Abstract
The stem-like cells of Glioblastoma multiforme (GBM) tumors (GSCs) are one of the important determinants of recurrence and drug resistance. The aims of the current study were to evaluate the anticancer effect of Olea europaea leaf extract (OLE) on GBM cell lines, the association between OLE and TMZ responses, and the effect of OLE and the OLE-TMZ combination in GSCs and to clarify the molecular mechanism of this effect on the expression of miRNAs related to cell death. The anti-proliferative activity of OLE and the effect of the OLE-TMZ combination were tested in the T98G, U-138MG and U-87MG GBM cell lines using WST-1 assay. The mechanism of cell death was analyzed with Annexin V/FITC and TUNEL assays. The effects of OLE on the expression levels of miR-181b, miR-153, miR-145 and miR-137 and potential mRNA targets were analyzed in GSCs using RT-qPCR. OLE exhibited anti-proliferative effects via apoptosis and necrosis in the GBM cell lines. In addition, OLE significantly induced the expression of miR-153, miR-145, and miR-137 and decreased the expression of the target genes of these miRNAs in GSCs (p < 0.05). OLE causes cell death in GBM cells with different TMZ responses, and this effect is synergistically increased when the cells are treated with a combination of OLE and TMZ. This is the first study to indicate that OLE may interfere with the pluripotency of GSCs by modulating miRNA expression. Further studies are required, but we suggest that OLE may have a potential for advanced therapeutic cancer drug studies in GBM.
Keywords: Olea europaea leaf extract, temozolomide, glioblastoma multiforme, cancer stem cell, MicroRNA
Introduction
Glioblastoma multiforme (GBM) is the most frequent primary brain tumor in adults and is characterized by a highly aggressive phenotype [1]. Over the last 10 years, only two chemotherapeutic agents, carmustine (1,3-bis(2-chloroethyl)-1-nitrosourea, BCNU implant) and temozolomide (TMZ), an imidazotetrazine derivative of dacarbazine, have received regulatory approval for treating malignant gliomas [2]. Despite progress in GBM treatment, some of these patients do not respond to different chemotherapeutics, and therefore, the prognosis is remains poor; the overall survival time of GBM patients is typically less than 1 year [3]. Recent studies showed that the presence of cancer stem cells (CSCs) in the heterogeneous cell populations of GBM tumors may be one of the important determinants of recurrence and drug resistance in these patients [4]. Because GBM therapy failure is primarily due to tumor recurrence, CSCs may be the key target for new drug investigations. Therefore, the aim of most ongoing studies is to identify drugs that are able to affect GBM CSC (GSC) biology [4].
Some medicinal herbs have become new frontiers for cancer therapy research due to their natural phytochemicals or phytonutrient compounds [5-7]. The olive tree, Olea europaea, grows widely in European and Mediterranean Countries, including Turkey. The constituents of Olea europaea leaf extract (OLE) have well-known benefits and metabolic healing properties [8]. However, although OLE is widely recognized with a phenolic- type, oleuropein, rich compound, which have antioxidant activity due to their ability to scavenge free radicals, the anti-cancer potential of OLE has not been adequately investigated [9-13]. Previously, the antitumor properties of OLE were revealed in human HL-60 promyelocytic leukemia cells [14], the Jurkat human leukemic cell line [15] and human colorectal adenocarcinoma HT29 and Caco-2 cell lines [16]. According to these studies, OLE may lead to protection against cancer via the induction of apoptotic pathways [14-16]. In addition, we have recently shown an anticancer effect of OLE on GBM T98G cells. Furthermore, we observed that OLE modulates the expression patterns of miRNAs that have been implicated in a number of cancer-associated metabolic pathways and biological processes [17]. According to our data, OLE modulates the expression of miR-181b, miR-153, miR-145, miR-137 and let-7d, which are related to anticancer activity in T98G cells and the response to TMZ [18]. Therefore, it was of interest to evaluate the anticancer effect of OLE in GBM cells which have different drug resistances.
The first aim of current study was to evaluate the anticancer effect of OLE in GBM cell lines that differ with respect to their responses to TMZ. Therefore, we analyzed the anticancer effect of OLE in the U-138MG and U-87MG cell lines and compared these effects with those observed in T98G cells. In addition, although GSCs do not respond well to chemotherapeutic agents, there have not been any studies evaluating the ability of plant extracts to overcome this resistance. Thus, the second aim of this study was to investigate the effect of OLE and the combination of OLE and TMZ in GSCs and to clarify the molecular mechanism of this effect by analyzing the expression of miRNAs before and after OLE treatment.
Materials and methods
OLE production
Standardized OLE (05.06.2007, 10-00014-00015-0) was kindly provided by Kale Naturel (Edremit-Balıkesir, Turkey) and prepared as described previously [18].
Determination of the active compound in OLE by HPLC analyses
An Agilent 1200 HPLC system (Waldbronn, Germany), consisting of a vacuum degasser, binary pump, autosampler and diode-array detector was used to identify the phenolic compounds in the OLE fractions. Chromatographic separations were carried out using an XBridge C18 (4.6×250 mm, 3.5 µm) column from Waters. The mobile phase consisted of 1% formic acid in water (solvent A) and acetonitrile (solvent B). The gradient conditions were as follows: 0-10 min, 13% B, 10-20 min, 41.5% B, 20-25 min, 70% B, 25-35 min, 10% B. The total run time was 35 min. The column was equilibrated for 10 min prior to each analysis at 25°C. The flow rate was 0.5 ml/min and the injection volume was 10 µl. The data acquisition and preprocessing were carried out with Chemstation for LC (Agilent). Oleuropein was monitored at a wavelength of 280 nm. The peak was identified on the basis of a comparison of the retention time and UV spectrum with an oleuropein standard.
Analysis of GBM cell lines
Cell line maintenance
The T98G, U-138MG and U-187MG human GBM cell lines were provided by the American Type Culture Collection (ATCC; Rockville, USA). The cells were grown in Dulbecco’s Modified Eagle’s Medium-F12 (DMEM-F12; HyClone, Utah, USA) containing L-glutamine supplemented with 10% fetal bovine serum (FBS, BIOCHROME, Berlin, Germany), 1 mM sodium pyruvate, 100 µg/ml streptomycin and 100 U/ml penicillin and incubated in a humidified 5% CO2 incubator at 37°C.
Determination of cytotoxicity and cell viability
As described previously for T98G cells, the cytotoxicity of ten different doses of OLE in U138MG and U-87MG was assayed using a Thoma chamber with 0.4% Trypan blue after 24 and 48 h incubation; a cell proliferation kit (WST-1, Roche Applied Sciences, Mannheim, Germany) was used to evaluate the viability of the cells [18]. All analyses were performed in quadruplicate. The results were expressed as a percentage of the untreated control. The absorbance of the untreated control cells was set to 100%, and the absorbance of OLE-treated cells was measured as the surviving percentage. The following formula was used to calculate the percent inhibition:
% inhibition =(1 - absorbance of sampie/absorbance of control) × 100
Measurement of the effect of OLE on apoptosis
Annexin-V-FITC/PI analysis
The percentage of apoptotic T98G, U-138MG and U87MG cells was assessed using a Annexin V-FITC/propidium iodide (PI) binding kit, (BD Pharmagen™ San Jose, CA, USA) using flow cytometry (FACSCanto, Becton Dickinson, USA) before and after culture with or without the addition of OLE according to the manufacturer’s specifications. Cells that stained only for Annexin-V were considered to be in early apoptosis, and those that stained for both Annexin-V and PI were considered to be in late apoptosis or necrosis. Cells that were negative for Annexin-V/PI were considered to be viable. Annexin-V-FITC/PI analysis was duplicated for all cell lines and OLE and/or TMZ treatments.
TUNEL assay
For TUNEL assays, T98G, U138MG and U-87MG cells were cultured in 4well chamber slides (Millicell EZ Slide, Millipore, USA). After 24 h of culture with or without the OLE, an ApopTag In Situ Apoptosis Detection Kit (Intergen, Purchase, NY, USA) was used to detect apoptotic cells according to the manufacturer’s instructions. Briefly, the cells were fixed with 1% paraformaldehyde, permeabilized by 0.3% Triton X-100, and then washed there times with PBS. DNA breaks were labeled by incubation (1 h, 37°C) with terminal deoxynucleotidyltransferase and a nucleotide mixture containing fluorescein isothiocyanate-conjugated dUTP. The cell nuclei were stained with PI, and the TUNEL-positive and total nuclei were observed under a fluorescent microscope (Nikon, Japan). More than 1500 nuclei were counted per field, and the experiment was repeated two times.
Determination of synergy or antagonism between OLE and TMZ
To determine the effective doses of TMZ (Sigma, USA) for U-138MG and U-87MG cell lines, the cells were seeded after standard trypsinization at a density of 2×104 cells/well in 96-well plates. After 24 h of culture in normal growth medium, the cells were exposed to graded concentrations of TMZ (300-500 µM), and WST-1 analyses were performed after 24 h and 48 h. For the T98G cell line, the effective doses of TMZ were determined previously [18].
