant diseases, including breast cancer, non–small cell lung cancer, gastric cancer, hepatocellular carcinoma, bladder urothelial carcinoma, and transitional cell carcinomas [7-9,15-17]. Previously, DACT1 was a novel functional tumor suppressor in gastric cancer through inhibiting NF-κB signaling pathway, and the promoter methylation of DACT1 was identified to be associated with tumor aggressiveness [7]. In this study, we demonstrated that mRNA and protein expression of DACT1 was inconsistently down-regulated in the 25 of 459 GC tissues. With the MSP analysis, we also found the different levels of DACT1 promoter methylation in above 25 GC tissues, including methylation, non-methylation, and partial methylation. Based on the above findings, we think that the methylated degree of DACT1 promoter should be preformed with a quantitative analysis to obtain the accurate data, which may evaluate whether methylation of DACT1 may serve as a biomarker for predicting the prognosis of GC patients. Therefore, we cloned the purified the bisulfite-treated DNA PCR products into the pUC18-T vector, and then we randomly selected five clones to sequence for detection the methylated status of CpG sites of DACT1 DNA promoter in all 459 GC patients. At present study, DACT1 was detected in 28.32% of 459 GC tissues, which is similar to the findings of 205 GC patients in the previous study [7]. Taking the expression differences of DACT1 in between GC tissues and normal gastric mucosal tissues into account, we considered that methylation of DACT1 promoter is GC specific.
So far, many investigators reported that the methylated levels or frequencies of genes were much higher in GC tissues than in normal gastric or non-adjacent tissues [18,19]. Although we initially could not found statistical survival differences between 130 GC patients with one or more methylated CpG sites and 329 GC patients without the methylated CpG sites of DACT1 promoter (P=0.995) with the BSP analysis, we thought that the optimal category of the methylated CpG sites for prediction the survival of 459 GC patients should be executed [20-23]. Owing to the quantitative detection of the CpG site methylation in this study, we considered the results of the methylated status of all CpG sites of DACT1 promoter should be analyzed in the large scale GC patients for the comparatively precise evaluation the prognostic prediction ability of the different methylated status of CpG sites.
Subsequently, we found that three methylated CpG sites (CpG -515, CpG -435, and CpG -430) of DACT1 promoter was significantly associated with the poor survival of 459 GC patients respectively. In addition, we also found the methylated CpG site count of DACT1 promoter was significantly associated with the survival of all GC patients. Although we failed to demonstrated that one of the methylated statuses of above three CpG sites (CpG -515, CpG -435, and CpG -430) of DACT1 promoter or the methylated CpG site count of DACT1 promoter was the independently prognostic predictor of 459 GC patients, we did think that the methylated status of DACT1 promoter was an potentially important biomarker for GC patients’ prognostic prediction owing to its wide practicability [7-9,15-17]. Eventually, we demonstrated that the methylated CpG site count of DACT1 promoter had the smaller AIC and BIC values than all three CpG site methylation statuses of DACT1 promoter in 459 GC patients, which represented that the methylated CpG site count of DACT1 promoter should be considered to be a potential variable for GC patients’ prognostic prediction.
To our knowledge, this study is the first investigation of large scale GC patients. There are some limitations to our study. Most tumor samples (453 of 459) are obtained from Chinese advanced stage GC patients in this study, which perhaps result in little bias of detection results comparing to the other reports. To obtain the convincing corroboration, we detected the significant protein and mRNA expression differences of DACT1 in between 25 of 459 GC tissues and 25 normal gastric mucosal tissues, and the protein and mRNA expression of DACT1 was inconsistent in 25 GC tissues. With the MSP analysis, we also found that the different levels of DACT1 promoter methylation in 25 GC tissues. Therefore, we believed that the quantitative detection of the methylation of DACT1 promoter should be executed. Eventually, we demonstrated that the methylated CpG site count of DACT1 promoter was the elaborate variable for precise evaluation the prognosis and a promising target for the further investigation of specific therapy. Future research should focus on the effects of the given CpG sites and the CpG site count contribution to the biological behaviors of GC cells and the targeted therapy to the given CpG sites of DACT1 promoter.
