Abstract
Internal ribosome entry site (IRES) elements are high‐order RNA structures that promote internal initiation of translation to allow protein synthesis under situations that compromise the general cap‐dependent translation mechanism. Picornavirus IRES elements are highly efficient elements with a modular RNA structure organization. Here we investigated the effect of Mg2+ concentration on the local flexibility and solvent accessibility of the foot‐and‐mouth disease virus (FMDV) IRES element measured on the basis of selective 2′‐hydroxyl acylation analyzed by primer extension (SHAPE) reactivity and hydroxyl radical cleavage. We have found that Mg2+ concentration affects the organization of discrete IRES regions, mainly the apical region of domain 3, the 10 nt loop of domain 4, and the pyrimidine tract of domain 5. In support of the effect of RNA structure on IRES activity, substitution or deletion mutants of the 10 nt loop of domain 4 impair internal initiation. In addition, divalent cations affect the binding of eIF4G, a eukaryotic initiation factor that is essential for IRES‐dependent translation that interacts with domain 4. Binding of eIF4G is favored by the local RNA flexibility adopted at low Mg2+ concentration, while eIF4B interacts with the IRES independently of the compactness of the RNA structure. Our study shows that the IRES element adopts a near‐native structure in the absence of proteins, shedding light on the influence of Mg2+ ions on the local flexibility and binding of eIF4G in a model IRES element.
Keywords: eIF4G‐binding, IRES‐dependent translation initiation, magnesium ions, RNA structure, SHAPE footprint
Abbreviations
- DMS
dimethyl‐sulfate
- Ebp1
ErbB3‐binding protein 1
- eIFs
eukaryotic initiation factors
- FMDV
foot‐and‐mouth disease virus
- IRES
internal ribosome entry site
- NMIA
N‐methylisatoic anhydride
- PTB
Polypyrimidine tract‐binding protein
- SHAPE
selective 2′‐hydroxyl acylation analyzed by primer extension
- WC
Watson‐Crick
Introduction
RNA structure plays a fundamental role in biological processes guided by RNA regulatory elements 1. As for most RNA regulatory elements, the RNA structure dictates the function of the internal ribosome entry site (IRES) 3. IRES elements, which were discovered in picornavirus genomic RNAs, are in charge of driving translation initiation in various RNA viruses and a subset of cellular mRNAs 6. Indeed, IRES‐driven translation initiation represents an alternative mechanism to start protein synthesis under situations that compromise cap‐dependent translation initiation, typically occurring in infected cells. However, under normal situations, most eukaryotic mRNAs initiate translation by a mechanism that depends on recognition of the m7GpppN residue (termed the cap) located at the 5′ end of most mRNAs 9.
IRES elements exhibit a wide diversity of nucleotide sequences, RNA secondary structures and trans‐acting factor requirements, as illustrated by the IRES elements present in the genome of picornaviruses, the intergenic region of dicistroviruses and hepatitis C virus 10. Earlier studies of the IRES element of foot‐and‐mouth‐disease virus (FMDV), a member of the Picornaviridae family, revealed a modular organization of domains, termed 1–5 in the 5′→3′ direction 11. These RNA domains contain specific structural motifs that are responsible for recruitment of eIFs (eIF4G, eIF4B and eIF3) and several IRES‐binding factors 12.
We have recently used selective 2′‐hydroxyl acylation analyzed by primer extension (SHAPE) to assess the RNA structure of IRES elements 3. This methodology has the advantage over other high‐resolution approaches of allowing analysis of long RNA molecules and providing information at single‐nucleotide resolution 18. Our studies have shown that domain 3 is a self‐folding region that harbors conserved motifs whose disruption impairs IRES activity. These motifs have been proposed to be involved in distant interactions on the basis that altering one of them affects the RNA structural organization of this domain, leading to defective IRES elements 19. However, three‐dimensional models of the picornavirus IRES RNA structure are not yet available. Although a high Mg2+ concentration inactivated the IRES‐dependent activity in cell‐free lysates of the closely related encephalomyocarditis virus 22, the influence of divalent cations on the IRES structure, and, in particular, whether the concentration of Mg2+ ions affects the binding of factors needed for IRES activity, are unknown.
Adoption of RNA tertiary structure is strongly dependent on Mg2+ ions, although a mixture of monovalent cations (K+ and Na+) with Mg2+ is required for efficient folding 23. In the case of the FMDV IRES, long‐distance RNA–RNA interactions were dependent on the concentration of the divalent ion in the folding buffer 19. Cations reduce the charge repulsion generated by phosphate anions of the backbone, stabilizing the folded conformation of RNA 26. It is well established that Mg2+, which has a high charge density, is the dominant divalent cation in biological systems 24. Moreover, Mg2+ ions perform both catalytic and structural roles, including mediation of helical packaging, promotion of long‐range interactions, and stabilization of RNA motifs 27.
In this study, we investigated the effect of Mg2+ concentration on the local flexibility and solvent accessibility of the IRES RNA measured by SHAPE reactivity and hydroxyl radical cleavage. We show that Mg2+ concentration affects the organization of discrete regions located in the apical region of domain 3, the 10 nt loop of domain 4, and the pyrimidine tract of domain 5. Based on these results, we analyzed the effect of divalent cations on the binding of eIF4G and eIF4B, two initiation factors that are involved in FMDV IRES‐dependent translation 12. We have found that eIF4B interacts with the IRES independently of the rigidity of the RNA structure, while binding of eIF4G is favored by RNA flexibility in this region. Collectively, this study provides new insights into the influence of Mg2+ ions on the local flexibility and protein binding of a picornavirus IRES, shedding light on the structure of a model IRES element.
Results
Magnesium ions determine the RNA structure of discrete IRES regions
This study was designed to investigate the conformation of the IRES element under various ionic conditions. To this end, in vitro‐synthesized RNA containing the functional FMDV IRES (nucleotides 1–462) upstream of the luciferase coding region (Fig. 1A) was denatured, snap‐cooled, and renatured in folding buffer under a range of MgCl2 concentrations, keeping the buffer concentration (HEPES pH 8, 100 mm) and monovalent cation concentrations (NaCl, 100 mm; KCl, 140 mm) constant. This procedure was followed to minimize intermolecular interactions, release mis‐folded molecules from kinetic traps and promote the most stable secondary structure. Refolded RNA was treated with N‐methylisatoic anhydride (NMIA), which preferentially acylates the 2′‐OH of riboses associated with conformational flexible nucleotides 29. Heavily modified positions correspond to nucleotides that are primarily single‐stranded, while non‐reactive nucleotides are involved in base pairs. Inflexible regions may also correspond to non‐WC base pairs, tertiary interactions, or a single stacking interaction in the C2′‐endo conformation 30. After NMIA treatment, the IRES region was examined by reverse transcriptase extension using primers spaced along the RNA sequence to obtain overlapping readings 3. Then, cDNA products derived from the untreated and NMIA‐treated RNAs were resolved in denaturing acrylamide urea gels in parallel with a sequence ladder obtained using the same primer (Fig. 1B). Gross modifications of SHAPE reactivity where observed in specific regions of domains 3 and 4 of the IRES folded at low Mg2+ concentration (0.5 mm) compared with RNA folded at high Mg2+ concentration (6 mm). Compared with the absence of Mg2+ in the folding buffer, the apical region of domain 3 showed a decreased SHAPE reactivity in residues 200–202 in the presence of 0.5 and 6 mm Mg2+ (Fig. 1B, left panel, black arrowheads). Likewise, a decrease in reactivity at positions 337–348 within domain 4 was observed at high Mg2+ concentration (Fig. 1B, right panel, black arrowheads).
