Abstract
Integral membrane proteins of the cell surface and most intracellular compartments of eukaryotic cells are assembled at the endoplasmic reticulum. Two highly conserved and parallel pathways mediate membrane protein targeting to and insertion into this organelle. The classical cotranslational pathway, utilized by most membrane proteins, involves targeting by the signal recognition particle followed by insertion via the Sec61 translocon. A more specialized posttranslational pathway, employed by many tail-anchored membrane proteins, is composed of entirely different factors centered around a cytosolic ATPase termed TRC40 or Get3. Both of these pathways overcome the same biophysical challenges of ferrying hydrophobic cargo through an aqueous milieu, selectively delivering it to one among several intracellular membranes and asymmetrically integrating its transmembrane domain(s) into the lipid bilayer. Here, we review the conceptual and mechanistic themes underlying these core membrane protein insertion pathways, the complexities that challenge our understanding, and future directions to over-come these obstacles.
Keywords: targeting, protein translocation
INTRODUCTION
Integral membrane proteins (IMPs) make up 20–30% of the eukaryotic proteome and are extremely diverse. They serve many essential cellular functions, such as functioning as signaling receptors, mediating intracellular trafficking, facilitating organelle biogenesis, and transporting a variety of molecules across cellular membranes. Structurally, IMPs range from having a single transmembrane domain (TMD) that simply anchors a soluble domain to the membrane to having tightly packed bundles containing more than 20 TMDs (Figure 1a). Most of the cell’s IMPs populate the plasma membrane and intracellular compartments of the secretory and endocytic pathways. All these proteins are initially assembled at the endoplasmic reticulum (ER). This is the site where an IMP’s TMD(s) are integrated into the membrane, final topology is determined, and tertiary and quaternary structures are achieved (Alder & Johnson 2004, Rapoport et al. 2004, Skach 2009). If these decisive steps in IMP biogenesis are successful, the IMP is subsequently sorted to its final location of function (Pryer et al. 1992, Rothman 1994). Otherwise, one of several quality-control pathways routes the IMP for degradation (Hegde & Ploegh 2010, Meusser et al. 2005).
Figure 1.
The structural diversity of integral membrane proteins. (a) Several types of integral membrane proteins are shown in different topologies. Most types of membrane proteins are inserted by a cotranslational pathway, although some use a posttranslational pathway. ER, endoplasmic reticulum. (b) Hydrophilicity plot of a G protein–coupled receptor (bovine rhodopsin) using the Kyte-Doolittle scale. The hydrophobic regions are below the axis and indicated in gold. Note that six of the seven transmembrane domains (TMDs) are easily recognizable by their hydrophobicity, but the seventh is more polar. Sequences of the first and last TMDs are shown; polar residues within each TMD are indicated by asterisks. (c) Three-dimensional structure of bovine rhodopsin showing that TMDs are highly variable in structure; some are tilted or kinked. TMD1 and TMD7 are shown in blue and red, respectively. The unusual sequence features of TMD7 (see panel b) correspond to its unusual structure in the membrane.
All IMPs destined to be inserted into the ER face a set of biophysical challenges that must be overcome by the targeting and insertion machinery. First, the hydrophobic TMDs of IMPs must be continuously shielded from the aqueous cytosol. This shielding is essential because the tremendously crowded cytosolic environment (~300 mg/ml protein) would promote potentially toxic aggregation and inappropriate interactions. Thus, TMDs need to be recognized as they emerge from the ribosome by the targeting machinery. Second, IMPs must be targeted to the appropriate organelle, which requires the cytosolic targeting factors to interface with specific membrane receptors. Third, the TMDs need a route of transport past the highly polar surface of the membrane into the hydrophobic core of the lipid bilayer. This TMD insertion must be asymmetric, with the final orientation consistent with the IMP’s final folded state. This means that the insertion machinery must recognize, orient, and provide a potential path into the membrane for a remarkably wide range of sequences.
The vast majority of IMPs are targeted and inserted into the ER as they are synthesized. This cotranslational pathway, discovered more than 30 years ago, utilizes the same machinery cells use to translocate soluble proteins across the ER membrane (Anderson et al. 1982, Lingappa et al. 1978, Rapoport 2007, Wickner & Schekman 2005). However, some IMPs whose TMDs emerge from the ribosome only after translation terminates cannot access the cotranslational pathway (Borgese et al. 2003, Kutay et al. 1993). Instead, these proteins insert posttranslationally by an entirely different pathway whose key components have only recently been discovered (Borgese & Fasana 2010). This review summarizes our current knowledge of these two IMP insertion pathways, highlights common principles that have emerged from their comparison, and identifies outstanding questions moving forward.
FEATURES AND DIVERSITY OF MEMBRANE PROTEINS
The current understanding of IMP insertion has been heavily informed by extensive studies inspecting, comparing, manipulating, and designing IMP substrates. These studies have identified statistical features of native IMPs and experimentally probed the relationship between IMP sequence, membrane insertion, and topology. Thus, we have a solid working framework regarding the common features of TMDs and the surrounding sequences that influence insertion. This framework also provides an approximate scope of the limits of sequences and structures that can be handled by the insertion machinery. An overarching conclusion from such studies is that the IMP insertion machinery must be remarkably flexible and highly dynamic. We first summarize these substrate-centric studies before moving on to a discussion of the insertion machinery and the nature of its interactions with membrane protein substrates.
The Transmembrane Domain
The general features of a TMD must be compatible with the biophysical constraints imposed by the lipid bilayer in which it resides (White & von Heijne 2008, White & Wimley 1999). For insertion into the ER, the “ideal” TMD is a ~20 residue α-helix composed of nonpolar, mostly hydrophobic side chains. This length is optimal for spanning the ~30-Å lipid bilayer of the ER membrane, and the α-helical structure maximizes hydrogen bonding of the hydrophilic peptide backbone to shield it from the hydrophobic acyl chains of the membrane phospholipids. Indeed, TMDs with the sole function of anchoring a soluble domain to the membrane adhere closely to this idealized TMD. Consequently, an appreciation of these principles has long been used to derive TMD prediction algorithms based primarily on hydrophobicity (Argos et al. 1982, Kyte & Doolittle 1982).
However, most TMDs not only are membrane anchors but also play other functional roles for the protein in which they are located. These functions occur within a local context that is usually distinct from the ER lipid bilayer in which the TMD is initially inserted. Many TMDs are part of a more complex IMP with other TMDs. Even the TMD of a single-spanning IMP often has additional functionality, such as the capacity to interact with other proteins or to be cleaved by intramembrane proteases (Urban & Freeman 2002). Furthermore, TMDs of different compartments have different lengths and sequence features that seem to reflect the vastly different lipid compositions, membrane asymmetry, and biophysical properties among cellular membranes (Sharpe et al. 2010). Yet, all TMDs must be inserted at the ER, and each TMD must be triaged into the lipid bilayer shortly after it is synthesized. Thus, the environment in which a TMD is initially inserted can be markedly different from the final context in which it functions. This necessarily means that TMD sequences have competing constraints: On the one hand, a TMD must possess certain features that allow it to be identified as such by the insertion machinery. On the other hand, it must also possess elements that allow its specific trafficking, assembly, and function in its final context.
For example, polytopic (or multispanning) IMPs often contain TMDs that are remarkably polar, unusually long or short, kinked, or of poor helical propensity (Elofsson & von Heijne 2007). Such TMDs can be entirely stable within the final IMP tertiary structure, but they can be difficult to recognize as TMDs when viewed in isolation (Figure 1b,c). Indeed, direct analysis of individual TMDs from polytopic IMPs reveals that they are often poor in targeting and insertion assays (Enquist et al. 2009). These considerations explain why hydropathy-based prediction algorithms more accurately predict TMDs of single-spanning IMPs than those of polytopic IMPs ( Jones 2007, Ott & Lingappa 2002). Thus, it is clear that in addition to sequence features of the TMD, its positional context within a more complex protein directly influences its integration into the membrane. How the translocation machinery interprets such contextual cues and balances it with local sequence features is only partially understood (possible models are discussed below).
Determinants of Transmembrane Domain Integration
Direct experimentation has quantified the relative free-energy contribution of each amino acid within an insulated 20-residue test sequence toward the insertion propensity of a TMD (Hessa et al. 2005a,b; Hessa et al. 2007, Lundin et al. 2008, White & von Heijne 2008). In these extensive analyses, the overall insertion efficiencies were measured for numerous engineered sequences that were systematically varied with respect to TMD orientation, amino acid composition, and amino acid position. These test sequences were carefully positioned to be housed uniformly within the translocation machinery, after which their net insertion efficiency was directly quantified (Figure 2a). Beyond confirming the expected strong dependency on hydrophobicity, position effects for each amino acid were identified and quantified. Remarkably, the position effects closely mirror the changing biophysical character across the lipid bilayer, ranging from the hydrophilic head groups to the central apposition of the acyl chains from each leaflet. The apparent free-energy contribution of each amino acid in any test sequence was found to be additive, with the sum determining the final insertion probability. The strong correlation of this “biological hydrophobicity scale” with biophysical partitioning experiments employing model peptides (Ulmschneider et al. 2005; Wimley & White 1996, 2000) suggests that, for an isolated TMD insulated by engineered sequences, insertion approximates a thermodynamically driven equilibration between an aqueous environment (presumably within the translocation channel) and the surrounding lipid bilayer.
Figure 2.
Assay systems used to probe sequence determinants of insertion and topology. (a) An integral membrane protein (IMP) is targeted to and docked at the translocon via earlier transmembrane domains (gray), and a later test sequence (red) is measured for insertion. The relative amounts of the two possible outcomes are assayed, typically by a modification such as glycosylation. (b) A single-spanning IMP and its flanking sequences are systematically varied, and the substrate is presented to the translocation apparatus. The final topology achieved by this combination targeting-translocation-insertion system is assayed, again typically using glycosylation as a readout.