To evaluate the synergistic or antagonistic effect of OLE on TMZ, cells were seeded at a density of 2×104 cells/well in 96-well plates. In the proliferation assays, the cells were exposed to two different concentrations of OLE, a dose of TMZ that was found to be effective in T98G, U-138MG and U-87MG cells, the combination of TMZ and OLE, or H2O2 at 24 or 48 h after plating. The dose-response curves of cell viability after treatment with OLE alone, TMZ alone or the combination for 24 h were analyzed. The nature of the interaction between TMZ and OLE was evaluated by WST-1 analysis. All analyses were performed in quadruplicate for all cell lines.
Analyses of GSCs
Characteristics of GSCs
Primary tumor cells from five patients, whose tumors were identified as GSC (+) based on CD133 and Nestin positivity and high mRNA expression levels of ITGA (NM_181501), VIM (NM_003380), CD44 (NM_000610) and OCT4 (NM_002701) in our previous study [19] were used in this study. The patients (4 women and 1 man) were aged 48-73 years at the time of diagnosis, and the median age was 58.6 ± 4.6 years. The primary tumors from 2 patients were localized to the brain’s parietal region, 2 primary tumors were located in the frontal region in, and 1 was localized to the thalamic region. Although none of the patients responded to TMZ treatment, and the median survival was 8.20 ± 2.4 years, the mean MGMT methylation rates of their tumors were 50% ± 0.0 [19]. The study was approved by the local ethics committee (2011-17/07) and conformed to the ethical standards of the Helsinki Declaration.
miRNA expression profiles of GSCs treated with OLE or TMZ
To evaluate the ability of OLE to modify miRNA expression in GSCs, 5 GSCs lines were treated with one of the following for 24 h: 1 or 2 mg/ml of OLE, 450 µM TMZ, or a combination of OLE and 450 µM TMZ in 6-well culture plates. The GSCs were then subjected to total RNA extraction using RNeasy kits (Qiagen, Germantown, MD). All of the RNA samples were assessed for RNA quantity and quality using a NanoDrop 2000 spectrophotometer. The protein and chemical contamination was determined by measuring the 260:280 and 260:230 ratios for each RNA sample. RNA samples with 260:280 ratios between 1.8 and 2.0, 260:230 ratios >1.8 and a total concentration between 200 and 400 ng/µl were selected for cDNA synthesis. Total RNA (5 ng) from the cells was rever-se-transcribed using the RT2 miRNA First Strand Kit (Qia-gen, Germantown, Maryland, USA). The samples were analyzed for the presence and differential expression of 5 cancer-related miRNAs, miR-181b (MIMAT0000257), miR153 (MIMAT0000439), miR-137 (MIMAT0000429), miR-145 (MIMAT0000437) and let-7d (MIMAT0000065), which are modulated by OLE in T98G cells according to a previous study [18] using RT2 miRNA primer assays (RT2 Profiler; Qiagen, Frederick Md, USA). The miRNA expression analyses were duplicated for each sample. The thermal cycling conditions for all assays were as follows: 95°C for 10 min, 45 cycles at 95°C for 15 s, and 60°C for 30 s, followed by melting curve analysis in a LightCycler 480II (Roche Diagnostics, Indianapolis, USA). The RNA input of miRNAs was normalized to the endogenous control SNORD 48, and the input of protein-coding genes was normalized to the TATA-binding protein. The initial copy number of the samples and the threshold cycle (Ct) for miRNA expression were determined using the Light Cycler 480II software (Roche Diagnostics, Indianapolis, USA). A miRNA reverse transcription control assay was used to test the efficiency of the miScript II Reverse Transcription Kit reaction; a specific primer set was used to detect a template synthesized from the kit’s built-in miRNA external RNA control. Positive PCR control assays were used to test the efficiency of the polymerase chain reaction chemistry and the instrument using a predispensed artificial DNA sequence and a primer set designed to detect this sequence. The 2^-ΔCt method was used to calculate the fold changes in miRNA expression between the tested samples [20].
Evaluation of the expression level of miRNA target genes
To evaluate the expression of the target genes of the significantly altered miRNAs depending on OLE treatment, RNAs were reverse transcribed using a cDNA Synthesis Kit (New England Biolabs, UK). The samples were then analyzed using RT-qPCR to profile the expression levels of TP53 (NM_000546), OCT4 (NM_002701), SOX2 (NG_009080.1), BCL2 (NM_000633) c-myc (NM_002467) and c-Met (NM_000245); we also evaluated the expression level of the human Beta Actin (ACTB) housekeeping gene. Gene expression analyses were performed in duplicate for each sample. Only samples with Ct values less than 35 were included in further analyses. PCR was carried out in a 20-µl reaction mixture that contained 5 µl of cDNA as a template, 10 µM specific oligonucleotide primer pairs, and SYBR Green qPCR master Mix (Qiagen, Germantown, MD). The cycle parameters were as follows: 95°C for 10 min, 45 cycles at 95°C for 15 s and 60°C for 60 s, followed by melting curve analysis in the LightCycler 480II (Roche Diagnostics, USA). The absence of genomic DNA contamination was confirmed by performing a no reverse transcription control with RNA samples using an ACTB RT-qPCR primer assay. The initial copy number of the samples and the threshold cycle (Ct) for mRNA expression was determined using the Light Cycler 480II software (Roche Diagnostics, Indianapolis, USA). The 2^-ΔCt method was used to calculate the fold change in mRNA expression between the tested samples [20].
Statistical analyses
One Way ANOVA and Tukey’s analyses were used to determine the statistical significance of the WST-1 data. RT² Profiler™ PCR Array Data Analysis was used to determine the statistical significance of changes in miRNA and mRNA expression. Values of P < 0.05 were considered to be statistically significant.
Results
Active compound in OLE
The amount of oleuropein was calculated as 9.36 ± 0.22 mg/ml (n=2) in standardized OLE according to HPLC/DAD analyses. The chromatogram of standardized OLE is shown in Figure 1.
Figure 1.
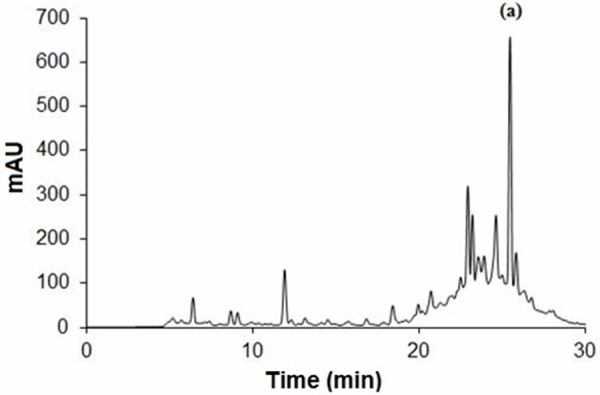
The HPLC chromatogram of standardized oleuropein. Oleuropein was present at 25.503 min. a: Oleuropein.
The effects of OLE on the viability and proliferation of U-138MG and U-87MG cells
U-138MG and U-87MG cells were seeded at a density of 2×104 cells/well in 96-well plates. After 24 h, the cells were treated with different concentrations of OLE. WST-1 assays were performed to study the proliferative and cytotoxic effects of OLE. The inhibitory concentration at which 50% of the cells die was identified (IC50). OLE decreased the viability of U-138MG and U-87MG cells. Optimal activity was observed at day 1. The percentage decrease in the proliferation of U-138MG cells at 1 and 2 mg/ml ranged from 55 to 62%, and the decrease in the proliferation of U-87MG cells at 1 and 2 mg/ml ranged from 43% to 48% at 24 h (Figure 2). When U-138MG and U87-MG cells were treated with H2O2, we observed 89.9% and 87% reductions in proliferation, respectively.
Figure 2.
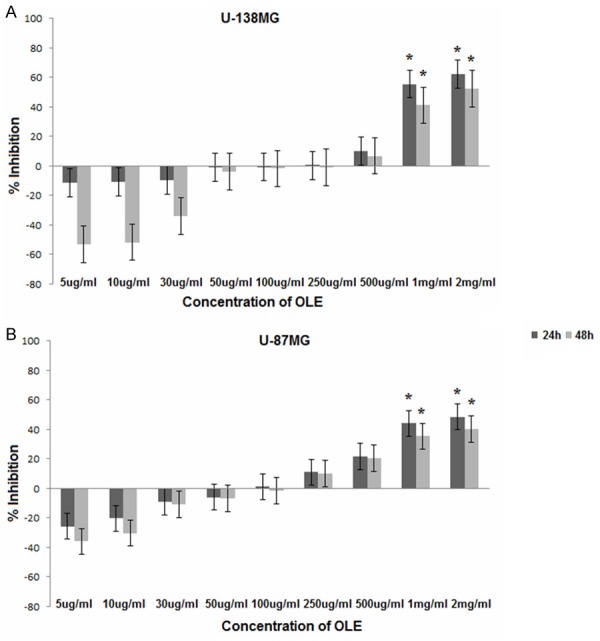
Inhibition of cell viability at different concentrations of OLE. (A) U-138MG (B) U-87MG; * P < 0.05; Evaluated using one-way ANOVA and Tukey’s tests using SPSS 16.00 software for Windows (IBM, Chicago, IL).