Figure 1.
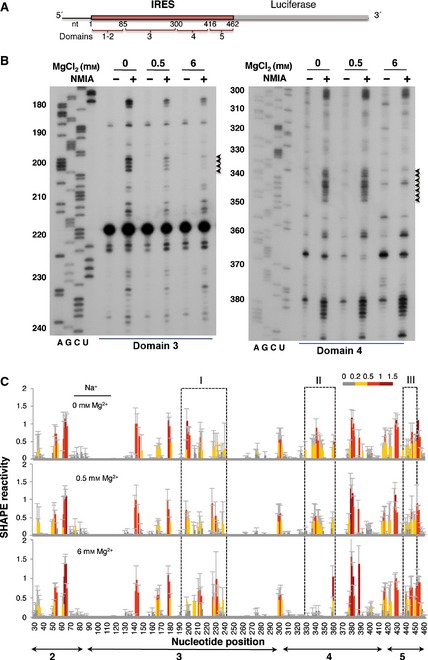
SHAPE reactivity of the IRES element as a function of the concentration of divalent ions in the folding buffer. (A) Schematic of the RNA harboring the FMDV IRES element. Numbers indicate the IRES nucleotides, with approximate locations of IRES structural domains 1–5. Domain 1 is not shown in subsequent figures because it corresponds to the right arm of the cre element and contributes very weakly to IRES activity. (B) Representative examples of primer extension assays corresponding to domains 3 and 4 of the IRES. The Mg2+ concentration and NMIA treatment are indicated at the top. Nucleotide positions are indicated on the left. AGCU indicate the sequence lanes used as ladders. Black arrowheads on the right indicate positions whose reactivity decreases upon increasing the concentration of Mg2+ in the folding buffer. (C) SHAPE reactivity profiles of the IRES at 0, 0.5 and 6 mm Mg2+. Values correspond to the mean SHAPE reactivity (±SD) for six independent assays. SHAPE reactivity is represented using a colored scale in which 0 indicates unreactive nucleotides and the mean intensity at highly reactive nucleotides is set to 1.0. Nucleotide positions are indicated on the x axis. Domains 2, 3, 4 and 5 of the IRES are indicated below the corresponding nucleotide positions. Boxes I, II and III indicate the regions that show the main reactivity changes.
To precisely measure the effect of varying Mg2+ concentration on the IRES structure, six independent assays were used to quantify SHAPE reactivity along the IRES region. The resulting profiles (Fig. 1C) revealed that the major reactivity changes were located in specific regions of domains 3, 4 and 5 (boxes I, II and III in Fig. 1C).
The local RNA flexibility displayed on the secondary IRES structure is shown in Fig. 2. Relative to the RNA folded in the absence of Mg2+ (Fig. 2A), changes in the IRES structure affecting several nucleotides under low and high Mg2+ concentrations (Fig. 2B,C) were located in three regions (boxes I, II and III in Fig. 2A–C). Specifically, nucleotides AAAA(199–202) (corresponding to the RAAA motif on the apical stem‐loop of domain 3) exhibited a decrease in reactivity upon Mg2+ addition (box I in Fig. 2A–C). Additionally, a moderately increased reactivity was observed in residues belonging to the bulges AACUCC(166–171) and CGCCCACACG(207–216), as well as the C‐rich loop UGCAACCCCAGCAC(230–243). On the other hand, nucleotides GUAACACGCGAC(337–348) corresponding to the 10 nt loop of domain 4 (box II in Fig. 2A–C) showed a significant decrease in reactivity under high Mg2+ concentration. Reactivity changes affecting more than one position were also observed at positions UUUCCU(440–445), corresponding to the pyrimidine‐rich tract of domain 5 (box III in Fig. 2A–C). Some changes of reactivity were also observed at individual residues (A361 and G389).
Figure 2.
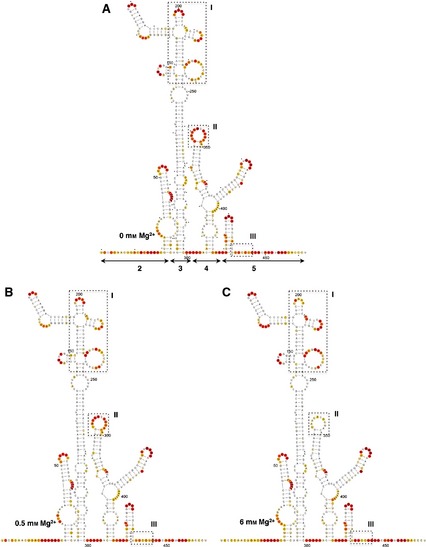
Effect of Mg2+ concentration on the secondary structure of the IRES. The SHAPE reactivity obtained using NMIA and 5′‐radiolabeled primers is represented using the colored scale shown in Fig. 1C. The RNA structure was visualized using VARNA 61. Boxes I, II and III represent the regions that display the main changes in the presence of 0 (A), 0.5 (B) or 6 mm (C) Mg2+ in the folding buffer. Nucleotides are numbered every 50 positions; a dot indicates the every 10th nucleotide; lines with arrowheads in (A) indicate the approximate location of IRES structural domains 2–5.
Independent measurements of the local RNA flexibility were performed using fluorescent primers and capillary electrophoresis (Fig. 3A). The results displayed on the IRES secondary structure (Fig. 3B) are largely coincident with those obtained using radiolabeled primers, in agreement with previous studies 32. In fact, only minor differences were observed in some positions, which may be due to the different methodology necessary to quantify the reverse transcriptase stop signals.
Figure 3.
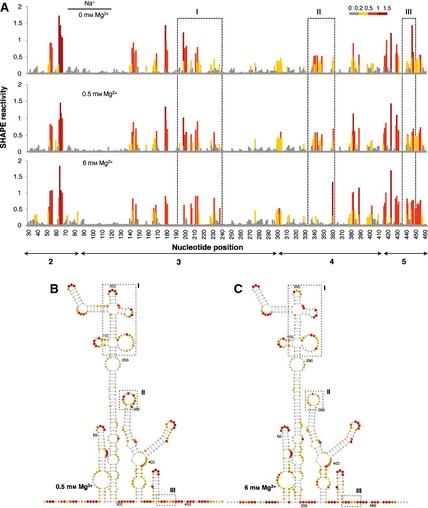
(A) SHAPE reactivity profiles of the IRES using fluorescent‐labeled primers and capillary electrophoresis. SHAPE reactivity is represented using a colored scale in which 0 indicates unreactive nucleotides and the mean intensity at highly reactive nucleotides is set to 1.0. Nucleotide positions are indicated on the x axis. Domains 2, 3, 4 and 5 of the IRES are indicated below the corresponding nucleotide positions. Boxes I, II and III indicate the regions that show the main reactivity changes. (B, C) The RNA structure at 0.5 Mg2+ (B) and 6 mm Mg2+ (C). The symbols used are described in the legend to Fig. 2.