Determinants of Topology
Just as the amino acid composition of TMDs dictates insertion probability, the primary sequence also influences the topology of transmembrane segments. The first identified and still overarching determinant of IMP topology is the positive-inside rule in which positive charges (from lysines and arginines) flanking a TMD are statistically found in the cytoplasm (Hartmann et al. 1989, von Heijne 1986b). This effect, which is most prominently observed for bacterial IMPs (von Heijne 1986b), is likely to be strongly influenced by the proton-motive force across the membrane (Cao et al. 1995) and a bilayer asymmetry that contains anionic phospholipids on the cytoplasmic face of the membrane (van Klompenburg et al. 1997). In the analogous eukaryotic ER, where there is no bilayer asymmetry or proton-motive force, the positive-inside statistical correlation to topology is less uniform ( Johansson et al. 1993; Sipos & von Heijne 1993; von Heijne 1986a, 1989; Wallin & von Heijne 1998). Nonetheless, it is still readily apparent for single-spanning IMPs and for the first TMD of polytopic IMPs. The basis of this effect is at least partially the result of electrostatic interactions of TMD flanking charges with the Sec61 translocon that may disfavor positive charges from translocating across the channel (Goder et al. 2004).
Studies examining how a single TMD near the N terminus of a protein inserts into the bilayer (Figure 2b) have identified additional factors that can also influence the orientation of a TMD (Higy et al. 2004, 2005). Longer or more hydrophobic TMDs favor a type I topology (N terminus facing the ER lumen, C terminus facing the cytosol), possibly because TMDs first enter the translocon in this orientation (Goder & Spiess 2003, Wahlberg & Spiess 1997), and a “stronger” TMD would partition into the lipid bilayer before reorientation. In contrast, a rapidly folding domain flanking the N terminus of the TMD tends to result in a type II topology (N terminus facing the cytosol, C terminus facing the ER lumen), presumably because the folded domain sterically hinders translocation through the translocon (Denzer et al. 1995). Similarly, an internal TMD encountered after the N terminus has already translocated into the ER lumen (via a cleavable N-terminal signal sequence) is constrained to a type I topology because the N terminus is precluded from coming back into the cytosol.
In polytopic IMPs, adjacent TMDs necessarily have alternate topologies, so insertion of the first TMD strongly constrains the orientation of the entire IMP (Wickner & Lodish 1985). However, this original determination is not absolute and clearly can be influenced by downstream events. In some proteins, the first TMD is insufficient to dictate insertion in a unique topology by itself and cooperates with the next TMD for insertion (Skach & Lingappa 1993). In other instances, a protein with two TMDs appears to sample multiple topologic states, as introducing a glycosylation site into the intervening sequence can bias the final outcome by forcing that region of polypeptide to remain in the ER lumen (Goder et al. 1999). Thus, at least some TMDs are likely to sample more than one topology before eventually inserting into the bilayer. Precisely when topology is irreversibly set remains unclear, as several examples of dynamic reorientation of polytopic IMPs have been described. For example, the protein Aquaporin-1 is initially made as a four-TMD protein but later acquires a six-TMD topology (Lu et al. 2000). More recently, a single charge mutation at the C terminus of a four-TMD protein was sufficient to completely invert the protein’s final topology (Seppala et al. 2010). This means that topology was not irrevocably set even by the last codon, indicating that very distant sequence elements can influence insertion and topology of TMDs synthesized far earlier.
Complexities and Limits of Prediction
The insertion and topology of marginally hydrophobic TMDs are especially influenced by both local and distant positional effects including flanking and downstream sequences, the fate of neighboring TMDs, and tertiary interactions, such as hydrogen bonding with an adjacent TMD (Enquist et al. 2009, Meindl-Beinker et al. 2006, Nilsson et al. 2000, Sato et al. 1998). Not only is this apparent in polytopic IMPs as noted above, but it can also be seen for a single TMD whose insertion can be influenced by clusters of charged amino acids more than 50 residues away (Fujita et al. 2010). Thus, the empirical rules elucidated from model substrates cannot necessarily be applied to a TMD inspected in isolation, particularly within a polytopic protein.
These effects may occur cotranslationally or may be the result of posttranslational readjustments during later stages of protein folding (Kauko et al. 2010). The full extent of how different sequence determinants in a polytopic IMP cooperate to dictate final topology is still not well understood, and it remains difficult to distinguish the contributions of local versus global effects, which have thus far been studied in only a few cases (Gafvelin & von Heijne 1994, Sato et al. 1998, Seppala et al. 2010, Skach 2009). As a result, highly reliable topology prediction of complex IMPs remains a challenge, and the consensus between different prediction methods remains relatively poor (Fagerberg et al. 2010, Jones 2007, Ott & Lingappa 2002). To better understand and predict the insertion of polytopic IMPs, expanding the number of available structures of IMPs will be essential. Only then can prediction algorithms that are derived from experimental validation using a small subset of model proteins be tested accurately. In addition, understanding how the insertion machinery interacts with and facilitates the integration of TMDs in molecular detail will likely provide more insights into how the primary amino acid sequences of IMPs are decoded into their final structure.
COTRANSLATIONAL MEMBRANE PROTEIN INSERTION
Most eukaryotic IMPs are inserted cotranslationally into the ER membrane. Genetic, biochemical, and phylogenetic analyses, including the functional reconstitution of model IMP insertion with purified components, have defined the minimal components of this insertion pathway (Deshaies & Schekman 1987, Görlich & Rapoport 1993, Görlich et al. 1992b, Jungnickel et al. 1994, Oliver et al. 1995, Poritz et al. 1990, Rapoport et al. 2004, Rothblatt et al. 1989, Stirling et al. 1992, Wickner & Schekman 2005). Both the targeting and insertion machinery are conserved in all organisms, and they are used not only for IMPs but also for the translocation of soluble proteins. Operationally, this pathway can be divided into two steps: recognition/targeting of a nascent IMP to an ER-localized translocon and translocation/insertion of the IMP into the ER membrane.
Recognition and Targeting
IMP targeting is mediated when the first hydrophobic element, either a cleavable N-terminal signal sequence or a TMD, emerges from a translating ribosome. This hydrophobic domain is recognized by the signal recognition particle (SRP) on the ribosome, taken to the ER via an interaction with the SRP receptor (SR), and transferred to the Sec61 translocon in a GTPase-dependent step (Grudnik et al. 2009, Keenan et al. 2001, Wild et al. 2004; Figure 3a). Mid-resolution (~8–12 Å) cryo-electron microscopy structures of a translating IMP substrate bound to an SRP (Halic et al. 2004), the ribosome-SRP-SR complex (Halic et al. 2006), and the ribosome-nascent chain (RNC)-Sec61 complex (Beckmann et al. 1997, Beckmann et al. 2001, Ménétret et al. 2000, Morgan et al. 2002) provide a general framework for how targeting is achieved (Halic & Beckmann 2005). This general framework has been substantially refined and informed by a wealth of biochemical studies and high-resolution crystal structures of several key domains of SRP and SR (Grudnik et al. 2009).
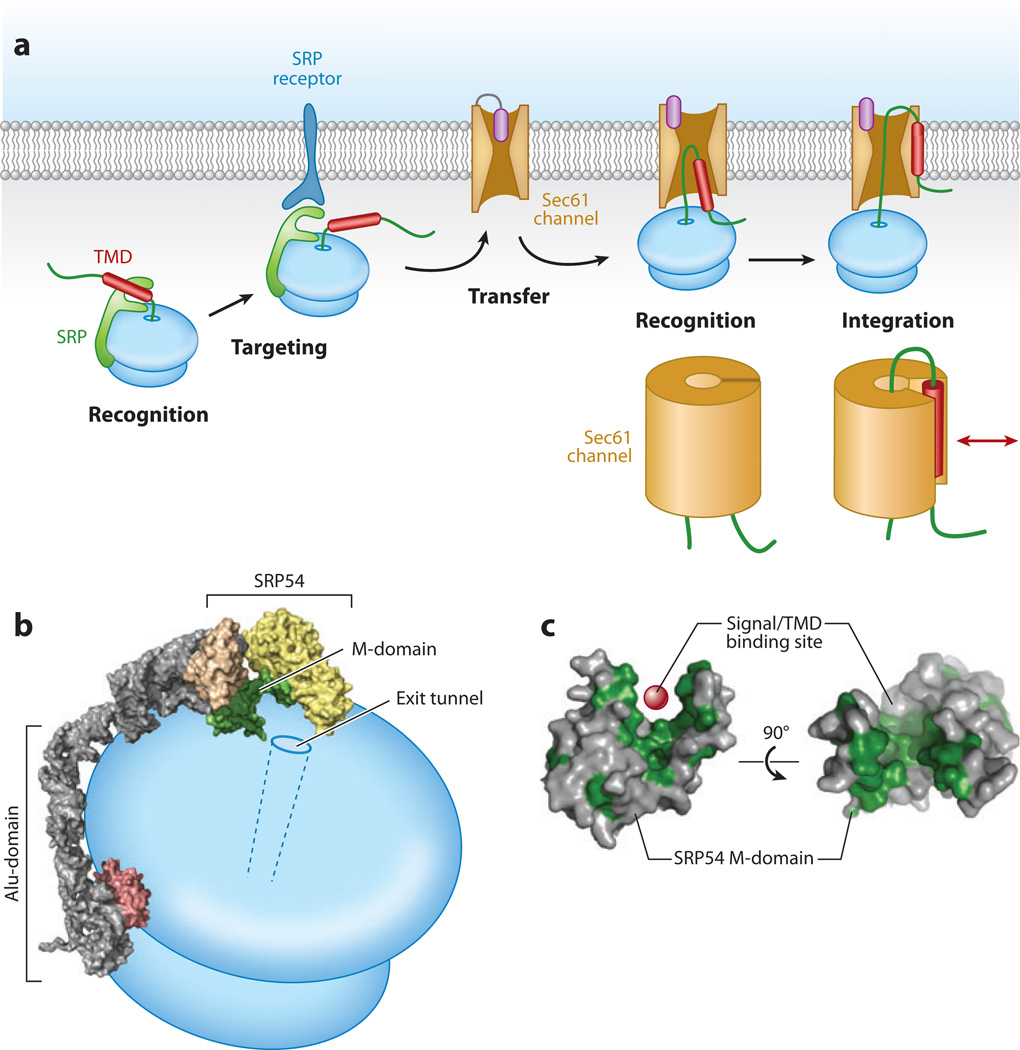
Figure 3.