OLE induces apoptosis in the T98G, U-138MG and U-87MG cell lines
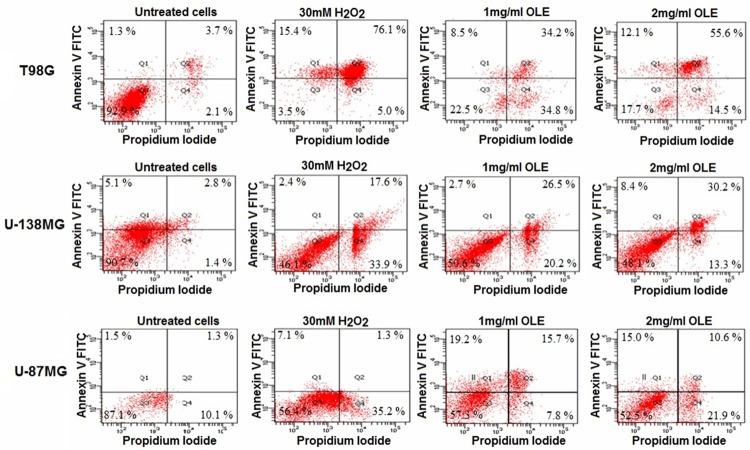
To determine the type of cell death (apoptosis or necrosis) induced by OLE, the cells were subjected to Annexin-V-FITC/PI and TUNEL assays before and after OLE treatment. T98G, U138MG and U-87MG cells were treated with 1 mg/ml and 2 mg/ml OLE for 24 hours. Based on Annexin V analyses, percentage of apoptotic T98G, U-138MG and U-87MG cells after treatment with 1 mg/ml OLE were 42.7%, 29.2% and 34.9%, respectively; after 2 mg/ml OLE treatment, 67.7%, 38.6% and 25.6% of cells were apoptotic, respectively (Figure 3). Furthermore, in TUNEL assays, the percentages of apoptotic cells observed in T98G, U-138MG and U-87MG cells treated with 1 mg/ml OLE were 43.5%, 28.5% and 36%, respectively; 2 mg/ml OLE resulted in 67%, 39.5% and 24.5% apoptosis, respsectively, compared to untreated cells (Figure 4).
Figure 3.
Apoptosis induced by 1 mg/ml and 2 mg/ml OLE at 24 h by Annexin V-FITC/PI assay.
Figure 4.
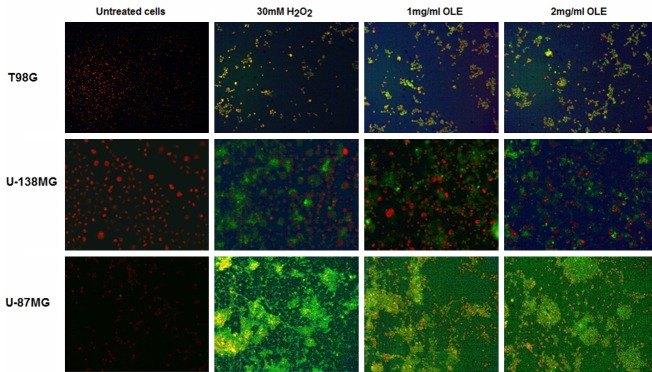
TUNEL assay performed 24 hours after the addition of OLE (x 10). The first column shows untreated control cells. The second column shows H2O2-treated (30 mM) positive control cells. The third and fourth columns show cells treated with 1 mg/ml and 2 mg/ml OLE, respectively.
The role of OLE on the effectiveness of TMZ
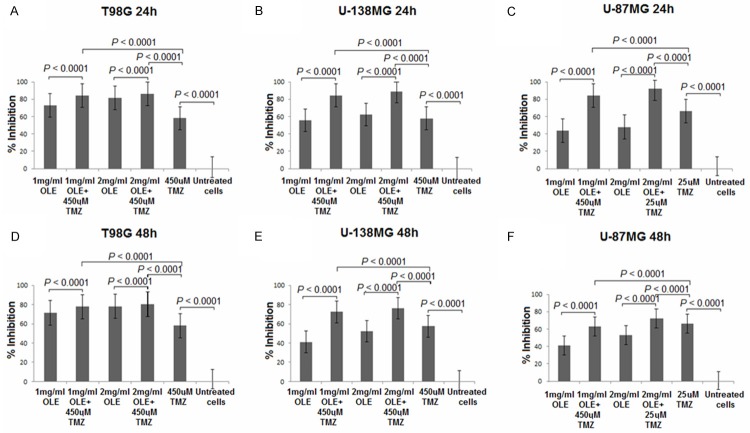
First, the cells were treated with a wide range of doses of TMZ, and WST-1 analyses was performed to define the commonly effective TMZ doses in T98G, U-138MG and U-87MG cells at 24 h and 48 h. The percentage decrease in proliferation of cells treated with 450 µM TMZ were 58.07%, 57.99% and 86.34% at 24 h and 57.90%, 57.36% and 83.00% at 48 h, respectively (Figure 5). Because all of the tested concentrations of TMZ were highly cytotoxic for U-87MG cells, 25 µM TMZ was used for the U-87MG cell line in the remainder of the study based on a literature search [21]. When U87MG cells were treated 25 µM TMZ, we observed a 66.25% decrease in proliferation at 24 h of incubation and a 66.31% decrease at 48 h of incubation.
Figure 5.
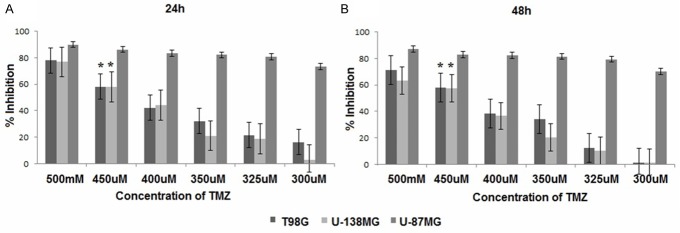
Antitumor effects of TMZ against T98G, U-138MG and U87MG cell lines. *P < 0.05; Evaluated using one-way ANOVA and Tukey’s tests using SPSS 16.00 software for Windows (IBM, Chicago, IL).
Second, we evaluated the potential interactions between OLE and TMZ in T98G, U-138 MG and U-87MG cell lines in the absence or presence of low-toxicity concentrations of OLE. WST-1 assays were performed after 24 h and 48 h of incubation, and the drug combinations were evaluated as reductions in proliferation (Figure 6). When T98G, U-138MG and U-87MG cells were treated with TMZ in the presence of 1 mg/ml OLE, the reduction in proliferation was determined to be 84.07, 84.47, and 84.18%, respectively; when the cells were treated with TMZ in the presence of 2 mg/ml OLE, the reduction in proliferation was determined to be 86.05, 88.70 and 92.12% at 24 h, respectively, compared to an untreated culture. This result indicated that OLE has synergistic effects with TMZ.
Figure 6.
Effect of OLE and TMZ concentration on the viability of T98G, U-138MG and U-87MG cells. Evaluated using one-way ANOVA and Tukey’s tests using SPSS 16.00 software for Windows (IBM, Chicago, IL).
The role of OLE in the modulation of miRNA expression in TMZ-resistant GSCs
Based on our analyses in the T98G, U-138MG and U-87MG cell lines, the optimal activity of OLE was determined to occur at concentrations of 1 mg/ml and 2 mg/ml on the 1st day of incubation. Therefore, we evaluated the effect of 1 mg/ml and 2 mg/ml OLE on miRNA expression in GSCs. The expression levels of mir-181b, miR-153, miR-137, miR-145 and let-7d were evaluated in 5 GSC lines treated with OLE and 450 µM TMZ. The results from untreated GSCs samples were compared to those obtained from cells treated with 1 mg/ml OLE, 2 mg/ml OLE, 1 mg/ml OLE and 450 µM TMZ, 2 mg/ml OLE and 450 µM TMZ. The expression of miR-137 was significantly up regulated after treatment with 1 mg/ml OLE in combination with 450 µM TMZ (p=0.03). After treatment with 2 mg/ml OLE and TMZ, miR-153, miR-145 and miR-137 were also significantly up regulated (p < 0.05; Table 1). In addition, when the samples were treated with OLE and TMZ, the expression level of miR-145 significantly increased compared to samples that were treated with TMZ alone (p < 0.05; Table 2). These results indicated that OLE and TMZ synergistically cause changes in the expression of these miRNAs in GSCs (Figure 7).
Table 1.