Further analysis of the effect of Mg2+ concentration on the IRES structure folded in the presence of K+, the predominant monovalent cation in the cell cytoplasm, indicated that the main change occurred at around 2 mm Mg2+ (Fig. 4A). Very few differences were found in individual positions, and these did not modify the overall pattern of SHAPE reactivity at the same Mg2+ concentration irrespective of the presence of Na+ or K+ ions (compare Fig. 4B,C with Fig. 2B,C). We therefore used 0.5 and 6 mm as the low and high concentrations of Mg2+ for further studies on IRES structure.
Figure 4.
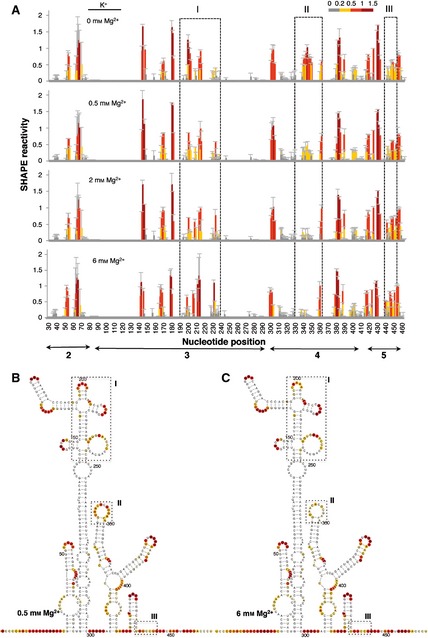
RNA structure analysis of the IRES renatured in the presence of the monovalent ion K+. (A). SHAPE reactivity profiles of the IRES in the presence of 140 mm KCl at increasing concentrations of Mg2+ (0, 0.5, 2 and 6 mm). Values correspond to the mean SHAPE reactivity (±SD). The SHAPE reactivity is represented using the colored scale shown in Fig. 1C. Boxes I, II and III indicate the regions that show the main reactivity changes. (B, C) The RNA structure at 0.5 Mg2+ (B) and 6 mm Mg2+ (C). The symbols used are described in the legend to Fig. 2.
Thus, we conclude that the Mg2+ concentration profoundly affects the organization of discrete IRES regions, i.e. the apical region of domain 3 and the 10 nt loop of domain 4, in addition to moderate reorganization of the pyrimidine tract of domain 5.
Mutational analysis reveals involvement of the 10 nt loop of domain 4 in IRES activity
Regarding the involvement in IRES function of the regions showing changes in local flexibility upon Mg2+ increase, previous data from our laboratory have shown that the apical loop of domain 3 is involved in IRES activity, and its sequence composition affects the RNA structure of the flanking stem‐loops 3. However, no information was available regarding the 10 nt loop of domain 4. Alignment of the IRES sequences from 180 FMDV isolates available in GenBank indicated that the sequence of the 10 nt loop at nucleotides 337–348 is conserved, with the exception of C343 and C345, which tolerated transitions and transversions (A or U and U or G, respectively) (Fig. 5A). Occasional substitutions were observed at positions U338 and A347. Indeed, a conserved motif GUAACANGNGAC(337–348) was inferred from sequence conservation. In contrast, flanking nucleotides that are engaged in base pairs according to SHAPE probing (Fig. 2A) were either invariant or covariant (Fig. 5A).
Figure 5.
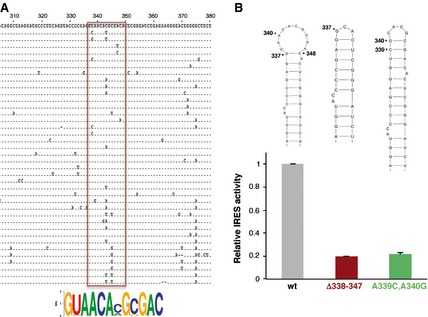
Contribution of the 10 nt loop of domain 4 to IRES activity. (A) Alignment of variant sequences within the 306–380 region of the FMDV IRES. A total of 180 sequences available in GenBank were aligned using clustalx. For simplicity, one representative of each variant is shown in the alignment. The pattern of nucleotide conservation (measured in bits) of the 10 nt loop of domain 4 is represented by a sequence logo obtained from alignment of nucleotides 337–348 (boxed in red). (B). Mutational analysis of the 10 nt loop of domain 4. The secondary structure of the wild‐type (wt) stem‐loop as well as the deletion mutant (Δ338–347) and the substitution mutant (A339C/A340G) are shown at the top. Relative IRES activity was determined as the ratio of luciferase to chloramphenicol acetyltransferase expressed from a bi‐cistronic construct in transfected BHK‐21 cells and normalized to the activity observed for the wild‐type IRES (set to 1). Values are means ± SD. The experiment was performed in triplicate and repeated at least three times.
Thus, to obtain information about the implication of the 10 nt loop at nucleotides 337–348 on IRES activity, we generated two mutants. The first one (Δ338–347) carried a deletion of the entire loop, and the second one (A339C/A340G) carried a double substitution that was intended to close the loop (Fig. 5B). Determination of the efficiency of internal initiation of translation of these mutants relative to the wild‐type IRES showed a decrease of activity of approximately 80% (Fig. 5B). These data demonstrate that the 10 nt loop of domain 4 contributes to IRES activity to a significant extent.
Assembly of eIF4G–IRES and eIF4B–IRES complexes reveals the protection of specific residues in domains 4 and 5
The IRES domains 4 and 5, which showed important changes in RNA structure upon folding at high concentration of Mg2+, provide the binding sites for eIF4G and eIF4B during internal initiation 6. Thus, we analyzed the influence of Mg2+ on the IRES–protein interaction on the basis of the SHAPE footprint, a methodology that allows the study of RNA molecules assembled in ribonucleoprotein complexes 18. In this assay, the protections correspond to RNA regions involved RNA–protein interactions or changes in RNA conformation induced by the RNA–protein interaction.
To determine the effect of Mg2+ ions on the interaction of eIF4G with the IRES, RNA folded at 0.5 mm Mg2+ was incubated with increasing amounts of purified His‐tagged eIF4G protein (0, 40 and 80 nm) prior to NMIA treatment. Primer extension analysis of the complexes assembled in vitro allowed measurement of the SHAPE reactivity of the IRES region in RNA incubated with or without eIF4G (Fig. 6A, lanes 2–4). Relative to the SHAPE reactivity observed with free RNA, eIF4G protein induces the protection of sequences that lie within domain 4 of the IRES. Specifically, nucleotides 400–418 were protected from NMIA attack upon addition of His‐tagged eIF4G at low Mg2+ concentration (blue bar/box in Fig. 6A,B). These results are in agreement with independent approaches, such as assembly of 48S pre‐initiation complexes and RNA–protein pull‐down and UV‐crosslink assays performed using a range of Mg2+ concentrations between 0.3 and 2.5 mm, which mapped the binding site of eIF4G to domain 4 of the encephalomyocarditis virus and FMDV IRES elements 16. In addition, nucleotides 340–350 were reorganized upon addition of the protein, with noticeable changes in the primer extension pattern (Fig. 6A,B, blue bar/box). However, residues 75, 293–303 and 447–454 were more reactive towards NMIA (Fig. 6B, red boxes), indicating modified organization of the regions flanking the protein‐binding site on the IRES structure. Reactivity changes around position 380 were observed independently of the RNA treatment. Additionally, nucleotides 53–66 within domain 2 were protected (Fig. 6B, blue box). However, none of the eIF4G‐induced protections observed at low Mg2+ concentration were detected for RNA folded at high Mg2+ concentration (Fig. 6C). These results led us to conclude that eIF4G–IRES interaction is sensitive to the RNA structure reorganization induced by divalent cations.