Basic steps in cotranslational membrane protein insertion. (a) An integral membrane protein (IMP) is recognized via a hydrophobic transmembrane domain (TMD) (or signal sequence) by the signal recognition particle (SRP) on the ribosome. This is targeted to the endoplasmic reticulum (ER) membrane via the SRP receptor (SR) and transferred to the Sec61 translocation channel. The transfer reaction is accompanied by a second recognition step, this time by the Sec61 channel. The TMD is then integrated into the membrane via lateral access to the lipid bilayer provided by the Sec61 channel. Below the last two diagrams are schematic representations of an early and late TMD integration step in which the TMD (red ) is leaving the Sec61 channel via a lateral opening. (b) Schematic diagram of the SRP positioned on the ribosome, taken from cryo-electron microscopy analyses. The signal sequence recognition domain of SRP54 (the M-domain) is indicated in green and is positioned adjacent to the exit tunnel on the ribosome. The portion of the Alu-domain that binds near the elongation factor site to slow translation is shown in pink. (c) The M-domain of SRP54 in two views with hydrophobic residues indicated in green. The red sphere represents the site of signal sequence (or TMD) binding within the hydrophobic groove.
In the prevailing model, the 54-kDa subunit of SRP (SRP54) is positioned at the exit tunnel of the ribosome and interacts with the hydrophobic substrate via its M-domain, whereas the Alu-domain of SRP binds near the elongation factor binding site on the ribosome to slow translation (Figure 3b). Upon SRP-SR interaction, the SRP-ribosome complex shifts to expose a portion of the ribosomal exit tunnel site to Sec61. Presumably, when Sec61 binds to this exposed site, the nascent chain is transferred from SRP to Sec61, the SRP-SR complex is disassembled, and the translational pause is relieved. These events are coordinated by the GTPase activities of SRP54, and the α and β subunits of SR, each of which are responsive to interactions with substrate, each other, the ribosome, and the translocon. Details of the targeting reaction and how individual steps are coordinated are reviewed extensively elsewhere (Grudnik et al. 2009, Keenan et al. 2001).
For the purposes of this review, several conceptual points are worth highlighting. First, the interaction of SRP with a hydrophobic signal or TMD occurs via a deep hydrophobic groove in the M-domain of SRP54 (Figure 3c). X-ray structures of this domain reveal that the hydrophobic groove is lined with many methionines that are thought to facilitate accommodation of diverse sequences via their flexible side chains ( Janda et al. 2010, Keenan et al. 1998). Thus, in the context of an RNC, SRP is relatively promiscuous and sequence independent, allowing a wide range of potential IMPs to be targeted successfully. Second, the interaction occurs precisely at the ribosomal exit tunnel (Halic et al. 2004) (Figure 3b), thereby completely precluding exposure of the TMD to the bulk cytosol. The maintenance of this interaction until a coordinated transfer to the Sec61 channel ensures constant shielding of the TMD during targeting. Third, SRP interaction with the first TMD transiently slows translation (Lipp et al. 1987, Walter & Blobel 1981). This increases the kinetic window for targeting (Lakkaraju et al. 2008, Mason et al. 2000) and prevents downstream TMDs from becoming exposed to the cytosol before docking at the translocon. Finally, the TMD is likely to be presented to the Sec61 channel in a coordinated and defined manner ( Jiang et al. 2008, Song et al. 2000), although this step is poorly understood. Coordinated transfer may be important because the putative signal sequence or TMD binding site on Sec61 is likely buried deep within its structure (van den Berg et al. 2004). Thus, the RNC has been hypothesized to prime the Sec61 channel to accept a signal sequence or TMD (Osborne et al. 2005), similar to how other translocation partners such as SecA in bacteria may prime SecY (the Sec61α homolog) (Zimmer et al. 2008). In this manner, TMDs are minimally exposed to the cytosol prior to integration, a theme that also appears to apply to posttranslational IMP insertion (as discussed below).
The Sec61-Protein-Conducting Channel
The ER translocon through which cotranslational substrates are inserted is centrally composed of the Sec61 complex, a heterotrimer composed of α, β, and γ subunits. Sec61α is unambiguously the component that forms the translocation channel and provides a lateral path for TMD insertion into the lipid bilayer. Early biochemical, biophysical, and cryo-EM studies illustrated that the translocon binds tightly to ribosomes through conserved and species-exchangeable binding sites and forms a channel continuous with the ribosomal tunnel through which the nascent chain emerges (Beckmann et al. 1997, Prinz et al. 2000). Electrophysiological measurements and environment-sensitive fluorescent probes showed that nascent proteins are initially in an aqueous environment when they first enter the translocon, and functional reconstitutions unequivocally identified the Sec61 complex as the core factor facilitating IMP insertion (Crowley et al. 1993,1994; Görlich & Rapoport 1993; Simon & Blobel 1991). Furthermore, cross-linking studies revealed that TMDs consistently interact with Sec61α early in the insertion process, before or simultaneously with their interactions with lipids (Do et al. 1996, Heinrich et al. 2000, Martoglio et al. 1995, Mothes et al. 1997), which suggests that the Sec61 complex forms an interface between proteinaceous and lipid environments for inserting TMDs. This is consistent with the simplest case of IMP insertion, in which the translocon passively facilitates TMD partitioning between its aqueous interior and the surrounding hydrophobic bilayer.
A mechanistic model for how lateral release of TMDs into the lipid bilayer occurs was suggested by high-resolution crystal structures of archaeal and bacterial homologs of the Sec61 complex (Tsukazaki et al. 2008, van den Berg et al. 2004) (Figure 4a,b). These structures showed that one Sec61 heterotrimer consists of a pseudosymmetric arrangement of the α subunit into two domains consisting of TMDs 1–5 and TMDs 6–10, respectively. These domains form an hourglass-shaped aqueous pore consisting of two symmetrical funnel-shaped cavities that converge into a constriction ring of hydrophobic residues in the middle of the bilayer. On the lumenal side of the pore, a short helix identified as the plug fills the center of that funnel. The γ subunit is composed of two helices: one that forms a TMD that diagonally crosses the interface between the two α subunit domains and one at the cytoplasmic surface of the membrane. The β subunit’s single TMD lines a third side of the α subunit, leaving the fourth side unobstructed for functioning as a putative lateral gate out of the central channel into the lipid bilayer. The interface of this lateral gate is complex and formed primarily by helices 2 and 7, with contributions from helices 3 and 8 (Figure 4a).
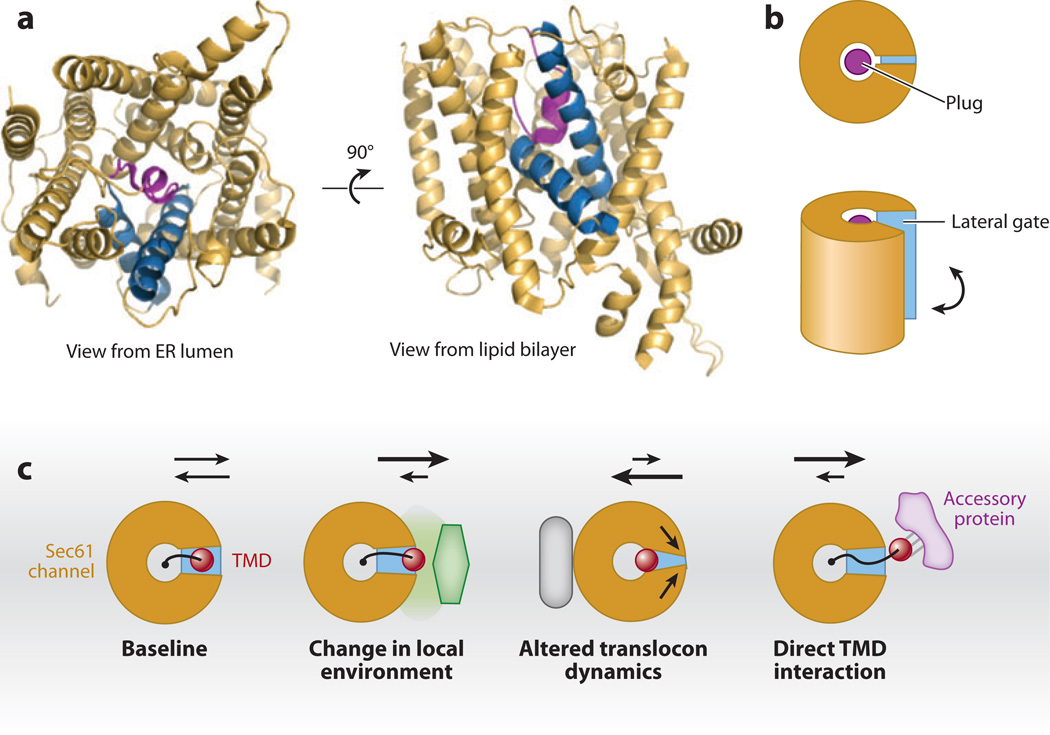
Figure 4.
The Sec61 translocon and potential accessory factors. (a) Structural representation of the archaeal homolog of the Sec61 complex as viewed from the endoplasmic reticulum (ER) lumen and from the plane of the membrane (rotated by 90°). The left and right halves show a pseudo twofold symmetry. The helices are organized around a central pore that is occluded by a short plug helix (purple). The two blue helices are helix 2 and helix 7, which form the so-called lateral gate. When these are separated, they provide a route of access to the lipid bilayer from the central pore. In the lipid bilayer view, note that the plug (which is in the central pore) is directly behind the lateral gate, illustrating how lipid access could be mediated when the gate helices move apart. (b) Schematic representation of the structures in panel a. (c) Schematic representation of how lateral exit of a transmembrane domain (TMD) from the Sec61 channel (as viewed from the ER lumen) could be influenced by trans-acting accessory factors. The baseline model shows the TMD (red ) tethered to a nascent chain (black line). The nascent chain is in the central channel, whereas the TMD is at the lateral gate. The arrows above the diagram indicate the partitioning of the TMD between the lateral gate and surrounding lipid bilayer ( gray plane). This partitioning could be biased toward the membrane if the local environment was altered by either a different lipid composition or a nearby protein. Conversely, a protein that binds to the translocon could influence its dynamic properties, such as opening of the lateral gate, thereby biasing against partitioning into the lipid bilayer. In the last diagram, an accessory protein such as TRAM (translocating-chain associating membrane protein) that directly binds to a TMD could also bias movement of the TMD out of the Sec61 channel.