Differential expression of miRNAs in GSCs treated with 450 μM TMZ in the presence or absence of 1 mg/ml and 2 mg/ml OLE
| mir-181b | miR-153 | miR-145 | miR-137 | let-7d | |
|---|---|---|---|---|---|
| Untreated | |||||
| 2^(-Avg.(Delta(Ct)) | 0.412367 | 0.000379 | 0.007922 | 0.000518 | 0.026388 |
| 1 mg/ml OLE | |||||
| 2^(-Avg.(Delta(Ct)) | 0.544876 | 0.004796 | 0.080772 | 0.004796 | 0.17776 |
| Fold Change | 1.3213 | 12.6582 | 1.1965 | 9.2663 | 6.7365 |
| 95% CI | (0.13, 2.51) | (0.00001, 42.64) | (0.00001, 37.00) | (0.00001, 31.01) | (0.00001, 32.83) |
| * P value | 0.6212 | 0.1375 | 0.1876 | 0.1754 | 0.6828 |
| 2 mg/ml OLE | |||||
| 2^(-Avg.(Delta(Ct)) | 0.713013 | 0.011296 | 0.034722 | 0.011296 | 0.136408 |
| Fold Change | 1.7291 | 29.8157 | 4.3832 | 21.8264 | 5.1694 |
| 95% CI | (0.05, 3.41) | (0.00001, 90.40) | (0.00001, 17.36) | (0.00001, 65.61) | (0.00001, 27.89) |
| * P value | 0.3557 | 0.0088 | 0.4323 | 0.0155 | 0.3847 |
| 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 0.088388 | 0.016289 | 0.080214 | 0.016289 | 0.046974 |
| Fold Change | 0.2143 | 42.9921 | 10.1261 | 31.4721 | 1.7802 |
| 95% CI | (0.00001, 0.43) | (0.00001, 132.10) | (0.00001, 37.69) | (0.00001, 95.91) | (0.00001, 8.58) |
| * P value | 0.4428 | 0.0118 | 0.2239 | 0.0155 | 0.2864 |
| 1 mg/ml OLE + 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 9.958994 | 0.028676 | 0.056563 | 0.018149 | 0.004588 |
| Fold Change | 24.1508 | 75.6884 | 7.1404 | 35.0660 | 0.1739 |
| 95% CI | (0.00001, 105.23) | (0.00001, 258.73) | (0.00001, 26.75) | (0.00001, 113.99) | (0.00001, 0.82) |
| * P value | 0.0842 | 0.1774 | 0.3209 | 0.0323 | 0.1360 |
| 2 mg/ml OLE + 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 1.503161 | 0.031034 | 0.583984 | 0.017027 | 0.143786 |
| Fold Change | 3.6452 | 81.9118 | 73.7208 | 32.8996 | 5.4490 |
| 95% CI | (0.72, 6.57) | (0.00001, 257.85) | (0.00001, 259.87) | (0.00001, 111.76) | (0.00001, 25.00) |
| * P value | 0.0227 | 0.0438 | 0.0005 | 0.0146 | 0.3762 |
Evaluated with the Independent sample T-test using RT2 Profiler PCR Array Data Analysis.
Table 2.
Fold differences in miRNAs between GSCs treated with 450 μM TMZ alone and in combination with 1 mg/ml and 2 mg/ml OLE
| mir-181b | miR-153 | miR-145 | miR-137 | let-7d | |
|---|---|---|---|---|---|
| 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 0.088388 | 0.016289 | 0.080214 | 0.016289 | 0.046974 |
| 1 mg/ml OLE + 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 9.958994 | 0.028676 | 0.056563 | 0.018149 | 0.004588 |
| Fold Change | 112.6732 | 1.7605 | 0.7051 | 1.1142 | 0.0977 |
| 95% CI | (0.00001, 492.25) | (0.00001, 4.51) | (0.00001, 1.79) | (0.00001, 2.59) | (0.00001, 0.26) |
| * P value | 0.0829 | 0.3133 | 0.7102 | 0.4842 | 0.1542 |
| 2 mg/ml OLE + 450 µM TMZ | |||||
| 2^(-Avg.(Delta(Ct)) | 1.503161 | 0.031034 | 0.583984 | 0.017027 | 0.143786 |
| Fold Change | 17.0063 | 1.9053 | 7.2803 | 1.0454 | 3.061 |
| 95% CI | (2.54, 31.47) | (0.00001, 3.99) | (0.00001, 15.22) | (0.00001, 2.68) | (0.00001, 7.35) |
| * P value | 0.0030 | 0.2173 | 0.0040 | 0.3944 | 0.4846 |
Evaluated with an independent sample T-test using RT2 Profiler PCR Array Data Analysis.
Figure 7.
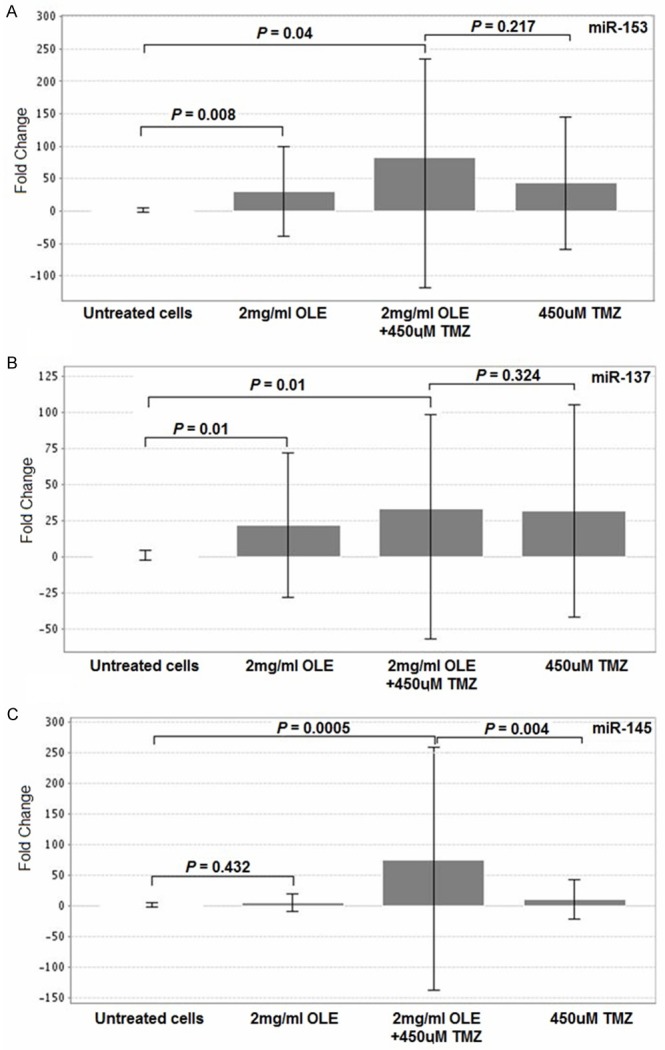
Changes in miRNA expression levels in GSCs after OLE or TMZ treatment. Evaluated with independent sample T-tests using RT2 Profiler PCR Array Data Analysis.
Significantly altered miRNAs fold changes were compared between GSCs treated 1 mg/ml or 2 mg/ml OLE and 450 µM TMZ combinations and 450 µM TMZ depend on their survival individually. All of the tested miRNAs had higher expression levels after OLE-TMZ treatment than treating with TMZ alone (Table 3).
Table 3.
Fold changes of GSCs in comparison with OLE-TMZ interactions and TMZ
| GSCs 1 | GSCs2 | GSCs 3 | GSCs 4 | GSCs 5 | |
|---|---|---|---|---|---|
| disease age | 58 | 73 | 64 | 50 | 48 |
| survival of cases | 5 months | > 48 months | 6 months | 16 months | 12 months |
| Fold change in compare to TMZ | |||||
| 1 mg/ml OLE+ TMZ | |||||
| miR-137 | 6.11 | 0.90 | 0.35 | 0.66 | 1.35 |
| 2 mg/ml OLE + TMZ | |||||
| miR-137 | 6.23 | 0.12 | 24.08 | 0.24 | 0.28 |
| miR-153 | 8.51 | 0.51 | 16.56 | 0.58 | 0.59 |
| miR-145 | 6.45 | 3.36 | 63.12 | 1.29 | 11.55 |
The target genes of the miRNAs that were significantly altered after OLE treatment (miR-145, miR-153 and miR-137) were identified by searching recent literature. The function of these genes was defined according to the NCBI-Gene database. The validated targets for these miRNAs are involved in signaling pathways related to cellular processes that include apoptotic regulation (Table 4).
Table 4.