Figure 6.
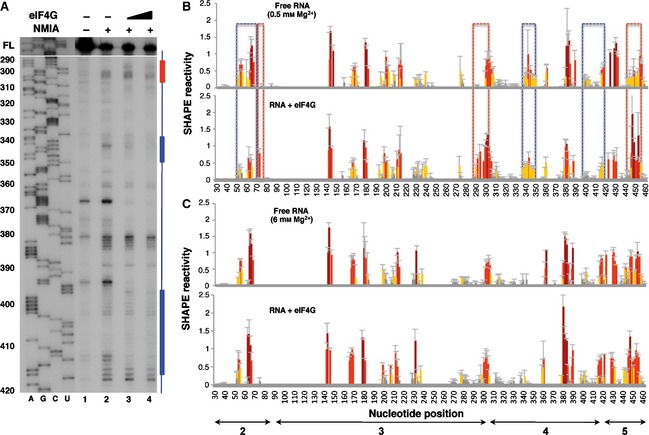
Impact of eIF4G on IRES SHAPE reactivity. (A) Primer extension analysis of RNA–protein complexes assembled at increasing concentrations of eIF4G (40 and 80 nm) using RNA folded at 0.5 mm Mg2+ treated (+) or untreated (–) with NMIA was performed using a 5′ end radiolabeled primer. Nucleotide positions are indicated on the left based on the sequencing lanes (AGCU) obtained using the same primer; cDNA full‐length (FL) products are shown at the top of each lane. Blue and red bars indicate protected or more reactive nucleotides, respectively. (B) RNA SHAPE reactivity for free RNA renatured in the presence of 0.5 mm Mg2+ (top) and upon addition of purified His‐tagged eIF4G (40 nm) (bottom). (C) RNA SHAPE reactivity for free RNA renatured in the presence of 6 mm Mg2+ (top) and upon addition of purified His‐tagged eIF4G (40 nm) (bottom). Values are means ± SD. Bars are colored according to reactivity based on the colored scale shown in Fig. 1C. Blue boxes indicate protected regions, and red boxes indicate regions with higher reactivity.
The RNA‐binding protein eIF4B interacts with picornavirus IRES elements, stimulating IRES activity 12. Interaction of eIF4B with the IRES element was first analyzed using RNA folded at 6 mm Mg2+ incubated with increasing amounts of purified His‐tagged eIF4B protein (0, 60, 300 and 500 nm). Of interest, a concentration of 300 nm (lane 4 in Fig. 7A) was sufficient to induce protection of residues belonging to domain 5, although the protection increased with protein concentration (blue bar in Fig. 7A). However, some nucleotides near the 3′ boundary of the IRES were more reactive in the presence of eIF4B (red bar in Fig. 7A).
Figure 7.
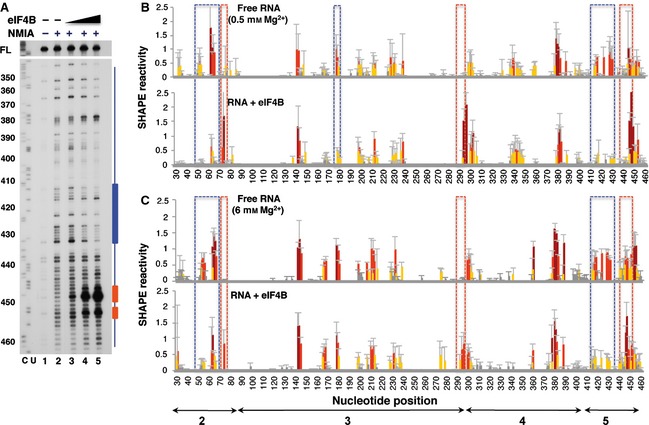
Impact of eIF4B on IRES SHAPE reactivity. (A) Primer extension analysis of RNA–protein complexes, assembled using RNA folded at 0.5 mm Mg2+ and increasing concentrations of eIF4B (60, 300 and 500 nm) and treated (+) or untreated (–) with NMIA, was performed with a 5′ end radiolabeled primer. Nucleotide positions are indicated on the left based on the sequencing lanes (CU) obtained using the same primer; cDNA full‐length products (FL) are shown at the top of each lane. Blue and red bars indicate protected or more reactive nucleotides, respectively. (B) RNA SHAPE reactivity for free RNA renatured in the presence of 0.5 mm Mg2+ (top) and upon addition of purified His‐tagged eIF4B (300 nm) (bottom). (C) RNA SHAPE reactivity for free RNA renatured in the presence of 6 mm Mg2+ (top) and upon addition of purified His‐tagged eIF4B (300 nm) (bottom). Values are means ± SD. Bars are colored according to reactivity based on the colored scale shown in Fig. 1C. Blue boxes indicate protected regions, and red boxes indicate regions with higher reactivity.
Next, the SHAPE reactivity values observed on free RNA at both 0.5 and 6 mm Mg2+ were compared with those obtained upon addition of eIF4B. Interestingly, the hairpin of domain 5 (nucleotides 416–433) was protected by eIF4B from NMIA attack at both low and high Mg2+ concentrations (blue boxes in Fig. 7B,C). In contrast, two short regions spanning nucleotides 293–297 (domain 3) and 447–454 (domain 5) became more reactive towards NMIA (red boxes in Fig. 7B,C). Residues belonging to domain 3 showed no gross changes in SHAPE reactivity with the exception of positions 179–180, while nucleotides 53–66 within domain 2 were protected. These results show that eIF4B specifically induces protection of residues belonging to domains 2 and 5.
It is worth mentioning that specific residues were differentially protected upon incubation of the IRES with eIF4G or eIF4B at low Mg2+ concentration (Fig. 8). For instance, while eIF4G–IRES complexes protect residues within domain 4 (blue‐shaded nucleotides in Fig. 8A), complexes assembled with eIF4B preferentially protect the hairpin of domain 5 (blue‐shaded nucleotides in Fig. 8B). However, in both cases, a reorganization of flanking sequences within the pyrimidine‐rich region of domain 5 and the basal region of domain 3 (red‐shaded nucleotides in Fig. 8A,B) was observed, suggesting that these residues are in close proximity in the three‐dimensional structure of the IRES.
Figure 8.
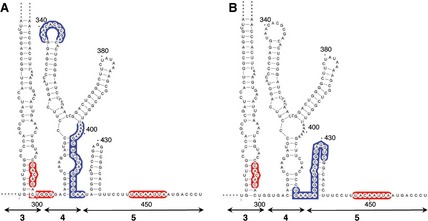
Summary of changes in NMIA attack upon incubation of the IRES with eIF4G (A) or eIF4B (B) at 0.5 mM Mg2+. Blue‐shaded nucleotides represent protected positions within domains 4 and 5, and red‐shaded nucleotides are those with higher reactivity.