Extensive cross-linking studies argue persuasively that the putative channel identified in the crystal structure houses the nascent chain during translocation and IMP integration (Cannon et al. 2005). For this to occur, interaction of the first hydrophobic signal sequence or TMD with the Sec61 complex presumably displaces the plug from the lumenal side of the pore and results in the opening of the channel (see the last steps in Figure 3a). The mechanism of plug displacement is not clear, but it may occur when the first TMD or signal sequence intercalates between lateral gate helices 2 and 7. Cross-linking studies in yeast and Escherichia coli systems using a signal sequence support such an interaction (Plath et al. 2004, Wang et al. 2004), and the juxtaposition of these two helices with the plug indicates a potential communication between them (van den Berg et al. 2004) (Figure 4a). Notably, mutants in the plug domain or constriction ring in E. coli and yeast have a signal-sequence suppressor, or prl, phenotype, which allows promiscuous channel engagement by partially defective signal sequences or TMDs, as would be expected from a channel biased toward an open state (Derman et al. 1993; Junne et al. 2006, 2007, 2010).
Once the plug is displaced and the channel is in an open state (a configuration for such structures remains elusive), it could house a nascent chain spanning the bilayer within its primarily hydrophilic channel while still providing a perpendicular point of access to the lipid bilayer via the lateral gate (Figure 4c). In this manner, a single Sec61 complex can permit translocation across or insertion into the membrane. Whether the single Sec61 heterotrimer is always the active state of a functioning translocon remains uncertain. For secretory and relatively simple (e.g., single-spanning) IMPs, additional complexities need not be invoked to explain satisfactorily their translocation and insertion (although this in no way precludes additional factors from playing key roles to maximize efficiency). However, the numerous observations with complex IMPs described below suggest that their correct insertion is very difficult to reconcile with a single ribosome-bound Sec61 heterotrimer as the sole machinery.
Accessory Factors
Although the Sec61 complex is the core factor essential for cotranslational IMP insertion, numerous accessory factors may aid in the biogenesis of IMPs in substrate-specific ways. Indeed, the broader translocon is an ill-defined ensemble of the Sec61 complex together with associated membrane proteins including enzymes (such as signal peptidase complex and oligosaccharyl transferase complex), putative accessory factors [such as the translocon-associated protein (TRAP) complex and translocating-chain associating membrane (TRAM) protein], membrane chaperones (such as Calnexin), and proteins of unclear function [such as ribosome-associated membrane protein 4 (RAMP4), Sec62, Sec63, p180, and others] (Görlich & Rapoport 1993, Meyer et al. 2000, Pina et al. 2011, Savitz & Meyer 1990, Tyedmers et al. 2000, Yamaguchi et al. 1999). The structures, stoichiometry, and functions of much of this machinery are poorly understood, and their roles in translocation or membrane insertion are essentially unexplored.
Some accessory factors, such as signal peptidase and oligosaccharyl transferase, are required for the proper folding and maturation of IMPs and, in some cases, could also influence topology (Goder et al. 1999, Yamagishi et al. 2011). Others, such as TRAP, are stably associated with membrane-bound ribosomes (Ménétret et al. 2000) and may directly influence the structure of the ribosome-translocon complex (Fons et al. 2003, Ménétret et al. 2000) and/or the functionality of the Sec61 translocon (Fons et al. 2003). And yet others, such as TRAM, can be cross-linked to a subset of nascent translocating proteins (Do et al. 1996, Görlich et al. 1992a, Heinrich et al. 2000, Mothes et al. 1994). What, if anything, could these factors be doing in the vicinity of the Sec61 translocon to influence IMP biogenesis?
At least three effects are possible (Figure 4c). First, the accessory factor could temporarily interact directly with parts of a nascent chain to influence directly its translocation, orientation, or membrane insertion. Second, an accessory factor could change the local environment that an inserting TMD encounters as it is leaving the Sec61 channel by sitting near the Sec61 lateral gate or perhaps influencing the local lipid composition. Third, the accessory factor could interact with the Sec61 complex to influence its dynamic properties, such as the mobility of its lateral gate. These mechanisms remain largely hypothetical, but each would influence TMD insertion by changing its translocation or partitioning behavior out of the Sec61 channel.
Of the various translocon components, TRAM has been best studied as a potential accessory factor in IMP biogenesis. TRAM is an integral ER protein with at least eight TMDs that is often located near the Sec61 complex and is necessary to reconstitute the integration of several model IMPs (Görlich & Rapoport 1993). Cross-linking studies show that TRAM can interact with both signal sequences (Görlich et al. 1992a, Voigt et al. 1996) and TMDs (Do et al. 1996, Heinrich et al. 2000, McCormick et al. 2003) in the vicinity of Sec61α, which supports a direct role in handling hydrophobic domains. Functional studies indicate that TRAM is especially stimulating for the translocation of proteins with weakly hydrophobic signal sequences or TMDs and that it enhances the integration of different substrates to different extents (Heinrich et al. 2000, Voigt et al. 1996). Consequently, TRAM may be necessary only in certain situations, such as those requiring a membrane chaperone to stabilize marginally hydrophobic TMDs at the threshold of being able to insert into the bilayer. TRAM could play a similarly important role in the insertion of polytopic IMPs by temporarily “holding” one or more TMDs before their coordinated partitioning into the bilayer (Do et al. 1996, Heinrich et al. 2000, Sadlish et al. 2005). This is supported by cross-linking studies showing that multiple TMDs of a polytopic IMP interact with Sec61α and/or TRAM at the same time before their apparent release into the bilayer (Heinrich & Rapoport 2003, Sadlish et al. 2005). Thus, by changing the local environment of the Sec61 channel, proteins such as TRAM may bias the intrinsic TMD hydrophobicity scale to favor lateral exit from the channel. In comparison, one could imagine that proteins restricting the dynamics of the Sec61 complex may restrict lateral gate opening to bias against insertion by narrowing the window of access to lipid for a translocating TMD.
COTRANSLATIONAL MEMBRANE PROTEIN TOPOGENESIS
With the above knowledge about the primary features of IMP substrates and the basic properties of the Sec61 complex, how can a holistic picture of TMD recognition and membrane topology determination be conceptualized? This question is complicated and, as discussed above, likely depends not only on the TMD but also on its flanking domains, position within a polypeptide, events that occurred prior to that point in biogenesis, and many other factors. As a simplified starting point, we first consider a generic potential TMD being synthesized by a translocon-bound ribosome (i.e., leaving targeting out of the picture for now) in which the preceding and subsequent sequences are left unspecified (Figure 5a).
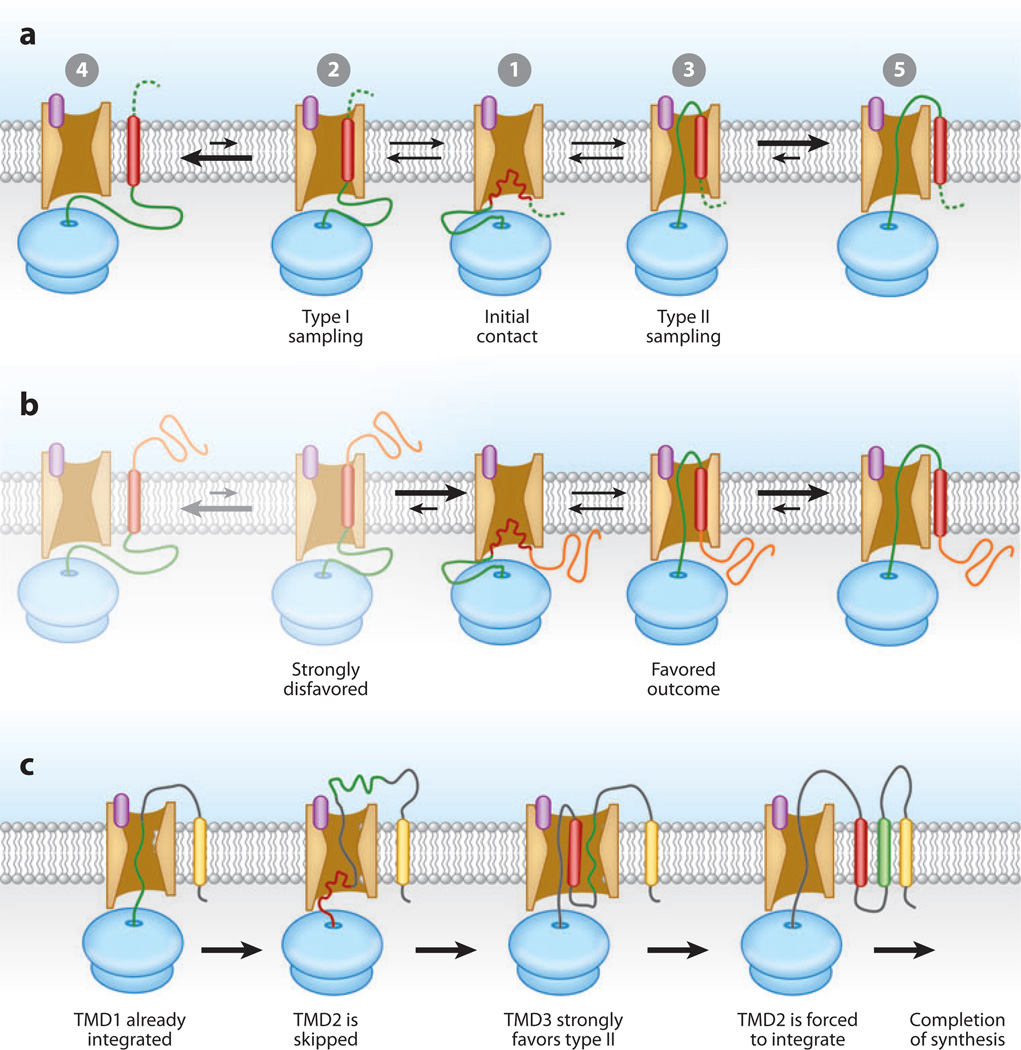
Figure 5.