Validated target genes and the related pathways associated with each miRNA
| miRNA | Target genes | |||
|---|---|---|---|---|
|
| ||||
| Symbol | Full name | Description (NCBI-Gene database) | Reference | |
| miR-153 | BCL2 | B-cell CLL/lymphoma 2 | This gene encodes an integral outer mitochondrial membrane protein that blocks the apoptotic death | [41] |
| Irs2 | insulin receptor substrate 2 | This gene encodes the insulin receptor substrate 2, a cytoplasmic signaling molecule that mediates effects of insulin, insulin-like growth factor 1, and other cytokines by acting as a molecular adaptor between diverse receptor tyrosine kinases and downstream effectors. The product of this gene is phosphorylated by the insulin receptor tyrosine kinase upon receptor stimulation, as well as by an interleukin 4 receptor-associated kinase in response to IL4 treatment | [41] | |
| MCL1 | myeloid cell leukemia sequence 1 | This gene encodes an anti-apoptotic protein, which is a member of the Bcl-2 family | [41] | |
| Akt1 | v-akt murine thymoma viral oncogene homolog 1 | AKT1 and the related AKT2 are activated by platelet-derived growth factor. In the developing nervous system AKT is a critical mediator of growth factor-induced neuronal survival. Survival factors can suppress apoptosis in a transcription-independent manner by activating the serine/threonine kinase AKT1, which then phosphorylates and inactivates components of the apoptotic machinery | [41] | |
| miR-137 | RTVP-1 | GLI pathogenesis-related 1 | This gene encodes a protein with similarity to both the pathogenesis-related protein (PR) superfamily and the cysteine-rich secretory protein (CRISP) family. Increased expression of this gene is associated with myelomonocytic differentiation in macrophage and decreased expression of this gene through gene methylation is associated with prostate cancer. The protein has proapoptotic activities in prostate and bladder cancer cells | [49] |
| c-MET | met proto-oncogene | The proto-oncogene MET product is the hepatocyte growth factor receptor and encodes tyrosine-kinase activity | [48] | |
| BCL2 | B-cell CLL/lymphoma 2 | This gene encodes an integral outer mitochondrial membrane protein that blocks the apoptotic death | [49] | |
| SOX2 | SRY (sex determining region Y)-box 2 | This intronless gene encodes a member of the SRY-related HMG-box (SOX) family of transcription factors involved in the regulation of embryonic development and in the determination of cell fate. The product of this gene is required for stem-cell maintenance in the central nervous system | [46] | |
| OCT-4 | POU class 5 homeobox 1 | This gene encodes a transcription factor containing a POU homeodomain that plays a key role in embryonic development and stem cell pluripotency. Aberrant expression of this gene in adult tissues is associated with tumorigenesis | [46] | |
| miR-145 | TP53 | tumor protein p53 | This gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism | [54] |
| c-MYC | v-myc avian myelocytomatosis viral oncogene homolog | The protein encoded by this gene is a multifunctional, nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation. It functions as a transcription factor that regulates transcription of specific target genes | [53] | |
| SOX9 | SRY (sex determining region Y)-box 9 | The protein encoded by this gene recognizes the sequence CCTTGAG along with other members of the HMG-box class DNA-binding proteins. It acts during chondrocyte differentiation and, with steroidogenic factor 1, regulates transcription of the anti-Müllerian hormone (AMH) gene | [53] | |
| BCL2 | B-cell CLL/lymphoma 2 | This gene encodes an integral outer mitochondrial membrane protein that blocks the apoptotic death | [55] | |
| CTGF | connective tissue growth factor | The protein encoded by this gene is a mitogen that is secreted by vascular endothelial cells. The encoded protein plays a role in cell adhesion in many cell types, and is related to platelet-derived growth factor | [50] | |
| SOX2 | SRY (sex determining region Y)-box 2 | This intronless gene encodes a member of the SRY-related HMG-box (SOX) family of transcription factors involved in the regulation of embryonic development and in the determination of cell fate. The product of this gene is required for stem-cell maintenance in the central nervous system | [52] | |
| OCT-4 | POU class 5 homeobox 1 | This gene encodes a transcription factor containing a POU homeodomain that plays a key role in embryonic development and stem cell pluripotency. Aberrant expression of this gene in adult tissues is associated with tumorigenesis | [52] | |
The expression levels of TP53, OCT-4, SOX2, BCL2, c-myc and c-Met, target genes of the significantly altered miRNAs after OLE treatment, were evaluated in 5 GSC lines treated with OLE and 450 µM TMZ. The fold changes of the samples in untreated GSCs were compared to those of cells treated with 1 mg/ml OLE, 2 mg/ml OLE, 1 mg/ml OLE and 450 µM TMZ, 2 mg/ml OLE and 450 µM TMZ (Table 5).
Table 5.
Changes in the expression levels of miRNA target genes in GSCs treated with 450 μM TMZ in the presence or absence of 1 mg/ml and 2 mg/ml OLE
| TP53 | OCT-4 | SOX2 | BCL2 | c-myc | c-met | |
|---|---|---|---|---|---|---|
| Untreated | ||||||
| 2^(-Avg.(Delta(Ct)) | 0.014418 | 0.02039 | 0.184795 | 0.042926 | 0.014458 | 0.011454 |
| 2 mg/ml OLE | ||||||
| 2^(-Avg.(Delta(Ct)) | 0.002418 | 0.002828 | 0.013831 | 0.010352 | 0.002418 | 0.002418 |
| Fold Change | 0.1677 | 0.1387 | 0.0748 | 0.2411 | 0.1672 | 0.2111 |
| 95% CI | (0.00001, 0.58) | (0.00001, 0.46) | (0.00001, 0.29) | (0.00001, 0.98) | (0.00001, 1.01) | (0.00001, 1.09) |
| * P value | 0.314169 | 0.227929 | 0.175984 | 0.351612 | 0.341963 | 0.302438 |
| 2 mg/ml OLE + 450 µM TMZ | ||||||
| 2^(-Avg.(Delta(Ct)) | 0.004743 | 0.002418 | 0.011502 | 0.007557 | 0.000975 | 0.000975 |
| Fold Change | 0.329 | 0.1186 | 0.0622 | 0.176 | 0.0675 | 0.0851 |
| 95% CI | (0.00001, 1.10) | (0.00001, 0.42) | (0.00001, 0.26) | (0.00001, 0.70) | (0.00001, 0.39) | (0.00001, 0.41) |
| * P value | 0.329112 | 0.220026 | 0.187845 | 0.353684 | 0.341256 | 0.29579 |
| 450 µM TMZ | ||||||
| 2^(-Avg.(Delta(Ct)) | 0.121751 | 0.121751 | 0.133046 | 0.155609 | 0.121751 | 0.121751 |
| Fold Change | 8.4444 | 5.9711 | 0.72 | 3.625 | 8.421 | 10.6295 |
| 95% CI | (0.00001, 28.07) | (0.00001, 20.13) | (0.00001, 2.91) | (0.00001, 14.44) | (0.00001, 50.50) | (0.00001, 53.82) |
| *P value | 0.209763 | 0.224161 | 0.23702 | 0.484775 | 0.371175 | 0.681602 |
Evaluated with an independent sample T-test using RT2 Profiler PCR Array Data Analysis.
Discussion
Despite progress in the treatment of GBM, these tumors are still incurable. The chemopreventive activity and preclinical antitumor effects of phytochemicals may herald new therapeutic approaches for patients with chemoresistance [22,23]. Olea europaea, which is native to the Mediterranean region, is one of these medicinal herbs [24]. The pharmacological properties of the oil, fruit and leaves of Olea europaea have been recognized as important components of medicine and a healthy diet due to their phenolic content [25]. In general, the most prominent phenolic compound of Olea europaea is oleuropein. Oleuropein can be found in all parts of the plant, but its concentration varies greatly between the various tissues [24]. In the current study, we evaluated the anti-carcinogenic activity of the leaves of this plant. Based on HPLC analyses, the standardized OLE that was used in this study contained 9.36 ± 0.22 mg/ml oleuropein.
In a previous study, we demonstrated that OLE modulates the expression of miR-181b, miR-153, miR-145, miR-137, and let-7d, which are related to anticancer activity and the response to TMZ in the T98G GBM cell line. The main conclusion drawn in this study was that OLE demonstrates an anti-proliferative effect in the T98G cell line and that the combination of OLE and TMZ increases the responsiveness of these cells to TMZ-mediated toxicity [18]. T98G is a TMZ-resistant GBM cell line that was derived from a 61-year-old Caucasian man; T98G cells have a fibroblast-like morphology and are heterozygous for MGMT methylation. Although this study described the potential anti-cancer effect of OLE in GBM tumors, there is a lack of knowledge about the effects of OLE on GBM cells with different characteristics that affect tumor aggressiveness, such as TMZ response, the morphology of the cells, and the age of the person from whom the tumor cells are derived. Thus, in the current study, we first evaluated the cytotoxic effect of OLE on the U-138MG and U-87MG cell lines using WST-1 analyses. U-138MG cells are highly resistant to TMZ. They are polygonal and were derived from a 47-year-old Caucasian male GBM patient with an unmethylated MGMT gene [http://www.lgcstandards-atcc.org/?geo_country=tr, 21]. Moreover, U-87MG cells are TMZ-sensitive and have an epithelial morphology. U-87MG cells were derived from a 44-year-old Caucasian male GBM patient whose MGMT gene is methylated [http://www.lgcstandards-atcc.org/?geo_country=tr, 21]. According to WST-1 analyses, the optimal activity of OLE was observed after 24 h. Because oleuropein, the active component of OLE, is a type of flavonoid, OLE may not maintain its anti-carcinogen activity for more than 24 h; Kurisawa et al demonstrated that flavonoids are only active for a few hours in the body [26]. Therefore, 24-hour OLE treatments were performed in the subsequent analyses. In addition, the IC50 of OLE for both U-138MG and U-87MG cells was 1 and 2 mg/ml; these results are similar to those obtained in T98G cells in a previous study [18]. At concentrations of 1 mg/ml and 2 mg/ml, OLE caused a significant decrease in the proliferation of U-138MG cells (ranging from 55% to 62%) and U-87 MG cells (ranging from 43% to 48%) at 24 h and 48 h (p < 0.05). Although the T98G, U-138MG and U-87MG cell lines differ with respect to their MGMT methylation status, the IC50s of OLE in these cell lines were similar. Therefore, we suggest that OLE may cause cell death via an MGMT-independent pathway.