Hydroxyl radical cleavage reveals a compact IRES structure at high concentrations of the divalent cation
The different patterns of reactivity observed in IRES complexes assembled with eIF4B or eIF4G proteins prompted us to assess the solvent accessibility of the IRES under low and high Mg2+ concentrations by hydroxyl radical cleavage footprinting, a methodology that reveals nucleotides buried within the folded RNA structure. In this assay, in situ‐generated hydroxyl radicals cleave locations on the RNA backbone in proportion to their solvent exposure 40. The solvent accessibility of the IRES was probed using Fe(II)–EDTA and H2O2 in a solution of RNA renatured in the presence or absence of Mg2+, followed by primer extension analysis with fluorescently labeled primers, and capillary electrophoresis was used to resolve the reaction products. The results yielded cleavage information at single‐nucleotide resolution, indicating solvent exposure of the 3′ end region of the IRES (Fig. 9A).
Figure 9.
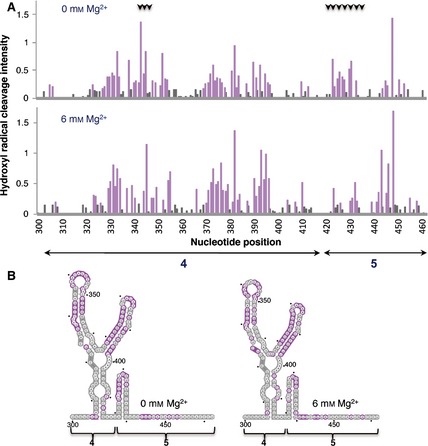
Hydroxyl radical probing of IRES domains 4 and 5. (A) Histograms of cleavage intensity versus nucleotide position in the absence or presence of Mg2+ determined using in situ‐generated hydroxyl radicals. Cleavages were determined by primer extension using fluorescent‐labeled primers and capillary electrophoresis. Gray bars indicate solvent‐inaccessible nucleotides in the folded RNA (intensities < half the mean), and solvent‐accessible residues are shown in purple. Black arrows at the top indicate positions whose accessibility decreases upon increasing concentration of Mg2+ in the folding buffer. (B) Predicted secondary RNA structure of domains 4 and 5 of the IRES imposing SHAPE constraints. Solvent‐inaccessible nucleotides are shown in gray; solvent‐accessible nucleotides are shown in purple.
Upon folding of the RNA in the presence of high concentrations of Mg2+, the hairpin of domain 5 became solvent‐inaccessible (Fig. 9B, gray nucleotides) indicating burial of this region due to structural compaction. Concomitantly, a change in accessibility to the solvent for residues within the 10 nt loop of domain 4 was observed (Fig. 9B, compare gray and purple nucleotides in left and right panels). Thus, the basal region of domain 4 and most of domain 5 remain solvent‐inaccessible at high Mg2+ concentration. These results, together with those shown in Figs 4, 5 and 6, suggest that eIF4B interaction with the IRES is independent of the compactness of the IRES structure, while binding of eIF4G is favored by RNA flexibility in this region, probably provided by a solvent‐accessible hairpin of domain 5.
Discussion
In this study, we show that the FMDV IRES structure is modulated by the concentration of Mg2+ ions. Using SHAPE methodology, we identified three discrete regions where the RNA structure is dependent on the cation concentration. Furthermore, the conformational changes induced by Mg2+ concentration differentially affected the binding of two host factors: eIF4G and eIF4B. Interestingly, the IRES–eIF4G complex was sensitive to the conformational changes induced by Mg2+ ions. Indeed, at high Mg2+ concentrations of the ion, no footprinting was observed on the IRES RNA, but it was readily observed at low Mg2+ concentration. Conversely, interaction of eIF4B with the IRES was detected at both low and high concentrations of the ion, leading to a similar pattern of reactivity irrespective of the concentration of Mg2+. The analysis of IRES–protein complexes revealed an increased reactivity in regions flanking eIF4G and eIF4B binding sites, indicating that assembly of these complexes generates secondary structure rearrangements in regions adjacent to the protein‐binding site.
In recent years, RNA has received much attention, and has been shown to be a major player in many cellular processes. Many of the novel biological functions identified depend on RNA structure. However, obtaining accurate three‐dimensional models of RNA molecules is hampered by the flexibility of RNA in solution and the relative low number of RNA three‐dimensional structures available so far 41. SHAPE probing methodology has the advantage over other high‐resolution approaches of allowing analysis of long RNA molecules in solution. In this regard, recent SHAPE structural studies have provided information on various RNA conformations, tertiary structures, long‐distance 3′→5′ interactions and ribonucleoprotein complexes of RNA in solution 42.
Based on the results shown here by measuring SHAPE reactivity using either radiolabeled primers and conventional denaturing gels or fluorescent primers and capillary electrophoresis, the IRES element adopts a near‐native conformation in the absence of proteins. In further support of these data, it is worth noting that the organization of the IRES in bulges and stems, as defined by the susceptibility towards NMIA attack under low (0.5 mm) Mg2+, is in agreement with that defined previously on the basis of DMS, and RNases T1 or T2 attack using a buffer containing 1 mm Mg2+ 11. Furthermore, all procedures used to probe the RNA structure of this IRES element indicated that the stem holding the apical region of domain 3 remains resistant to NMIA, DMS, and RNases T1 or T2 attack, and mutations that disrupt some of these base pairs led to defective IRES elements 46.
With regard to the changes induced by different Mg2+ concentrations on the local RNA flexibility of the apical region of domain 3, it is worth noting that this region harbors three conserved motifs (GNRA, RAAA and the C‐rich loop). Disruption of these motifs or destabilization of the corresponding stems led to a defective IRES and induced changes in the RNA structure of this region 3. The SHAPE reactivity changes observed upon Mg2+ increase in the folding buffer are consistent with a strong influence of the divalent cation on the structural organization of this RNA domain.
As Mg2+ ions dictate RNA tertiary folding 26, a potential outcome of our work was identification of nucleotides involved in tertiary interactions within the IRES element. Attempts to identify positions involved in distant interactions made use of RNA structure prediction programs that incorporate SHAPE reactivity data 49. This was performed as an unbiased method to detect base pairs between nucleotides that resulted in modified reactivity upon Mg2+ incubation. However, no gross changes in stem‐loop composition were detected irrespective of whether the full‐length IRES, individual domains or combinations of its structural domains (4–5, 3–4–5 or 2–4–5) were used. This result shows that the secondary RNA structure is stable even in the absence of Mg2+, which, in turn, appears to indicate that Mg2+ contributes to tertiary interactions.