Models of transmembrane domain (TMD) insertion. (a) Generic model for TMD-translocon interactions. Upon its initial emergence from the ribosome, a TMD (red) enters the cytosolic vestibule of the Sec61 channel (➀). This initial metastable state allows the nascent chain to sample either of two orientations (➁, ➂). These states are envisioned to be interconvertible for a limited period of time, until the TMD laterally moves into the lipid bilayer in one or the other orientation (➃, ➄). (b) Depending on any of various parameters, the sequence of events in panel a can be biased toward one or the other outcome. In the example shown, the N-terminal domain is lengthy and partially folded (orange) by the time the TMD emerges from the ribosome. This strongly disfavors its translocation into the lumen, a requirement for sampling the type I topology. Consequently, its sampling of and eventual insertion in the type II orientation is the favored outcome. (c) An example of nonsequential insertion of TMDs in a polytopic integral membrane protein (IMP). Starting from the left, a polytopic IMP begins insertion. At the stage shown, TMD1 (yellow) has already inserted, and TMD2 (green) is entering the translocation channel. However, owing to sequence features of TMD2, it is not recognized by the translocon and is skipped, resulting in its translocation into the endoplasmic reticulum (ER) lumen. TMD3 (red) is then made and interacts with the translocon in a manner similar to that shown in panel a. Its local sequence features heavily favor the type II orientation (same as TMD1), thereby forcing segments of the previously translocated part of the protein (including TMD2) back into the translocation channel. The marginally hydrophobic TMD2 can now insert into the membrane accompanied by TMD3.
Transmembrane Domain–Translocon Interactions
Spectroscopic and cross-linking studies suggest that, as a candidate TMD completes synthesis and resides within the ribosomal tunnel, it can interact with proteins lining the tunnel and be induced into a more compact (possibly α-helical) structure (Woolhead et al. 2004). Presumably, such an event is not necessary for every TMD, but it appears to represent the first point of TMD recognition (Liao et al. 1997). Interestingly, a similar sensing of TMDs or other motifs inside the ribosome has been observed in the cytosol, with effects on ribosome structure (e.g., via recruitment of factors to its surface) (Berndt et al. 2009, Mariappan et al. 2010) and function (such as translational stalling or delayed termination) (Mariappan et al. 2010, Nakamura et al. 1996, Yanagitani et al. 2011). Whether analogous effects are also observed for RNCs at the translocon is not entirely clear, but this may involve changes to the architecture of the ribosome-translocon complex and possibly to translocon composition (Liao et al. 1997, Pool 2009, Woolhead et al. 2004). Such structural changes could prepare the translocon for dealing with the TMD upon its emergence from the ribosome.
Next, the candidate TMD exits the ribosomal tunnel, presumably into the cytosolic vestibule of the Sec61 complex (Figure 5a). The secondary structure of the TMD at this point is poorly understood but may be highly dynamic given the ability of at least some TMDs to sample multiple conformations and topologic orientations (Goder & Spiess 2003, Goder et al. 1999). The TMD (and immediately flanking regions) may be partially stabilized by interactions with residues of the Sec61 channel (Goder et al. 2004; Junne et al. 2006, 2010; McCormick et al. 2003) or nearby membrane proteins such as TRAM (Do et al. 1996, Heinrich et al. 2000). With further synthesis of at least 20 residues beyond the TMD, it would have the potential freedom to sample both type I and type II topologies.
In the type I configuration, the downstream polypeptide would reside in the cytosolic vestibule and/or spool out between the ribosome and translocon (Figure 5a, left). In the type II configuration, the downstream polypeptide would reside within the Sec61 channel (Figure 5a, right). The position and secondary structure of the TMD during this sampling of topologic orientations is not clear but presumably depends at least partially on its amino acid composition and hydrophobicity. The most attractive possibility is that the TMD is α-helical and poised at the lateral gate, where it could sample the lipid environment and potentially exit the channel (Hessa et al. 2005a, 2007). Cross-linking studies provide evidence that the TMD makes protein-protein interactions with the Sec61 channel from a relatively rigid location (McCormick et al. 2003, Sadlish et al. 2005). Some TMDs have been observed to cross-link to Sec61α simultaneously with TRAM and/or lipids (Do et al. 1996, Heinrich et al. 2000, Martoglio et al. 1995, Mothes et al. 1997, Sadlish et al. 2005), which suggests that they are positioned at the lateral gate. If there are no other constraints, the TMD settles on an orientation based on local sequence features such as TMD hydrophobicity, length, and flanking charges. With further synthesis, the candidate TMD can laterally partition into the bilayer (presumably via the lateral gate) (Heinrich et al. 2000).
It is attractive to posit that most, if not all, TMDs that are synthesized on a membrane-bound ribosome are handled in this generic manner in which they make certain stereotypic interactions with the ribosome, Sec61 complex, and accessory factors. Where each TMD would be unique is in the constraints imposed on its ability to sample the different topologic configurations and/or the lipid bilayer. These constraints could be imposed by local sequence features (TMD hydrophobicity, amino acid composition, flanking charges), positional context (the location of the TMD within a protein), and trans-acting factors. The net effect of these constraints on TMD dynamics would be to bias the topology and insertion of each TMD in a highly contextual manner. With this framework in mind, let us now consider how it applies to different types of proteins, ranging from simple to complex.
Single-Spanning Membrane Proteins
The generic mechanism readily explains single-spanning IMPs containing their only TMD within ~40 residues of the N terminus (Higy et al. 2004). In these cases, the N-terminal domain is sufficiently short and unstructured so as not to preclude sampling of each topology by the TMD. In the type II orientation, the N-terminal domain is cytosolic, whereas sampling the type I orientation requires the N-terminal domain to translocate through or reside within the aqueous Sec61 channel. Integration of the TMD secures the final topology, and the remainder of the protein completes synthesis on the membrane-bound ribosome.
The situation changes when the N-terminal domain preceding the TMD is long or structured (Kida et al. 2001, 2005). Now, the ability of the TMD to sample the type I topology is disfavored because the N-terminal domain cannot be pulled easily through the narrow Sec61 channel (Figure 5b). Thus, sampling of the type II orientation is favored, thereby biasing the final topology upon integration of the TMD. The N-terminal domain length at which the type I topology is substantially disfavored depends somewhat on the features of the N-terminal sequence, but it seems to be approximately 50–60 residues on the basis of examination of natural proteins (Wallin & von Heijne 1995).
To make a type I protein with a longer N-terminal domain, a cleavable N-terminal signal sequence is typically needed to initiate translocation of the N terminus before the TMD is synthesized. Once the N terminus is committed to the lumen, the TMD emerges from the ribosome and presumably goes through the same generic steps outlined above, but without being able to sample readily the type II topology (because the already translocated N terminus would need to be pulled back through the Sec61 channel to accommodate this). This constraint therefore biases insertion in the type I topology.
Thus, topology of even these simple IMPs is determined by a combination of local sequence features, such as hydrophobicity and flanking charges, and more global constraints, such as the length and structure of the N-terminal domain. If the global constraint is severe, unfavorable local features can be bypassed. For example, a type II IMP with a large N-terminal domain may not need to obey the positive-inside rule to ensure efficient insertion into the correct topology, given that the alternative is highly disfavored. Thus, TMD context, sometimes even more than local sequence elements, can play a very large role in insertion and topology; this principle becomes especially important for the polytopic IMPs considered next.
Polytopic Membrane Proteins
The earliest models for polytopic IMP insertion, proposed more than 30 years ago, envisioned the successive integration of each TMD as each emerges from the ribosome (Blobel 1980, Wessels & Spiess 1988). This sequential-insertion model is clearly feasible and necessitates that every TMD and its local sequence elements be sufficiently robust to drive both its translocon recognition and membrane insertion. Because TMDs in a polytopic IMP need to alternate orientations, each TMD should contain key topology determinants such as asymmetric flanking charges. This together with constraints imposed by the preceding TMD would facilitate proper biogenesis. This may well occur for some proteins, and it is feasible that sequential insertion is relatively common in bacterial systems where the positive-inside rule is much more rigorously observed and the insertion machinery appears to be simpler.
As discussed above, eukaryotic polytopic IMPs are far more structurally diverse, and their TMDs are less stereotypic than are prokaryotic IMPs. This may be a consequence of their need to be more functionally dynamic with respect to diverse interactions and regulation, thereby imposing considerable sequence constraints that can clash with those needed for insertion. These same features may also be why eukaryotic IMPs are poorly expressed in heterologous systems and difficult to crystallize relative to their prokaryotic counterparts. These and other considerations mean that poorly recognizable TMDs that in isolation are not able to insert must nonetheless be inserted in the context of the native IMP (Enquist et al. 2009, Lu et al. 2000). One way to imagine how this occurs is if the local contributions that would normally disfavor insertion of a TMD can be overcome by strong constraints imposed by other parts of the protein (Fujita et al. 2010, Kauko et al. 2010, Nilsson et al. 2000, Yamagishi et al. 2011).
As an example (Figure 5c), suppose that the second TMD (TMD2) of a polytopic IMP is poorly hydrophobic and therefore not recognized as a TMD when it is first synthesized. Rather than inserting into the membrane, this segment of polypeptide is translocated into the ER lumen. Produced later in synthesis would be TMD3, which has strong local topology determinants and is suitably hydrophobic for efficient membrane insertion. TMD3 would enter the translocon, presumably sample the different orientations and states, and laterally insert into the membrane. If TMD3 strongly favors the same topology as the already inserted TMD1, a segment of intervening polypeptide would be forced to span the membrane. When constrained in this manner, the energetically least costly outcome would be for the previously skipped marginal TMD2 to insert into the membrane. Thus, in this and related scenarios, TMDs can be inserted nonsequentially, with some being temporarily skipped and residing briefly in the ER lumen or cytosol. This has been demonstrated for artificially constructed polytopic TMDs, as which weakly recognized TMDs have been observed in both the ER lumen and cytosol (Goder et al. 1999, Kauko et al. 2010, Yamagishi et al. 2011). Even in vivo, TMDs of native IMPs at least transiently access the cytosol and/or ER lumen during polytopic IMP biosynthesis (Cheng & Gilmore 2006).