To evaluate the molecular mechanism of the action of OLE on tumor cell viability, Annexin-V-FITC/PI and TUNEL assays were performed. These analyses verified the similarity of the cytotoxic effect of OLE in T98G, U-138MG and U-87MG cells. However, based on the Annexin-V-FITC/PI assay, both 1 mg/ml and 2 mg/ml OLE cause cell death via late apoptosis or necrosis. Mijatovic et al previously reported similar data regarding the action of OLE in the B16 melanoma cell line. They suggest that a continuous and intensive decrease of Bim and a temporary decrease in p53 triggered by OLE, together with Bcl-XL amplification, were most likely responsible for the inactivation of caspase-3, the inhibition of caspase-3, -8 and -9 transcription and the subsequent deviation from the typical apoptotic process with a shift towards necrosis [6]. In a recent study, Reyes-Zurita et al reported an effect of maslinic acid, a phenolic component of OLE, on the extrinsic, and later the intrinsic, apoptotic pathways in Caco-2 human colon-cancer cells using Annexin-V-FITC/PI staining. Additionally, the induction of apoptosis was linked to higher levels of Bid cleavage and the early activation of caspase-8 and caspase-3 [27]. Our findings support the results of Mijatovic et al; treating cells with only the phenolic compounds of Olea europaea may alter the direction of cell death toward apoptosis. Although necrosis is not generally a preferred type of cell death, in our previous study, we demonstrated a very low cytotoxic effect of 1 mg/ml and 2 mg/ml OLE (16 and 22%, respectively) on fresh human mononuclear lymphocytes as a non-tumor cell model [18]. Therefore, we suggest that these findings strengthen the evidence supporting the potential of OLE as a candidate in further anti-cancer drug studies.
Previously, evaluating the effect of OLE in combination with TMZ, we analyzed the effect of TMZ on T98G, U-138MG and U-87MG cell lines. Similar to previous studies, we indicated that the IC50 of TMZ for cell viability in the T98G and U-138MG cell lines is higher than that of U-87MG cells [21]. 450 µM TMZ caused 58.07% and 57.99% inhibition of T98G and U-138MG cells, respectively, and 25 µM TMZ caused 66.25% inhibition of U-87MG cells in 24 h. Evaluating the potential interaction between OLE and TMZ, we found that while 1 mg/ml OLE caused 84.07, 84.47, and 84.18% inhibition in T98G, U-138MG and U-87MG cells, respectively, 2 mg/ml OLE caused 86.05, 88.70 and 92.12% inhibition, respectively in combination with TMZ. Therefore, we suggest that OLE may synergistically affect the toxicity of TMZ. The ability of OLE to modulate the effectiveness of some cytostatic drugs was shown in a previous study [6]. Mijatovic et al demonstrated the synergistic effect of OLE on cisplatin and paclitaxel. However, they also reported an antagonistic interaction between OLE and TMZ, in contrast to our present findings [6]. One of the TMZ resistance mechanisms relies on high levels of O6-alkylguanine DNA alkyltransferase, a DNA repair protein that selectively removes the methyl adduct from the MGMT gene’s O6 position of guanine [28]. Thus, the expression level of MGMT in tumors may affect the success of TMZ therapy. B16 mouse melanoma cells do not show MGMT expression [29]. Thus, B16 cells may respond well to TMZ. Similarly, in the current study, while the combination of 2 mg/ml OLE and TMZ inhibited cell proliferation 1.48- and 1.52-fold more than TMZ alone in T98G and U-138MG cells, this inhibition rate was 1.39-fold more in compare to TMZ alone in U-87MG cells, which have methylated MGMT. We therefore suggest that although the action of OLE may be independent from the expression status of MGMT, the interaction between OLE and TMZ may be associated with the level of MGMT. Nonetheless, although MGMT expression is a well-known mechanism underlying the response to TMZ, recent studies have reported that it is not the only mechanism. While patients with unmethylated tumors experience unexpected favorable outcomes after receiving radiochemotherapy, some patients with a methylated promoter do not benefit from concomitant and adjuvant TMZ treatment [30,31]. Additionally, the majority of patients with GSCs cannot benefit from TMZ as needed, and no correlation was reported between stemness properties and MGMT protein expression levels in GBM tumors [32]. In our previous study, the survival time of patients with GSCs was significantly shorter than that of patients without GSCs, although MGMT methylation rate of GSC (+) tumors was greater than that of GSC (-) tumors. Thus, there may be other mechanisms such as miRNAs that play crucial roles in all steps of tumorigenesis, function in response to TMZ therapy. To this end, we determined the relation between TMZ resistance and altered expression of miR-181b and miR-455-3p in GSCs [19]. For this reason, evaluating the TMZ response in GSCs with this approach has become remarkable.
In the current study, we also evaluated the effect of OLE and the combination of OLE and TMZ on miRNA expression level in GSCs. According to our previous study, the expression levels of miR-181b, miR-153, miR-145, miR-137, and let-7d were significantly altered in T98G cells after OLE treatment. In addition, the combination of OLE and TMZ caused a significant increase in the expression levels of these miRNAs compared to TMZ treatment alone [18]. The levels of miR-153, miR-137 and miR-145 in GSCs is also significantly modulated after treatment with 2 mg/ml OLE in the present study (p=0.008; 0.015 and 0.432, respectively). One of these miRNAs, miR-153, has a tumor suppressor function in various cancers such as breast, ovarian, prostate, colon, rectal and GBM [33-39]. In a recent study, the expression level of miR-153 was observed to differ in GSCs and their more differentiated progeny. Based on this study, the expression of miR-153 was reported to be lower in CD133 (+) tumors than in CD133 (-) tumors [40]. In the present study, treating GSCs with 450 µM TMZ caused a 42.99-fold increase in the expression of miR-153, and after treatment with 1 mg/ml and 2 mg/ml OLE, the expression of miR-153 in GSCs was 12.6- and 29.81-fold up-regulated (p=0.1375 and 0.008, respectively). Furthermore, after treating cells with a combination of 1 mg/ml or 2 mg/ml OLE and 450 µM TMZ, the level of miR-153 was increased 75.68- and 81.91-fold (p=0.177 and 0.04, respectively). These expression levels were 1.76 and 1.90-fold higher than those after treatment with 450 µM TMZ alone (p=0.313 and 0.217, respectively). According to Xu et al, miR-153 decreases the expression of both Bcl-2 and Mcl-1 protein, which play crucial roles in regulating apoptosis in GBM cell lines [34]. In the current study, we determined that treatment with 2 mg/ml OLE or a combination of 2 mg/ml OLE and TMZ reduced the expression level of BCL2 in GSCs 4.14- and 5.68-fold, respectively (p=0.351 and 0.353). This implies that the induction of miR-153 by OLE may play a role in the activation of apoptosis by modulating BCL2 gene regulation. In addition, Xu et al demonstrated in their recent study that miR-153 may also inhibit the protein kinase B (PKB/Akt) pathway by decreasing the protein level of Irs-2 [41]. The deregulation of the Akt pathway is observed very frequently in GBM. The activation of this pathway leads to defects in the control of cell growth and survival that induce metastatic potential and resistance to chemotherapy and radiation [42]. Therefore, we suggest that miR-153 may overcome resistance to TMZ by also targeting the Akt pathway. Moreover, when we evaluated the expression level of miR-153 in individual GSCs, we observed that the synergistic effect of OLE and TMZ differed between cases. Treating the cells with OLE and TMZ induced miR-153 expression, but the extent of this expression depended on the TMZ response of patients. Similar to the data obtained from U-87MG cells that respond well to TMZ, the effect of the OLE-TMZ combination was lower in cases with shorter survival and higher in cases with longer survival after TMZ therapy. Because the MGMT methylation rates of the cases included in the study were similar (50.00 ± 00) [19], we suggest that the TMZ response of the GSCs may be modulated by other mechanisms.