Candidate nucleotides to form base pairs were also manually analyzed by pairwise comparison of the changes in reactivity and solvent accessibility. Attempts to identify candidate base pairs involving residues of the 10 nt loop of domain 4 did not detect a second site at which reactivity decreased to the same extent under high Mg2+ concentration. Given the difficulties of predicting non‐canonical base pairs in the RNA structure, it is possible that residues involved in long‐range interactions were missed if either non‐canonical base pairs or internal bulges were involved. Although we cannot rule out the possibility that a high Mg2+ concentration stabilizes intra‐loop interactions 52, potential pairing of nucleotides GUAACACGCGAC(337–348) with positions CUCGGUUUA(390–398) was identified. Interestingly, this is partially coincident with the protection of residues UUUAAAAAAG(397–403) by eIF4G at 0.5 mm Mg2+ concentration but not at 6 mm. Thus, recognition of the eIF4G‐binding site at low mm Mg2+ may be at least in part favored by a more flexible RNA structure due to lack of this putative tertiary interaction.
The contribution of the 10 nt loop of domain 4 to IRES activity was analyzed in transfection assays using two mutants, consisting of a full deletion of the loop and a double mutant carrying the substitutions A339C and A340G (designed to base pair with C345 and G346), mimicking the conformation observed at high Mg2+ concentration. Both mutants showed a reduction in internal initiation activity of approximately 80% relative to the wild‐type IRES, suggesting that A339 and A340 perform critical roles in the contribution of this loop to IRES activity. In support of these data, we have found that this particular region is reorganized upon addition of eIF4G, and that the eIF4G–IRES interaction is sensitive to the RNA structure reorganization induced by divalent cations. This result has important implications given that the concentration of Mg2+ in the cellular cytoplasm is approximately 0.5–1 mm 54. Interestingly, although K+ is the predominant monovalent cation in the intracellular environment, we found that Mg2+ concentration determines RNA structure irrespective of which monovalent ion (Na+ or K+) is used, in agreement with previous data 23. Furthermore, the low RNA‐binding affinity of eIF4G 55 may explain the moderate protection of eIF4G in the absence of other factors such as eIF4A, PTB and Ebp1 16.
Because of the relevance of the IRES structural organization for picornavirus protein synthesis, we compared the IRES solvent accessibility measured by hydroxyl cleavage with the inhibitory capacity of 2′‐O‐methyl oligoribonucleotides targeting the entire IRES region in mammalian cells 56. Interestingly, while most oligoribonucleotides complementary to domain 4 were non‐inhibitory, those complementary to the 10 nt loop of domain 4 and the hairpin of domain 5 were inhibitory. This positive correlation exemplifies the importance of understanding RNA structure for rational development of small molecules with potential antiviral activity.
Collectively, our results reveal the critical role of divalent cations for IRES structure, that, in turn, affects the interaction of the IRES with eIF4G, an essential factor for translation initiation driven by this IRES element. Future studies are necessary to dissect the implications of divalent cations on assembly of competent IRES–protein complexes in living cells. Additionally, as the FMDV IRES is a long, very efficient element that may serve as model for RNA studies of complex IRES elements, understanding the principles controlling its RNA structure has potential implications for other ribonucleoprotein complexes controlling internal initiation of translation.
Experimental procedures
Constructs and RNA synthesis
The construct expressing the FMDV IRES has been described previously 3. Mutagenesis of the IRES element was performed using the Quikchange mutagenesis procedure (Agilent Technologies, Santa Clara, CA, USA), using oligonucleotides 5′‐CAGGTACCCCGAGGCACTCGGGATCTGA‐3′ (forward) and 5′‐TCAGATCCCGAGTGCCTCGGGGTACCTG‐3′ (reverse) to generate the mutant Δ338–347, and 5′‐AGGTACCCCGAGGTCGCACGCGACACTCGG‐3′ (forward) and 5′‐CCGAGTGTCGCGTGCGACCTCGGGGTACCT‐3′ (reverse) for the mutant A339C/A340G. All plasmids were sequenced to ensure correctness (Macrogen Europe, Amsterdam, The Netherlands).
Prior to in vitro transcription, plasmids were linearized using SphI. RNA synthesis was performed for 2 h at 37 °C using T7 RNA polymerase 3 and the linearized DNA template in 40 mm Tris/HCl, 50 mm dithiothreitol, 0.5 mm rNTPs, as described previously 17.
SHAPE analysis
RNAs (2 pmol) were treated with NMIA (Invitrogen, Eugene, OR, USA) 29 using a range of MgCl2 concentrations (0–6 mm) in the folding buffer. Prior to primer extension, 0.5 pmol of treated and untreated RNAs were incubated with 2 pmol of the antisense 5′ end fluorescently‐labeled primer 5′‐TAGCCTTATGCAGTTGCTCTCC‐3′ at 65 °C for 5 min and 35 °C for 5 min, and then chilled on ice for 2 min 57. Primer extension reactions were performed in a final volume of 16 μL containing reverse transcriptase buffer (50 mm Tris/HCl pH 8.3, 3 mm MgCl2, 75 mm KCl, 8 mm dithiothreitol) and 1 μm of each dNTP. The mixture was heated at 52 °C for 1 min prior to addition of 100 units of SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA), and incubated at 52 °C for 30 min. A sequencing ladder was generated using untreated RNA in the presence of 0.1 mm ddCTP. NED fluorophore (Applied Biosystems, Warrington, UK) was used for both NMIA‐treated and untreated samples, while VIC fluorophore (Applied Biosystems, Warrington, UK) was used for the sequencing ladder. cDNA products were resolved by capillary electrophoresis.
For primer extension using radiolabeled primers, equal amounts of NMIA‐treated and untreated RNAs (0.5 pmol) were incubated with 2 pmol each of the antisense 5′ end 32P‐labeled primers 5′‐CTACGAAGCAACAGTG‐3′, 5′‐CCCGGGTGTGGGTACC‐3′, 5′‐GGAATGGGATCCTCGAGCTCAGGGTC‐3′ and 5′‐GGCCTTTCTTTATGTTTTTGGCG‐3′ in separate reactions, as described previously 17. Primer extension was performed as above. After alkaline hydrolysis of the RNA template 17, cDNAs were precipitated and fractionated in 6% acrylamide/7 m urea gels in parallel to a sequence ladder obtained using the same primer as in the primer extension reaction.
SHAPE RNA probing in the presence of IRES‐binding factors
RNA–protein complexes were assembled in folding buffer containing a high Mg2+ concentration (100 mm HEPES pH 8.0, 6 mm MgCl2, 100 mm NaCl) or folding buffer containing a low Mg2+ concentration (100 mm HEPES pH 8.0, 0.5 mm MgCl2, 100 mm NaCl) in the presence of increasing concentration of His‐tagged eIF4B (60, 300 and 500 nm) or His‐tagged eIF4G (40 and 80 nm) for 10 min at room temperature. Then, RNA alone or RNA pre‐incubated with the protein of interest was treated with NMIA and analyzed by primer extension analysis using radiolabeled primers as described in the previous paragraph.