Several variations on this theme can be further envisaged. First, rather than being skipped entirely, candidate TMDs may be stored temporarily at or near the site of integration without necessarily committing to complete release into the membrane (Do et al. 1996, Sadlish et al. 2005). Such TMDs could be held by accessory proteins or part of the Sec61 complex. Such provisional TMDs could at a later time, by the same mechanism outlined above, integrate or even reorient contingent on downstream events (Kauko et al. 2010, Lu et al. 2000). In more elaborate scenarios (building on essentially the same principles), multiple TMDs could be stored or skipped temporarily, only to reorient and integrate well after their initial encounter with the translocon. In the extreme case, reorientation and insertion of a subset of TMDs would occur completely posttranslationally in a process analogous to soluble protein folding (Kauko et al. 2010, Lu et al. 2000, Skach 2009). How such posttranslational IMP folding (and possible reorientation) occurs remains completely unexplored. In each of these various scenarios of nonsequential or posttranslational insertion, chaperones in the cytosol, membrane, and/or lumen may be needed to stabilize intermediates to minimize off-pathway interactions.
The above framework is developed from studies with the simplest model proteins that have explored mechanistic issues of TMD-translocon interactions combined with parallel studies of complex proteins that illustrate the types of gymnastics that a nascent IMP can undergo. However, beyond the Sec61 complex and possibly TRAM, the machinery required for these proposed processes remains very poorly explored. Indeed, only robustly inserting model type I and type II single-spanning IMPs have been reconstituted to any reasonable efficiency with purified Sec61 complex and TRAM (Görlich & Rapoport 1993, Oliver et al. 1995). Thus, although we can roughly explain how TMDs of considerably varied sequence and biophysical properties may be accommodated by the insertion machinery, a molecular-level understanding remains to be rigorously developed.
CHALLENGES TO UNDERSTANDING THE COTRANSLATIONAL PATHWAY
In the above, we propose an amalgamated model for cotranslational IMP insertion that balances contributions from purely thermodynamic effects with other factors that can modify or constrain TMD insertion in several ways. The observation that single TMDs analyzed in isolation insert by their net “biological hydrophobicity” (Hessa et al. 2007) suggests an underlying aqueous-lipid partitioning mechanism that fits nicely with a laterally gated core Sec61 channel deduced from structural studies (van den Berg et al. 2004). However, this base principle must necessarily be modified by several additional variables as revealed by instances of TMD prediction failure for complex IMPs, numerous noncanonical TMDs observed in native proteins, and considerable diversity of insertion behavior from studies of both engineered and native IMPs. To account for these myriad observations, our working model proposes that a baseline stereotypic mechanism for handling a newly made hydrophobic element is dynamically biased by a combination of cis-acting local sequence elements, more distant global constraints, and trans-acting accessory factors.
Our current understanding of cis-acting sequence determinants (both local and distant) is rather extensive, and further refinements are easily obtainable as desired. However, a mechanistic picture of how these sequences are interpreted by the translocon is limited. Although the core machinery for cotranslational IMP insertion at the ER has been identified and structural information is now emerging, functional details remain obscure. Although the remarkably elegant reconstitution of model IMP insertion with purified membrane factors was achieved long ago (Görlich & Rapoport 1993), its subsequent application to understanding either detailed mechanism or complex IMP insertion has been technically challenging. At present, essentially all in vitro insertion studies utilize crude ER microsomes, whose membrane factors are both ill-defined and almost completely refractory to manipulation. Hence, a major goal moving forward will be the development of a robust but highly malleable insertion system reconstituted from purified recombinant proteins.
Developing methods to efficiently recapitulate and accurately assay complex IMP insertion into proteoliposomes reconstituted from solubilized ER membrane extracts is an important immediate goal. The eventual plan would be to replace key factors (e.g., the Sec61 complex or TRAM) with those produced from recombinant sources, thereby allowing detailed structure-function analysis. For mechanistic insights into an essential cellular process such as IMP insertion, this biochemical strategy (combined with parallel structural studies) will be integral to success, as indirect effects and compensatory changes would be very difficult to control with knockdown strategies. For more nuanced aspects of IMP biogenesis such as insertion of ambiguous TMDs or ensuring high-fidelity topology determination (Junne et al. 2007, Tipper & Harley 2002), carefully designed genetic screens or precise assays combined with large-scale mutant collections may help identify roles for putative accessory factors.
POSTTRANSLATIONAL MEMBRANE PROTEIN INSERTION
Although the majority of ER IMPs are targeted and inserted cotranslationally, a subpopulation inserts posttranslationally. Even though Sec61 in complex with other factors (such as Sec62 and Sec63) can mediate posttranslational translocation of soluble proteins in yeast (Rapoport 2007, Wickner & Schekman 2005), a role in posttranslational membrane protein insertion has not been found. This suggests another way that a subset of membrane proteins might be inserted into the ER membrane. Posttranslationally inserted proteins would include any IMPs that contain TMD(s) that are not exposed to the cytosol long enough during translation for efficient recognition by ribosome-bound SRP. Translational termination of such proteins makes them poor SRP substrates, necessitating their recognition, targeting, and insertion by a purely posttranslational mechanism.
Examples of such proteins include extremely small IMPs of either orientation and tail-anchored (TA) proteins that contain their only TMD within ~40 residues of the C terminus (Borgese & Fasana 2010, Chi et al. 1996, Coïc et al. 2005, Navarre et al. 1994, Wawrzynow et al. 1992) (see Figure 1a). The number of small IMPs remains unclear, largely because they are difficult to identify by bioinformatics. Nonetheless, several studies suggest that small proteins, many of which are predicted to be IMPs, are considerably underrepresented in genome annotations (Frith et al. 2006, Gerstein et al. 2010, Hemm et al. 2008, Ingolia et al. 2009, Roy et al. 2010).
By comparison, TA proteins are well annotated, and bioinformatics studies suggest they encompass ~3–5% of all IMPs in most eukaryotic genomes (Beilharz et al. 2003, Kalbfleisch et al. 2007). In mammals, this corresponds to ~300–400, most of which are targeted to the ER for insertion (Wattenberg & Lithgow 2001). They are functionally diverse, and members are involved in intracellular trafficking (e.g., essentially all SNAREs), protein translocation (e.g., Sec61β and γ), protein maturation, degradation, organelle structure, and lipid homeostasis (Borgese et al. 2003). Thus, their correct insertion is of basic cellular importance across all eukaryotes.
The logistical incompatibility of engaging the SRP pathway by TA proteins was appreciated long ago (Kutay et al. 1993). Studies shortly thereafter rigorously established that for a model TA protein, posttranslational insertion neither depends on nor efficiently engages the SRP-Sec61 system (Brambillasca et al. 2005, Kutay et al. 1995, Steel et al. 2002, Yabal et al. 2003). Although many subsequent studies established that an ATP- and protein-dependent process was involved (Abell et al. 2007; Kim et al. 1997, 1999; Masaki et al. 2003), specific machinery or a coherent framework for any TA-protein insertion pathway remained obscure until recently. Because a posttranslational IMP insertion pathway would need to overcome the same biophysical obstacles faced by the cotranslational pathway, concepts established in the cotranslational pathway proved crucial in elucidating the posttranslational pathway.
TRC40: A Tail-Anchored-Protein Targeting Factor
In the cotranslational pathway, SRP serves both a chaperoning and a targeting role, ensuring complete shielding of its hydrophobic cargo during cytosolic transit until the translocon is reached. Reasoning that TA proteins must be similarly chaperoned, biochemical strategies searched for TMD-dependent interaction partners that remain associated until successful membrane targeting (Favaloro et al. 2008, Stefanovic & Hegde 2007). This led to the identification of a highly conserved, essential cytosolic ATPase that was named TRC40 for TMD recognition complex of 40 kDa (Stefanovic & Hegde 2007). TRC40 is evolutionarily related to the bacterial arsenite transport factor ArsA (hence its original annotation as Asna-1), although its function in eukaryotes has clearly diverged to play a role in TA insertion.
Unbiased biochemical studies in an in vitro mammalian system established TRC40 as the primary interaction partner of most (but not all) newly synthesized ER-destined TA proteins in the cytosol via a TMD-dependent association (Favaloro et al. 2008, 2010; Stefanovic & Hegde 2007). Importantly, TRC40 did not interact with a TMD emerging from a translating ribosome but favored ribosome-released proteins containing a TMD near the C terminus (Stefanovic & Hegde 2007). Conversely, SRP interaction with a ribosome-released TMD is typically inefficient, presumably because SRP-substrate interactions are highly favored by the precise positioning of SRP relative to the ribosomal exit tunnel. These observations helped to explain why the TRC40 and SRP pathways do not compete or interfere with each other despite recognizing otherwise similar TMDs. TRC40 was further shown to have putative protein receptors at the ER membrane that stimulated substrate release in a manner dependent on the ATPase activity of TRC40. Concomitant with release from TRC40, the TA protein was inserted into the membrane. Thus, the identification of TRC40 as a cytosolic chaperone that specifically recognizes and facilitates the insertion of TA proteins provided a rudimentary framework for posttranslational TA-protein insertion (Stefanovic & Hegde 2007; Figure 6a).