Another miRNA that was significantly affected in GSCs by treatment with OLE was miR-137. There is a lack of miR-137 expression in GBM [43]. The low expression of miR-137 in GBM may be associated with promoter methylation [44]. In our previous study, we observed 1.41- and 1.86-fold up-regulation of miR-137 expression after 1 mg/ml and 2 mg/ml OLE treatment in T98G cells [18]. Moreover, in the current study, we observed 9.26- and 21.82-fold inc=reases in the expression level of this miRNA after treatment of GSCs with 1 mg/ml and 2 mg/ml OLE. In recent studies, the effect of curcumin, a well-studied medical plant, on epigenetic mechanisms such as histone modifications, DNA methylation, and miRNAs was identified. According to these studies, curcumin plays a role in cancer death and progression by selectively activating or inactivating gene expression via the induction of epigenetic modifications [45]. Because both curcumin and oleuropein are natural polyphenols, we suggest that OLE may also have the ability to modulate methylation. Thus, further studies are necessary to clarify this activity of OLE. When GSCs were treated with 450 µM TMZ alone, the regulation of miR-137 was 31.47-fold up-regulated. These data indicate that the magnitude of the effect of OLE on miR-137 regulation is similar to that of TMZ. After treating GSCs with the combination of 1 mg/ml or 2 mg/ml OLE and 450 µM TMZ, the expression level of miR-137 was increased 35.06- and 32.89-fold, respectively, compared to untreated controls (p=0.03 and 0.01, respectively). These expression levels were 1.11- and 1.04-fold higher than the expression level observed after treatment with 450 µM TMZ alone (p=0.484 and 0.394, respectively). miR-137 functions in the G0/G1 cell cycle arrest and decreases the expression of the cell cycle proteins CDK6 and phosphorylated Rb [43]. In recent studies, miR-137 was also reported to decrease the self-renewal ability of GSCs, which is one of the major features of cancer stem cells and rapid tumor progression, and the expression of the stem cell-associated proteins Nanog, Shh, Oct4 and Sox2 were determined [46]. In the current study, after treating with 2 mg/ml OLE or a combination of 2 mg/ml OLE and TMZ, we observed decreased expression levels of OCT-4 (7.21 and 8.43-fold) and SOX2 (13.36 and 16.06-fold), which are target genes of miR-137. In addition, according to recent studies, the activation of MET signaling leads to an increase in the expression of the stemness transcriptional regulator Oct4 [47]. In a study of melanoma, it was revealed that miR-137 regulates multiple targets including c-Met [48]. In the present study, we also determined that treating GSCs with 2 mg/ml OLE or a combination of 2 mg/ml OLE and TMZ caused 4.73- and 17.21-fold reductions in the expression level of c-Met, respectively. Therefore, OLE may decelerate the aggressiveness of GSCs by reducing the stemness properties of tumors by modulating miR-137. Additionally, one of the targets of miR-137 is RTVP-1, which is highly expressed in astrocytic tumors in a grade-dependent manner [44,49]. The protein product of RTVP-1 also causes resistance to tumor necrosis factor-related apoptosis by activating c-Jun-NH2-kinase and increasing the expression of BCL2 in GBM [49]. In the current study, we also defined the effect of OLE on the regulation of BCL2 (a 4.14-fold decrease). Therefore, these data make our findings concerning the inhibitor effect of OLE on cell proliferation in GSCs and their stemness properties stronger. In addition, when we evaluated the expression level of miR-137 in individuals, we observed that although all cases had undergone TMZ therapy and their MGMT methylation rates were similar, the combination of OLE and TMZ caused 6.23-fold up-regulation of miR-137 in the case with the shortest survival (5 months) and it caused an 8.3-fold decrease in miR-137 levels in the case with longest survival (> 48 months). These data also support our hypothesis regarding the action of OLE and TMZ; the interaction depends on both the status of MGMT and another mechanism related to TMZ response.
miR-145 is the third miRNA that is significantly altered after OLE treatment in GSCs. In recent studies, the expression of miR-145 was reported to be down-regulated in glioma cell lines and GSCs compared to astrocytes and neural progenitor cells, respectively [50]. Similarly, in our previous study, we observed a lower expression of miR-145 in GSC (+) tumors compared to GSC (-) tumors [19]. In the current study, after treatment with 1 mg/ml and 2 mg/ml OLE, the expression level of miR-145 was increased 1.2- and 4.38-fold (p=0.187; 0.432, respectively). Further, when GSCs were treated with 2 mg/ml OLE in combination with 450 µM TMZ, miR145 expression was 73.72-fold elevated (p=0.0005). This expression level was 7.28-fold higher than that observed after treatment with 450 µM TMZ alone (p=0.004). OCT4 and SOX2 are two of the pluripotency factors of GSCs [51]. Because of the target sites of OCT4 and SOX2 genes 3’ UTR regions, miR-145 may also be involved in the regulation of the stemness properties of GBM tumors [52]. In the current study, the expression levels of OCT4 and SOX2 were reduced 7.21- and 13.36-fold in GSCs, respectively, after treatment with 2 mg/ml OLE. On the other hand, when GSCs were treated with the combination of 2 mg/ml OLE and 450 µM TMZ, these genes were 8.43- and 16.06-fold decreased, respectively. In addition, according to Rani et al, miR-145 targets also another SOX family member, SOX9, and functions as a tumor suppressor RNA in glioma cells; thus, reduced levels of miR-145 may lead to neoplastic transformation and malignant progression in glioma [53]. Taking together, these data suggest that OLE may act as an inhibitor of pluripotency factors in GSCs via the induction of miR-145. Additionally, Rani et al demonstrated that the silencing of miR-145 leads to the overexpression of molecules associated with cell proliferation, such as cyclin D1, c-myc, and N-myc [53]. According to the present study, treatment with 2 mg/ml OLE resulted in a 4.38-fold increase in miR-145 expression and 5.97-fold decrease in c-myc expression (p=0.341). Moreover, when cells were treated with 2 mg/ml OLE in combination with 450 µM TMZ, c-myc expression was 14.82-fold decreased (p=0.341). On the other hand, when individual cases were evaluated for the possible association between their response to TMZ and miR-145 expression levels after 2 mg/ml OLE and 450 µM TMZ treatment, the effect of the OLE-TMZ combination was smallest (3.36-fold up-regulation of miR-145) in the case with longest survival and larger (6.45-fold) in the case with shortest survival. Moreover, for the other 3 cases, the expression level of miR-145 was not associated with survival. Therefore, we suggest that although the combination of OLE and TMZ induces miR-145 expression in all GSCs, the rate of induction may be affected by other factors.
In summary, in the present study, treatment with 1 mg/ml and 2 mg/ml OLE caused cytotoxicity in U-138MG and U-87MG cells. These doses are similar to the defined doses of OLE in T98G cells in our previous study [18]. Although OLE causes cell death via both apoptosis and necrosis, we demonstrated in our previous study that the cytotoxic activity of OLE is negligible in normal cells [18]. In addition, when we evaluate the effect of OLE on the regulation of miRNAs in GSCs, we found the effect of 2 mg/ml OLE alone to be very similar to the effect of 450 µM TMZ. Furthermore, although the magnitude of the changes differ depending on the MGMT status and/or other mechanisms, we found that using OLE in combination with TMZ raised the rate of inhibition of cell proliferation in GBM and increased the expression levels of miR-153, miR-137 and miR-145 in GSCs compared to cells treated with only TMZ. Therefore, taking all the data together, we believe that OLE may synergistically affect the activity of TMZ with less cytotoxic activity in non-tumor cells. On the other hand, to a better understanding of the anti-carcinogenic activity of OLE, evaluation of the effects of the active compounds in OLE on GBM tumors are required. Furthermore, based on the current results in GBM cell lines, MGMT methylation status is a factor that impacts the effectiveness of the OLE and TMZ combination. However, also based on our results, the mechanism of action of OLE-TMZ and the relationship between drug resistances in individual GSCs is an open avenue for future investigation.
In conclusion, to the best of our knowledge, our data are the first to demonstrate that OLE causes cell death in GBM cells with different responses to TMZ and that this effect is synergistically increased in combination with TMZ. In addition, although we determined how OLE altered miRNAs in GBM cells in a previous study, the current study uniquely demonstrates the effect of OLE on changes to these miRNAs in GSCs. Our findings are also the first to indicate that OLE may interfere with the pluripotency of GSCs by modulating miRNA expression. Further studies and validation are needed, but we suggest that OLE may be a strong candidate for studies of therapeutic cancer drugs.
Acknowledgements
This study was supported by a grant from the Scientific Research Projects Foundation (BAP) of the Uludag University of Turkey [Project No. OUAP (T)-2012/17].
Disclosure of conflict of interest
None.