Hydroxyl radical footprinting
RNA (2 pmol) was denatured, chilled on ice and folded in folding buffer (40 mm MOPS pH 8.0, 80 mm KOAc, and 0, 0.5 or 6 mm MgCl2). Fe(II)–EDTA (7.5 mm Fe(SO4)2(NH4)2·6H2O and 11.25 mm EDTA, pH 8.0), 0.3% hydrogen peroxide and 150 mm sodium ascorbate solutions were freshly prepared before each experiment 58. Hydroxyl radical cleavage was initiated by adding 1 μL of the Fe(II)–EDTA complex, 1 μL of sodium ascorbate and 1 μL of hydrogen peroxide solution to the reaction. A control reaction lacking Fe(II)–EDTA was performed in parallel. After incubation at 37 °C for 30 s, reactions were quenched and precipitated by addition of 75% glycerol (one‐third volume), 1 μL of 20 mg·mL−1 glycogen, 1 μL of 3 m NaCl, 2 μL of 0.5 m EDTA and 2.5 volumes of ice‐cold ethanol. RNAs were re‐suspended and reverse‐transcribed. Primer extension reactions were performed using fluorescent primers as described above for SHAPE reactivity. The NED fluorophore was used for both treated and untreated samples, while the VIC fluorophore was used for the sequencing ladder. cDNA products were resolved by capillary electrophoresis.
SHAPE reactivity and hydroxyl radical cleavage data analysis
Quantitative SHAPE reactivity for individual datasets obtained with radiolabeled primers was normalized to a scale spanning 0–1.5, in which 0 indicates unreactive nucleotides and the mean intensity at highly reactive nucleotides is set to 1.0 3. For each dataset, the intensity of the untreated RNA was subtracted from that of the treated RNA to obtain the net reactivity. Then, a normalization factor was determined by excluding the 2% most‐reactive peak intensities, followed by calculating the mean for the next 8% of peak intensities. Reactivity values were then divided by this mean 58. Data from at least three independent assays were used to calculate the SHAPE reactivity (mean ± SD).
SHAPE electropherograms were analyzed using qushape software 59. Quantitative SHAPE reactivities for individual datasets were normalized to a scale spanning 0–1.5, in which 0 indicates an unreactive nucleotide, and the mean intensity at highly reactive nucleotides is set to 1.0.
Hydroxyl radical cleavage intensities were normalized to a scale from 0 to 1.5, where 1.0 is defined as the mean intensity of highly reactive residues. Nucleotides with reactivity values lower than half the mean reactivity are defined as solvent‐inaccessible 60.
RNA structure analysis
The secondary RNA structure was visualized using VARNA 61. RNAsc, RNAstructure and shapeknots software 49 were used to predict the secondary RNA structure including SHAPE reactivity values.
Nucleotide sequence analysis
Alignment of 180 FMDV IRES nucleotide sequences available in GenBank was performed using clustalx with default parameters (http://www.clustal.org). Polymorphic positions in the region 306–380 were determined relative to the FMDV C‐S8 isolate (GenBank accession number AF274010.1). The sequence logo (http://weblogo.berkeley.edu) was generated from alignment of the 180 IRES sequences. The overall height of each stack indicates the sequence conservation at that position (measured in bits), and the height of symbols within the stack reflects the relative frequency of the corresponding nucleic acid at that position.
Expression and purification of proteins
Escherichia coli BL21 was transformed with plasmids expressing His‐tagged eIF4G 62 and His‐tagged eIF4B 63, and grown in Luria–Bertani medium at 37 °C. Protein expression was induced using isopropyl‐β‐d‐thiogalactopyranoside for 2 h. Bacterial cell lysates were prepared in binding buffer (20 mm NaH2PO4, 500 mm NaCl, 20 mm imidazole), and cell debris was eliminated by centrifugation twice at 16 000 g for 30 min at 4 °C. The lysate was loaded into His‐GraviTrap columns (GE HealthCare, Little Chalfont, UK), and the recombinant protein was eluted using 500 mm imidazole. Proteins were dialyzed against phosphate buffer, pH 6.8, containing 1 mm dithiothreitol, and stored at −20 °C in 50% glycerol.
IRES activity assays
Relative IRES activity was quantified as the expression in transfected BHK‐21 cell monolayers of luciferase normalized to that of chloramphenicol acetyltransferase from bi‐cistronic mRNAs as described previously 47. Experiments were performed in triplicate wells, and each experiment was repeated at least three times.
Author contributions
GL, NF, EMS Planned experiments; GL, NF Performed experiments; GL, EMS Analyzed data; EMS Wrote the paper.
Acknowledgements
We are grateful to I.N. Shatsky [Department of Chemistry and Biochemistry of Nucleoproteins, Belozersky Institute of Physico‐Chemical Biology, Lomonosov Moscow State University, Moscow, Russia)] and A. Nieto [Department of Molecular and Cellular Biology, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (C.S.I.C.), Madrid (Spain)] for provision of His‐tagged eIF4B and His‐tagged eIF4G expression vectors, respectively, J. Ramajo [Department of Genome Dynamics and Function, Centro de Biología Molecular Severo Ochoa, CSIC‐Universidad Autónoma de Madrid, Madrid (Spain)] for excellent technical assistance, and C. Gutierrez [Department of Genome Dynamics and Function, Centro de Biología Molecular Severo Ochoa, CSIC‐Universidad Autónoma de Madrid, Madrid (Spain)], I. Dotu [Biology Department, Boston College, Massachusetts (USA)], C. Briones [Department of Molecular Evolution, Centro de Astrobiología (INTA‐CSIC), Torrejón de Ardoz, Madrid (Spain)] and R. Diaz‐Toledano[Department of Genome Dynamics and Function, Centro de Biología Molecular Severo Ochoa, CSIC‐Universidad Autónoma de Madrid, Madrid (Spain)] for helpful comments on the manuscript. This work was supported by grants BFU2011‐25437, CSD2009‐00080 (Ministerio de Economía y Competitividad) and an institutional grant from Fundación Ramón Areces.