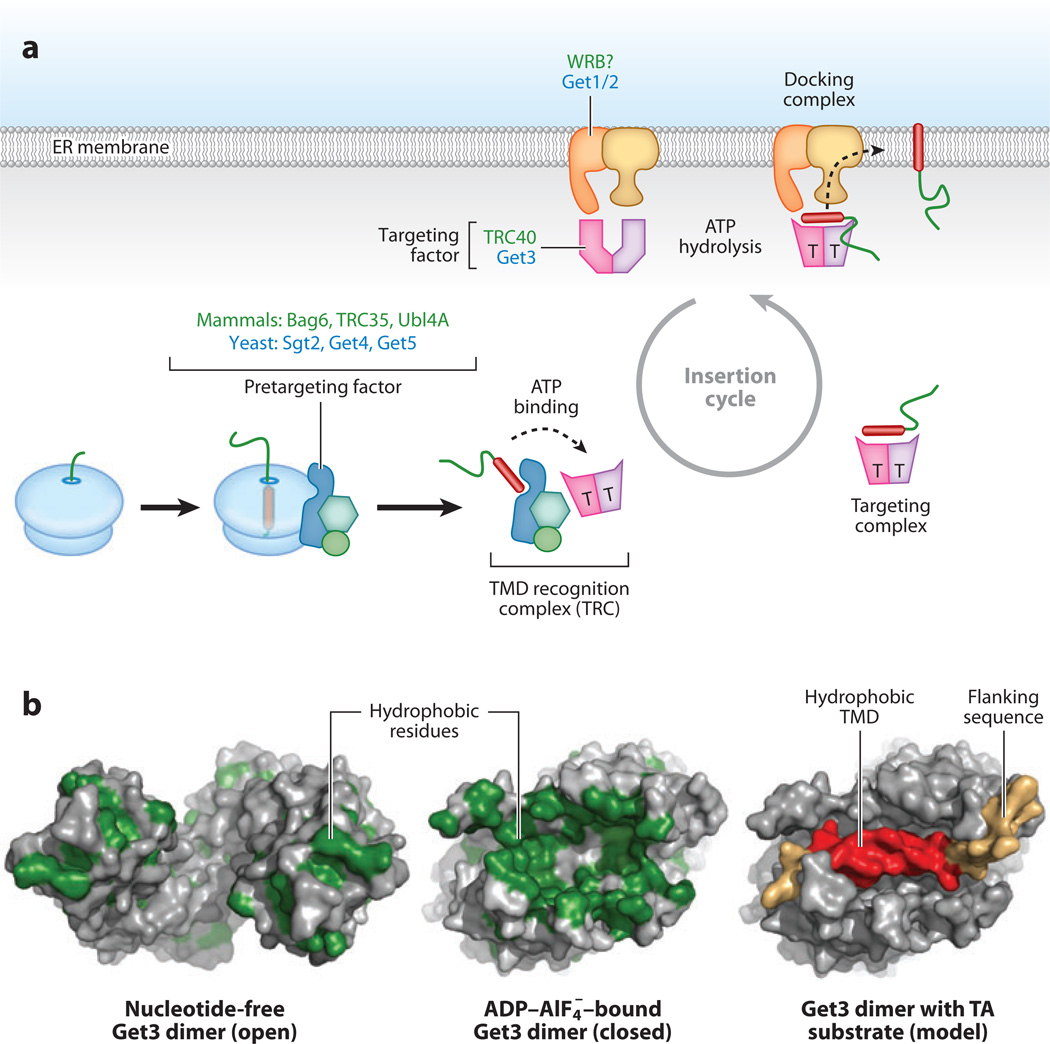
Figure 6.
Tail-anchored (TA) protein insertion. (a) Schematic model of the known components and steps mediating posttranslational insertion of a TA protein. When the transmembrane domain (TMD) of a TA protein is synthesized, it favors recruitment of a pretargeting factor to the ribosomal surface. This is composed of Bag6, TRC35, and Ubl4A in mammals. The analogous complex in yeast is formed by Sgt2, Get4, and Get5 as well as other chaperones. Its location near the ribosome would favor capture of the TA protein upon its release. The pretargeting factor together with the targeting factor (TRC40 in mammals, Get3 in yeast) ( pink) form the TRC. This is thought to be a transient complex that facilitates sorting, recognition, and loading of the TA protein onto the targeting factor. The targeting factor is an ATPase, and its substrate-bound form is thought to be ATP-bound (indicated by a T). This is delivered to the endoplasmic reticulum (ER) membrane via a receptor composed of Get1 and Get2 in yeast (a mammalian homolog of Get1 may be WRB). The docking complex of Get1-2-3 somehow facilitates substrate release and insertion in a step that depends on ATP hydrolysis by Get3. The now-vacant Get3 (which is in a different open conformation) is recycled to the cytosol to complete the insertion cycle. (b) Structural representations of the Get3 dimer in the open conformation (lacking nucleotide) and closed conformation (with bound ADP-AlF4). In the left two structures, hydrophobic residues are shown in green, illustrating that the closed conformation contains a large hydrophobic groove. The right panel shows a hypothetical model for the closed conformation bound to the TMD region of a model TA protein. The hydrophobic TMD (19 residues) is shown in red, with flanking sequences in gold.
The GET Pathway of Yeast
Progress on this basic framework was greatly accelerated by the high evolutionary conservation of TRC40. The Saccharomyces cerevisiae homolog (originally called Arr4, again on the basis of its sequence homology to ArsA) had been associated with a wide range of seemingly disparate phenotypes that are all membrane-associated processes (Auld et al. 2006, Shen et al. 2003). Its vesicular trafficking defects, which emerged from a large-scale genetic interaction study of secretory mutants (Schuldiner et al. 2005), placed it as part of a Golgi complex to ER trafficking pathway (hence the term Get3), although many of the earlier phenotypes remained unexplained. This was elegantly resolved when Get3 was found to play a key role in yeast TA-protein biogenesis; each of the various phenotypes is secondary to a different TA protein (Schuldiner et al. 2008). Get3 interacts with the TMDs of TA proteins, and its deletion results in the mislocalization of ER TA proteins to the cytosol or mitochondria ( Jonikas et al. 2009, Schuldiner et al. 2008). Thus, Get3 is now part of the GET pathway for the guided entry of TA proteins into the ER (Figure 6a).
The original and subsequent synthetic genetic array analysis of yeast genes (Battle et al. 2010; Costanzo et al. 2010; Jonikas et al. 2009; Schuldiner et al. 2005, 2008) clustered Get3 with five other genes whose products are the membrane proteins Get1 and Get2 and the cytosolic proteins Get4, Get5, and Sgt2. As expected from their similar genetic interactions, phenotypes of each of their deletions are similar, and all show evidence of TA-protein misinsertion and mislocalization (Costanzo et al. 2010, Jonikas et al. 2009, Schuldiner et al. 2008). Get3 has strong physical and genetic interactions with the ER-localized Get1 and Get2 (Auld et al. 2006), which suggests that these form the membrane receptor for Get3 targeting to the ER (Schuldiner et al. 2008). Without Get1 and/or Get2, Get3 is cytosolic, often in detergent-insoluble aggregates with TA proteins. In vitro reconstitutions showed that Get1/2 recruit Get3 to the ER membrane in an ATP-dependent manner (Schuldiner et al. 2008). Thus, Get1 and Get2 are thought to form an ER-localized complex that serves as a receptor for Get3-mediated TA-protein targeting (Figure 6a). Thus far, mammalian ortholog(s) of Get2 have not been identified, and the mammalian homolog of Get1, tryptophan-rich basic protein (WRB), has been proposed only recently to have a possible role in TA-protein insertion (Vilardi et al. 2011).
Mechanism of Transmembrane Domain Recognition
The structural basis of signal sequence binding by SRP revealed how highly diverse sequences sharing only general biophysical properties all could be recognized (Janda et al. 2010, Keenan et al. 1998). The identification of an entirely different TMD recognition factor (Stefanovic & Hegde 2007) afforded a unique opportunity to expand on these principles. Indeed, almost simultaneously, several groups reported crystal structures of various Get3 homologs (Bozkurt et al. 2009, Hu et al. 2009, Mateja et al. 2009, Suloway et al. 2009, Yamagata et al. 2010). These studies established that Get3 is a Zn2+-coordinated homodimer, each monomer of which contains an ATPase domain and a dynamic a-helical domain whose conformation changes in a nucleotide-dependent manner. In the nucleotide-free (or ADP-bound) open conformation, the helical domains of each Get3 monomer are spread apart from each other and folded to bury most hydrophobic surfaces (Figure 6b). In sharp contrast, the closed conformation containing ADP-AlF4− (an ATP hydrolysis transition state mimic) contains juxtaposed helical domains rearranged to reveal a deep hydrophobic groove rich in methionine residues, much like the M-domain of SRP54 (Mateja et al. 2009) (Figure 6b, compare with Figure 3c).
The hydrophobic groove of Get3 is large enough to accommodate an α-helix of ~20 residues (modeled in Figure 6c, right panel), consistent with the features of a typical TMD. By contrast, the analogous SRP groove is significantly smaller, presumably because it also must bind signal sequences containing fairly short (~7 residue) hydrophobic regions. Mutational and functional analyses map the Get3 groove residues as being important for substrate interaction and functionally linked to the switch regions of the ATPase domain (Mateja et al. 2009). Hydrogen-deuterium exchange experiments further support a direct role for the helical domain in TA-protein binding (Bozkurt et al. 2009). Thus, the current model is that binding and release of TA proteins from TRC40/Get3 is tightly coordinated with its cycle of ATP binding and hydrolysis (Figure 6a). However, a mechanistic framework remains to be established for how the events of substrate loading and unloading are spatially and temporally coordinated in the context of the rest of the TA targeting pathway components.
Pretargeting Events
The SRP targeting paradigm highlights the importance of constant TMD chaperoning through the cytosol. Yet, neither Get3 nor TRC40 associate with ribosomes, suggesting that upstream pretargeting factors may exist to bridge the ribosome and these targeting factors. Meanwhile, the poorly understood Get4 and Get5 were observed to form a ternary complex with Get3, and they had been found in an earlier proteomic analysis of ribosomes (Fleischer et al. 2006, Jonikas et al. 2009). Thus, Get4 and Get5 were speculated to play some role in ensuring efficient substrate capture by Get3 ( Jonikas et al. 2009), a model recently supported by two parallel studies in the yeast and mammalian systems (Mariappan et al. 2010, Wang et al. 2010).