Acknowledgements
Supported in part by grants from the National Basic Research Program of China (973 Program) 2010CB529301, the Anticancer Major Projects of Tianjin Municipal Science and Technology Commission 12ZCDZSY16400, and the Science Found Program of Tianjin Medical University 2012KYM01.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Meyer HJ, Wilke H. Treatment strategies in gastric cancer. Dtsch Arztebl Int. 2011;108:698–705. doi: 10.3238/arztebl.2011.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Yang Y, Lu M, Shen L. Predictive value of serum CEA, CA19-9 and CA72.4 in early diagnosis of recurrence after radical resection of gastric cancer. Hepatogastroenterology. 2011;58:2166–2170. doi: 10.5754/hge11753. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 5.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 6.Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell. 2002;2:449–461. doi: 10.1016/s1534-5807(02)00140-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Kang W, Go MY, Tong JH, Li L, Zhang N, Tao Q, Li X, To KF, Sung JJ, Yu J. Dapper homolog 1 is a novel tumor suppressor in gastric cancer through inhibiting the nuclear factor-κB signaling pathway. Mol Med. 2012;18:1402–1411. doi: 10.2119/molmed.2012.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yau TO, Chan CY, Chan KL, Lee MF, Wong CM, Fan ST, Ng IO. HDPR1, a novel inhibitor of the WNT/β-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene. 2005;24:1607–1614. doi: 10.1038/sj.onc.1208340. [DOI] [PubMed] [Google Scholar]
- 9.Yang ZQ, Zhao Y, Liu Y, Zhang JY, Zhang S, Jiang GY, Zhang PX, Yang LH, Liu D, Li QC, Wang EH. Downregulation of HDPR1 is associated with poor prognosis and affects expression levels of p120-catenin and beta-catenin in nonsmall cell lung cancer. Mol Carcinog. 2010;49:508–519. doi: 10.1002/mc.20622. [DOI] [PubMed] [Google Scholar]
- 10.Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106:483–493. doi: 10.1002/cncr.21657. [DOI] [PubMed] [Google Scholar]
- 11.Nitsche U, Maak M, Schuster T, Künzli B, Langer R, Slotta-Huspenina J, Janssen KP, Friess H, Rosenberg R. Prediction of prognosis is not improved by the seventh and latest edition of the TNM classification for colorectal cancer in a single-center collective. Ann Surg. 2011;254:793–800. doi: 10.1097/SLA.0b013e3182369101. [DOI] [PubMed] [Google Scholar]
- 12.Cho YK, Chung JW, Kim JK, Ahn YS, Kim MY, Park YO, Kim WT, Byun JH. Comparison of 7 staging systems for patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Cancer. 2008;112:352–361. doi: 10.1002/cncr.23185. [DOI] [PubMed] [Google Scholar]
- 13.Smith DD, Schwarz RR, Schwartz RE. Impact of total lymph node count on staging and survival after gastrectomy for gastric cancer: data from a large US-Population Database. J. Clin. Oncol. 2005;23:7114–7124. doi: 10.1200/JCO.2005.14.621. [DOI] [PubMed] [Google Scholar]
- 14.Katoh M, Katoh M. Identification and characterization of human DAPPER1 and DAPPER2 genes in silico. Int J Oncol. 2003;22:907–913. [PubMed] [Google Scholar]
- 15.Yin X, Xiang T, Li L, Su X, Shu X, Luo X, Huang J, Yuan Y, Peng W, Oberst M, Kelly K, Ren G, Tao Q. DACT1, an antagonist to Wnt/β-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15:R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng H, Deng Z, Wang Z, Zhang W, Su J. The role of aberrant promoter hypermethylation of DACT1 in bladder urothelial carcinoma. J Biomed Res. 2012;26:319–324. doi: 10.7555/JBR.26.20110099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng H, Lu M, Mao LJ, Wang JQ, Li W, Wen RM, Chen JC. Relationships among MTHFR a1298c gene polymorphisms and methylation status of Dact1 gene in transitional cell carcinomas. Asian Pac J Cancer Prev. 2012;13:5069–5074. doi: 10.7314/apjcp.2012.13.10.5069. [DOI] [PubMed] [Google Scholar]
- 18.Leung WK, Yu J, Ng EK, To KF, Ma PK, Lee TL, Go MY, Chung SC, Sung JJ. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91:2294–2301. [PubMed] [Google Scholar]
- 19.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 20.Colaneri A, Wang T, Pagadala V, Kittur J, Staffa NG Jr, Peddada SD, Isganaitis E, Patti ME, Birnbaumer L. A minimal set of tissue-specific hypomethylated CpGs constitute epigenetic signatures of developmental programming. PLoS One. 2013;8:e72670. doi: 10.1371/journal.pone.0072670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartley I, Elkhoury FF, Heon Shin J, Xie B, Gu X, Gao Y, Zhou D, Haddad GG. Long-lasting changes in DNA methylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS One. 2013;8:e77859. doi: 10.1371/journal.pone.0077859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi T, Matsuda Y, Yamashita S, Hattori N, Kushima R, Lee YC, Igaki H, Tachimori Y, Nagino M, Ushijima T. Estimation of the fraction of cancer cells in a tumor DNA sample using DNA methylation. PLoS One. 2013;8:e82302. doi: 10.1371/journal.pone.0082302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YK, Jin S, Duan S, Lim YC, Ng DP, Lin XM, Yeo GSh, Ding C. Improved reduced representation bisulfite sequencing for epigenomic profiling of clinical samples. Biol Proced Online. 2014;16:1. doi: 10.1186/1480-9222-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