References
- 1.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J & Wise JA (2009) A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol 16, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K & Luhrmann R (2013) RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J 32, 2804–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez N, Fernandez‐Miragall O, Ramajo J, Garcia‐Sacristan A, Bellora N, Eyras E, Briones C & Martinez‐Salas E (2011) Structural basis for the biological relevance of the invariant apical stem in IRES‐mediated translation. Nucleic Acids Res 39, 8572–8585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perard J, Leyrat C, Baudin F, Drouet E & Jamin M (2013) Structure of the full‐length HCV IRES in solution. Nat Commun 4, 1612. [DOI] [PubMed] [Google Scholar]
- 5.Jan E & Sarnow P (2002) Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324, 889–902 [DOI] [PubMed] [Google Scholar]
- 6.Martinez‐Salas E, Pacheco A, Serrano P & Fernandez N (2008) New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol 89, 611–626 [DOI] [PubMed] [Google Scholar]
- 7.Filbin ME & Kieft JS (2009) Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spriggs KA, Bushell M & Willis AE (2010) Translational regulation of gene expression during conditions of cell stress. Mol Cell 40, 228–237 [DOI] [PubMed] [Google Scholar]
- 9.Sonenberg N & Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez‐Salas E (2008) The impact of RNA structure on picornavirus IRES activity. Trends Microbiol 16, 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez‐Miragall O, Lopez de Quinto S & Martinez‐Salas E (2009) Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res 139, 172–182 [DOI] [PubMed] [Google Scholar]
- 12.Lopez de Quinto S, Lafuente E & Martinez‐Salas E (2001) IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7, 1213–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N & Martinez‐Salas E (2009) A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 37, 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stassinopoulos IA & Belsham GJ (2001) A novel protein–RNA binding assay: functional interactions of the foot‐and‐mouth disease virus internal ribosome entry site with cellular proteins. RNA 7, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI & Hellen CU (2000) A cell cycle‐dependent protein serves as a template‐specific translation initiation factor. Genes Dev 14, 2028–2045 [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Abaeva IS, Marintchev A, Pestova TV & Hellen CU (2011) Common conformational changes induced in type 2 picornavirus IRESs by cognate trans‐acting factors. Nucleic Acids Res 39, 4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez N, Garcia‐Sacristan A, Ramajo J, Briones C & Martinez‐Salas E (2011) Structural analysis provides insights into the modular organization of picornavirus IRES. Virology 409, 251–261 [DOI] [PubMed] [Google Scholar]
- 18.Duncan CD & Weeks KM (2010) Nonhierarchical ribonucleoprotein assembly suggests a strain‐propagation model for protein‐facilitated RNA folding. Biochemistry 49, 5418–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez‐Miragall O & Martinez‐Salas E (2003) Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9, 1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez‐Miragall O, Ramos R, Ramajo J & Martinez‐Salas E (2006) Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 12, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung S & Schlick T (2013) Candidate RNA structures for domain 3 of the foot‐and‐mouth‐disease virus internal ribosome entry site. Nucleic Acids Res 41, 1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenvi CL, Dong KC, Friedman EM, Hanson JA & Cate JH (2005) Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations – implications for the study of ribosome dynamics. RNA 11, 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan ZJ & Chen SJ (2011) Importance of diffuse metal ion binding to RNA. Met Ions Life Sci 9, 101–124 [PMC free article] [PubMed] [Google Scholar]
- 24.Draper DE (2004) A guide to ions and RNA structure. RNA 10, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos R & Martinez‐Salas E (1999) Long‐range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES). RNA 5, 1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowman JC, Lenz TK, Hud NV & Williams LD (2012) Cations in charge: magnesium ions in RNA folding and catalysis. Curr Opin Struct Biol 22, 262–272 [DOI] [PubMed] [Google Scholar]
- 27.Behrouzi R, Roh JH, Kilburn D, Briber RM & Woodson SA (2012) Cooperative tertiary interaction network guides RNA folding. Cell 149, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer K, Petersen A, Niepmann M & Beck E (1995) Interaction of eukaryotic initiation factor eIF–4B with a picornavirus internal translation initiation site. J Virol 69, 2819–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson KA, Merino EJ & Weeks KM (2006) Selective 2′–hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 30.Weeks KM & Mauger DM (2011) Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res 44, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortimer SA & Weeks KM (2009) C2′‐endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci USA 106, 15622–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortimer SA & Weeks KM (2009) Time‐resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one‐second snapshots and at single‐nucleotide resolution. Nat Protoc 4, 1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez de Quinto S & Martinez‐Salas E (1997) Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol 71, 4171–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pineiro D, Fernandez N, Ramajo J & Martinez‐Salas E (2013) Gemin5 promotes IRES interaction and translation control through its C–terminal region. Nucleic Acids Res 41, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez de Quinto S & Martinez‐Salas E (2000) Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6, 1380–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolupaeva VG, Pestova TV, Hellen CU & Shatsky IN (1998) Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 273, 18599–18604 [DOI] [PubMed] [Google Scholar]
- 37.Clark AT, Robertson ME, Conn GL & Belsham GJ (2003) Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol 77, 12441–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassili G, Tzima E, Song Y, Saleh L, Ochs K & Niepmann M (2004) Sequence and secondary structure requirements in a highly conserved element for foot‐and‐mouth disease virus internal ribosome entry site activity and eIF4G binding. J Gen Virol 85, 2555–2565 [DOI] [PubMed] [Google Scholar]
- 39.Pestova TV, Hellen CU & Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16, 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangan P, Masquida B, Westhof E & Woodson SA (2003) Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci USA 100, 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruz JA, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cao S, Das R, Ding F, Dokholyan NV, Flores SCet al (2012) RNA‐Puzzles: a CASP‐like evaluation of RNA three‐dimensional structure prediction. RNA 18, 610–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao F, Gulay SP, Kasprzak W, Dinman JD, Shapiro BA & Simon AE (2013) The kissing‐loop T–shaped structure translational enhancer of pea enation mosaic virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J Virol 87, 11987–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B, Grigull J, Ore MO, Morin S & White KA (2013) Global organization of a positive‐strand RNA virus genome. PLoS Pathog 9, e1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lusvarghi S, Sztuba‐Solinska J, Purzycka KJ, Pauly GT, Rausch JW & Grice SF (2013) The HIV–2 Rev‐response element: determining secondary structure and defining folding intermediates. Nucleic Acids Res 41, 6637–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero‐Lopez C, Barroso‐Deljesus A, Garcia‐Sacristan A, Briones C & Berzal‐Herranz A (2014) End‐to‐end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X‐tail region. Nucleic Acids Res 42, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serrano P, Ramajo J & Martinez‐Salas E (2009) Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology 388, 221–229 [DOI] [PubMed] [Google Scholar]
- 47.Martinez‐Salas E, Regalado MP & Domingo E (1996) Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J Virol 70, 992–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez N, Buddrus L, Pineiro D & Martinez‐Salas E (2013) Evolutionary conserved motifs constrain the RNA structure organization of picornavirus IRES. FEBS Lett 587, 1353–1358 [DOI] [PubMed] [Google Scholar]
- 49.Zarringhalam K, Meyer MM, Dotu I, Chuang JH & Clote P (2012) Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One 7, e45160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuter JS & Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH & Weeks KM (2013) Accurate SHAPE‐directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci USA 110, 5498–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chauhan S & Woodson SA (2008) Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc 130, 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bindewald E, Wendeler M, Legiewicz M, Bona MK, Wang Y, Pritt MJ, Le Grice SF & Shapiro BA (2011) Correlating SHAPE signatures with three‐dimensional RNA structures. RNA 17, 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunther T (2006) Concentration, compartmentation and metabolic function of intracellular free Mg2+. Magnes Res 19, 225–236 [PubMed] [Google Scholar]
- 55.Lomakin IB, Hellen CU & Pestova TV (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol 20, 6019–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fajardo T Jr, Rosas MF, Sobrino F & Martinez‐Salas E (2012) Exploring IRES region accessibility by interference of foot‐and‐mouth disease virus infectivity. PLoS One 7, e41382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dotu I, Lozano G, Clote P & Martinez‐Salas E (2013) Using RNA inverse folding to identify IRES‐like structural subdomains. RNA Biol 10, 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGinnis JL, Duncan CD & Weeks KM (2009) High‐throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 468, 67–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karabiber F, McGinnis JL, Favorov OV & Weeks KM (2013) QuShape: rapid, accurate, and best‐practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chakraborty S, Mehtab S, Patwardhan A & Krishnan Y (2012) Pri‐miR–17–92a transcript folds into a tertiary structure and autoregulates its processing. RNA 18, 1014–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darty K, Denise A & Ponty Y (2009) VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J & Nieto A (2000) Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol 20, 6259–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC & Shatsky IN (2003) Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol Cell Biol 23, 8925–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]