References
- 1.Piperi C, Themistocleous MS, Papavassiliou GA, Farmaki E, Levidou G, Korkolopoulou P, Adamopoulos C, Papavassiliou AG. High incidence of MGMT and RARbeta promoter methylation in primary glioblastomas: association with histopathological characteristics, inflammatory mediators and clinical outcome. Mol Med. 2010;16:1–9. doi: 10.2119/molmed.2009.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Rocca RV, Mehdorn HM. Localized BCNU chemotherapy and the multimodal management of malignant glioma. Curr Med Res Opin. 2009;25:149–160. doi: 10.1185/03007990802611935. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Würth R, Barbieri F, Florio T. New Molecules and Old Drugs as Emerging Approaches to Selectively Target Human Glioblastoma Cancer Stem Cells. Biomed Res Int. 2014;2014:126586. doi: 10.1155/2014/126586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HH, Yang LL, Wang CC, Hu SY, Chang SF, Lee YH. Differential effects of natural polyphenols on neuronal survival in primary cultured central neurons against glutamate and glucose deprivation induced neuronal death. Brain Res. 2003;986:103–113. doi: 10.1016/s0006-8993(03)03197-4. [DOI] [PubMed] [Google Scholar]
- 6.Mijatovic SA, Timotijevic GS, Miljkovic DM, Radovic JM, Maksimovic-Ivanic DD, Dekanski DP, Stosic-Grujicic SD. Multiple antimelanoma potential of dry olive leaf extract. Int J Cancer. 2011;128:1955–1965. doi: 10.1002/ijc.25526. [DOI] [PubMed] [Google Scholar]
- 7.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 8.Gilani A, Khan A, Shah A, Connor J, Jabeen Q. Blood pressure lowering effect of olive is mediated through calcium channel blockade. Int J Food Sci Nutr. 2005;56:613–620. doi: 10.1080/09637480500539420. [DOI] [PubMed] [Google Scholar]
- 9.Owen RW, Haubner R, Würtele G, Hull E, Spiegelhalder B, Bartsch H. Olives and olive oil in cancer prevention. Eur J Cancer Prev. 2004;13:319–326. doi: 10.1097/01.cej.0000130221.19480.7e. [DOI] [PubMed] [Google Scholar]
- 10.Cragg GM, Newman DJ, Yang SS. Natural product extracts of plant and marine origin having antileukemia potential. The NCI experience. J Nat Prod. 2006;69:488–498. doi: 10.1021/np0581216. [DOI] [PubMed] [Google Scholar]
- 11.Escrich E, Moral R, Grau L, Costa I, Solanas M. Molecular mechanisms of the effects of olive oil and other dietary lipids on cancer. Mol Nutr Food Res. 2007;51:1279–1292. doi: 10.1002/mnfr.200700213. [DOI] [PubMed] [Google Scholar]
- 12.Cottet V, Touvier M, Fournier A, Touillaud MS, Lafay L, Clavel-Chapelon F, Boutron-Ruault MC. Postmenopausal breast cancer risk and dietary patterns in the E3N-EPIC prospective cohort study. Am J Epidemiol. 2009;170:1257–1267. doi: 10.1093/aje/kwp257. [DOI] [PubMed] [Google Scholar]
- 13.Bosetti C, Pelucchi C, La Vecchia C. Diet and cancer in Mediterranean countries: carbohydrates and fats. Public Health Nutr. 2009;12:1595–1600. doi: 10.1017/S1368980009990425. [DOI] [PubMed] [Google Scholar]
- 14.Abaza L, Talorete T, Yamada P, Kurita Y, Zarrouk M, Isoda H. Induction of growth inhibition and differentiation of human leukemia HL-60 cells by a Tunisian Gerboui olive leaf extract. Biosci Biotechnol Biochem. 2007;71:1306–1312. doi: 10.1271/bbb.60716. [DOI] [PubMed] [Google Scholar]
- 15.Fares R, Bazzi S, Baydoun SE, Abdel-Massih RM. The antioxidant and anti-proliferative activity of the Lebanese Olea europaea Extract. Plant Foods Hum Nutr. 2011;66:58–63. doi: 10.1007/s11130-011-0213-9. [DOI] [PubMed] [Google Scholar]
- 16.Reyes F, Centelles J, Lupianez J, Cascante M. (2a, 3b)-2, 3-Dihydroxyolean-12-en-28-oic acid, a new natural triterpene from Olea europea, induces caspase dependent apoptosis selectively in colon adenocarcinoma cells. FEBS Lett. 2006;580:6302–6310. doi: 10.1016/j.febslet.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 17.Arola-Arnal A, Bladé C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS One. 2011;6:e25982. doi: 10.1371/journal.pone.0025982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tunca B, Tezcan G, Cecener G, Egeli U, Ak S, Malyer H, Tumen G, Bilir A. Olea europaea leaf extract alters microRNA expression in human glioblastoma cells. J Cancer Res Clin Oncol. 2012;138:1831–1844. doi: 10.1007/s00432-012-1261-8. [DOI] [PubMed] [Google Scholar]
- 19.Tezcan G, Tunca B, Bekar A, Preusser M, Berghoff AS, Egeli U, Cecener G, Ricken G, Budak F, Taskapılıoglu MO, Kocaeli H, Tolunay S. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol Neurobiol. 2014;34:679–92. doi: 10.1007/s10571-014-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Yoshino A, Ogino A, Yachi K, Ohta T, Fukushima T, Watanabe T, Katayama Y, Okamoto Y, Naruse N, Sano E, Tsumoto K. Gene expression profiling predicts response to temozolomide in malignant gliomas. Int J Oncol. 2010;36:1367–1377. doi: 10.3892/ijo_00000621. [DOI] [PubMed] [Google Scholar]
- 22.Craig WJ. Health-promoting properties of common herbs. Am J Clin Nutr. 1999;70:491–495. doi: 10.1093/ajcn/70.3.491s. [DOI] [PubMed] [Google Scholar]
- 23.Setzer WN, Setzer MC. Plant-derived triterpenoids as potential antineoplastic agents. Mini Rev Med Chem. 2003;3:540–556. doi: 10.2174/1389557033487854. [DOI] [PubMed] [Google Scholar]
- 24.Omar SH. Oleuropein in olive and its pharmacological effects. Sci Pharm. 2010;78:133–154. doi: 10.3797/scipharm.0912-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visioli F, Poli A, Gall C. Antioxidant and other biological activities of phenols from olives and olive oil. Med Res Rev. 2002;22:65–75. doi: 10.1002/med.1028. [DOI] [PubMed] [Google Scholar]
- 26.Kurisawa M, Chung JE, Uyama H, Kobayashi S. Enzymatic synthesis and antioxidant properties of poly(rutin) Biomacromolecules. 2003;4:1394–1399. doi: 10.1021/bm034136b. [DOI] [PubMed] [Google Scholar]
- 27.Reyes-Zurita FJ, Rufino-Palomares EE, Medina PP, Leticia García-Salguero E, Peragón J, Cascante M, Lupiáñez JA. Antitumour activity on extrinsic apoptotic targets of the triterpenoid maslinic acid in p53-deficient Caco-2 adenocarcinoma cells. Biochimie. 2013;95:2157–2167. doi: 10.1016/j.biochi.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 29.Humbert O, Fiumicino S, Aquilina G, Branch P, Oda S, Zijno A, Karran P, Bignami M. Mismatch repair and differential sensitivity of mouse and human cells to methylating agents. Carcinogenesis. 1999;20:205–214. doi: 10.1093/carcin/20.2.205. [DOI] [PubMed] [Google Scholar]
- 30.Everhard S, Tost J, El Abdalaoui H, Crinière E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Thillet J. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakrishnan V, Kushwaha D, Koay DC, Reddy H, Mao Y, Zhou L, Ng K, Zinn P, Carter B, Chen CC. Post-transcriptional regulation of O(6)-methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark. 2011-2012;10:185–193. doi: 10.3233/CBM-2012-0245. [DOI] [PubMed] [Google Scholar]
- 32.Melguizo C, Prados J, González B, Ortiz R, Concha A, Alvarez PJ, Madeddu R, Perazzoli G, Oliver JA, López R, Rodríguez-Serrano F, Aránega A. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10:250. doi: 10.1186/1479-5876-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology. 2010;57:734–743. doi: 10.1111/j.1365-2559.2010.03686.x. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–1035. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Chen R, Huang S, Wu Y, Li G, Zhang B, Liu Q, Yin D, Liang Y. miR-153 sensitized the K562 cells to As2O3-induced apoptosis. Med Oncol. 2012;29:243–247. doi: 10.1007/s12032-010-9807-6. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 37.Kheirelseid EA, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G, Kerin MJ. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int J Colorectal Dis. 2013;28:247–260. doi: 10.1007/s00384-012-1549-9. [DOI] [PubMed] [Google Scholar]
- 38.Anaya-Ruiz M, Cebada J, Delgado-López G, Sánchez-Vázquez ML, Pérez-Santos JL. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev. 2013;14:2983–2986. [PubMed] [Google Scholar]
- 39.Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J, Thomas GJ, Packham G, Mirnezami AH. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73:6435–6447. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, Zhang D, Kang J, Wu Z. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep. 2013;40:2789–2798. doi: 10.1007/s11033-012-2278-4. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Liao X, Lu N, Liu W, Wong CW. Chromatin-modifying drugs induce miRNA-153 expression to suppress Irs-2 in glioblastoma cell lines. Int J Cancer. 2011;129:2527–31. doi: 10.1002/ijc.25917. [DOI] [PubMed] [Google Scholar]
- 42.Narayan RS, Fedrigo CA, Stalpers LJ, Baumert BG, Sminia P. Targeting the Akt-pathway to improve radiosensitivity in glioblastoma. Curr Pharm Des. 2013;19:951–957. [PubMed] [Google Scholar]
- 43.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, de Carvalho AC, Slavin S, Jacoby E, Yalon M, Toren A, Mikkelsen T, Brodie C. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4:665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teiten MH, Dicato M, Diederich M. Curcumin as a regulator of epigenetic events. Mol Nutr Food Res. 2013;57:1619–1629. doi: 10.1002/mnfr.201300201. [DOI] [PubMed] [Google Scholar]
- 46.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 47.Jun HJ, Bronson RT, Charest A. Inhibition of EGFR induces a c-MET-driven stem cell population in glioblastoma. Stem Cells. 2014;32:338–348. doi: 10.1002/stem.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmüller SB. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2012;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 49.Ziv-Av A, Taller D, Attia M, Xiang C, Lee HK, Cazacu S, Finniss S, Kazimirsky G, Sarid R, Brodie C. RTVP-1 expression is regulated by SRF downstream of protein kinase C and contributes to the effect of SRF on glioma cell migration. Cell Signal. 2011;23:1936–1943. doi: 10.1016/j.cellsig.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E, Yalon M, Toren A, Rempel SA, Brodie C. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 53.Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15:1302–1316. doi: 10.1093/neuonc/not090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaudhry MA, Omaruddin RA. Differential regulation of microRNA expression in irradiated and bystander cells. Mol Biol (Mosk) 2012;46:634–643. [PubMed] [Google Scholar]