In yeast, biochemical dissection and structural studies of the genetically identified Get3, Get4, Get5, and Sgt2 suggested that the stable Get4-Get5 subcomplex is a scaffold that recruits Sgt2 (via Get5) and Get3 (via Get4) (Bozkurt et al. 2010, Chang et al. 2010, Chartron et al. 2010, Wang et al. 2010). Sgt2 further interacts with various chaperones via its TPR motifs and, importantly, also directly interacts with the TMDs of ER-destined TA proteins (Wang et al. 2010). This suggests that the Get4-Get5 subcomplex bridges the two TMD-specific chaperones Sgt2 and Get3 to form the TRC (Figure 6a). On the basis of biochemical analysis, TA substrate bound to Sgt2 could be efficiently transferred to Get3 in a Get4-Get5-dependent manner (Wang et al. 2010). Together with genetic analysis suggesting that Sgt2, Get4, and Get5 act earlier in the pathway than does Get3 (Battle et al. 2010, Costanzo et al. 2010, Jonikas et al. 2009), these findings suggest a pretargeting function for the Sgt2-Get4-Get5 subcomplex in loading substrates onto the targeting factor Get3. The selectivity of Sgt2 for ER but not mitochondrial TA proteins further suggests a role in sorting among these destinations (Wang et al. 2010).
In the mammalian system, TA-protein capture by TRC40 requires additional factors that biochemical fractionation identified as a three-protein subcomplex composed of Bag6 (also called Bat3 or Scythe), TRC35, and Ubl4A (Mariappan et al. 2010). TRC35 and Ubl4A are recognizable homologs of Get4 and Get5, respectively, whereas Bag6 was independently identified in pull-down experiments as a TMD-specific TA-protein interactor (Leznicki et al. 2010, Mariappan et al. 2010, Stefanovic & Hegde 2007). Additional interaction studies showed that the Bag6 complex can interact with TRC40 (Mariappan et al. 2010), presumably via TRC35, on the basis of yeast homology. Thus, the TRC35-Ubl4A subcomplex appears to bridge the two TMD-interacting chaperones Bag6 and TRC40 to form the mammalian TRC, analogous to the situation in yeast. Evidence for such a function comes from the observation that depletion of the Bag6 subcomplex results in inefficient TA-protein capture by TRC40 (Mariappan et al. 2010) and reduced insertion into ER microsomes (Leznicki et al. 2010). Thus, the mammalian Bag6-TRC35-Ubl4A subcomplex and yeast Sgt2-Get4-Get5 subcomplex are analogous (and partially homologous) pretargeting factors that directly bind and transfer TA-protein substrates to TRC40 and Get3, respectively (Figure 6a). Whether mammalian homologs of Sgt2 (called SGTA and SGTB) also play a role in pretargeting steps remains to be determined. Of note, however, is the observation that SGTA interacts with Bag6 (Winnefeld et al. 2006), indicating that it is part of the TRC in mammalian as well as yeast systems.
The need for a pretargeting step involving a seemingly complex mechanism is somewhat unclear, but it may increase the fidelity of sorting, provide a point of regulation or quality control, facilitate loading of substrate onto TRC40, or provide a bridge between TRC40 and the ribosome to minimize substrate exposure to the cytosol. In support of the last notion, the Bag6 complex preferentially associates with ribosomes synthesizing hydrophobic domains, even when such domains are still within the ribosome tunnel, which provides a plausible mechanism for how TA proteins are routed into their targeting pathway despite numerous other chaperone systems in the cell (Mariappan et al. 2010). Bag6-complex recruitment may be facilitated by the several-fold delay in the translational termination of a TA protein (Mariappan et al. 2010). It is therefore possible that the Bag6 complex may relieve this termination block to coordinate TA-protein release with Bag6 ribosomal recruitment.
Furthermore, the Bag6 complex does seem to provide additional functionality to the TA pathway by acting as a triage factor. Not only can it recruit TRC40 for the purposes of productive targeting, but it can also recruit ubiquitination machinery to facilitate substrate degradation (Hessa et al. 2011). This multifunctionality seems to be important for quality control in the cytosol, where membrane proteins whose insertion fails must necessarily be degraded efficiently. Understanding how this triage function works, and whether it is also used to regulate the levels of TA protein insertion under different conditions, remains to be studied.
CHALLENGES TO UNDERSTANDING THE POSTTRANSLATIONAL PATHWAY
Although our understanding of neither the mammalian nor the yeast system is complete, a consideration of the two together provides a coherent path from the ribosome to the ER membrane (Figure 6a). Nonetheless, this TA-insertion pathway has been painted in broad brushstrokes, and a refined understanding of most steps is lacking. Starting at the ribosome, a key issue is precisely how a TA substrate is first captured. Although the Bag6 complex appears to be recruited to the ribosome (Mariappan et al. 2010), its binding site and mechanism of interaction with either the ribosome or substrate are unknown. More specifically, the nature of the Bag6 substrate interaction domain and its location relative to the ribosome exit tunnel needs to be determined. An attractive idea is if the N-terminal soluble domain of a TA protein binds to ribosome-associated Hsp70 (Gautschi et al. 2001, Nelson et al. 1992). In this view, the Hsp70 interaction would temporarily tether the TA protein so that upon translational termination it cannot escape into bulk cytosol. An interaction between Hsp70 and Bag6 (or Sgt2 in yeast) (Kaye et al. 2000, Thress et al. 2001, Wang et al. 2010) may then facilitate capture of the TMD, even if Sgt2 or Bag6 are not poised precisely at the exit tunnel.
After initial capture by the Bag6 or Sgt2 complex, the subsequent step is substrate transfer to TRC40 or Get3, respectively. A mechanistic understanding (at least in yeast) seems to be within reach given the already detailed mapping of interactions among the components (Chang et al. 2010, Wang et al. 2010), knowledge of structures of several of the factors (Bozkurt et al. 2009, Chang et al. 2010, Hu et al. 2009, Mateja et al. 2009, Suloway et al. 2009, Yamagata et al. 2010), and the ability to reconstitute this event with recombinant proteins (Wang et al. 2010). Presumably, a nucleotide-dependent interaction between the Sgt2 complex (specifically Get4) and Get3 (Chartron et al. 2010) will facilitate a conformational change in the latter that coordinates exposure of the hydrophobic groove with substrate transfer. Some degree of coordination is presumably needed to avoid premature substrate release or inappropriate exposure of a large hydrophobic surface on Get3.
The final targeting and insertion steps currently await detailed mechanistic analysis that first requires reconstitution with recombinant factors. Once this is accomplished, insight into the mechanism of ATPase cycle coordination with targeting, substrate release, and insertion can be investigated. At present, Get1 and Get2 seem to be the only membrane factors for insertion (Schuldiner et al. 2008), but this has not been rigorously demonstrated. It is further unclear whether insertion of the TMD into the lipid bilayer simply requires its release from Get3/TRC40 in the vicinity of the membrane or whether a more active role for protein factors is required. The ability of at least some TMDs to insert unassisted (Brambillasca et al. 2005, 2006) makes the former model attractive, but relying on a spontaneous process in vivo would seem highly risky given the potential for aggregation of exposed TMDs. Thus, the precise roles of Get1 and Get2 in targeting, stimulation of ATPase activity, substrate release, and insertion remain to be elucidated. In organisms outside of yeast, the identity of the TRC40 receptor remains to be defined. However, given the high conservation of TRC40, a weakly homologous sequence that eventually proves to be structurally similar would not be surprising.
Finally, the structural basis of this pathway remains an area of intense investigation. As with the biochemical analysis, the most challenging will be the earliest steps involving the ribosome and the last step involving the membrane. With considerable recent advances in eukaryotic ribosome structures (Armache et al. 2010, Ben-Shem et al. 2010) and increasing success in IMP expression and crystallization (Granseth et al. 2007), there is reason for optimism on these two fronts. Importantly, the nonessentiality of the GET pathway in yeast (Schuldiner et al. 2005, 2008) considerably eases the technical obstacles of testing insights derived from in vitro studies in vivo. Thus, it seems possible that without the complicating feature of ongoing protein synthesis, the comparatively simpler posttranslational insertion pathway will yield its secrets more readily than the longer-studied cotranslational pathway.
CONCLUSIONS
Eukaryotic cells have evolved remarkably coordinated and regulated mechanisms to immediately shield a TMD as soon as it emerges from the ribosome until its insertion into the lipid bilayer. This coordination involves not only precise spatial regulation (such as SRP positioning relative to the exit tunnel), but also temporal regulation via effects on translation elongation and termination. There is little doubt that all the substrate capture and transfer reactions in membrane insertion pathways will prove to be highly coordinated, but we currently have limited molecular insight for them. For example, the transfer of substrate from SRP to the Sec61 complex remains very poorly understood despite its critical role in ensuring efficient translocation and insertion. Thus, understanding how an intrinsically insoluble domain such as a TMD manages to navigate through an aqueous cytosol poses a fascinating general biochemical problem.
An explanation for this problem likely will also reveal how the many different membrane-protein targeting pathways (to mitochondria, peroxisomes, and chloroplasts in addition to the ER) manage to stay distinct in spite of the fact that TMDs are the main recognition element in each case. For example, how does SRP avoid a mitochondrial membrane protein emerging from the ribosome, or why do mitochondrial TA proteins not engage the ER pathway? The answers likely lie not only in the intrinsic specificities of the recognition factors but also in their spatiotemporal coordination. A hint of how this can be achieved is seen with the lack of cross talk between SRP and TRC40; the former gains an advantage with its ribosome localization, whereas the latter enjoys much higher cellular concentrations. In general, however, the mechanisms to ensure high fidelity among targeting pathways that recognize generally similar substrates remain to be fully elucidated.
Because fidelity is unlikely to be perfect, mechanisms exist to deal with failed IMP targeting (Hessa et al. 2011). One may anticipate that cytosolic quality-control pathways need to be tightly linked to targeting pathways given the inability of TMD-containing proteins to reside freely in the cytosol. It is therefore very intriguing that, in the TA pathway, both Bag6 and Ubl4A contain ubiquitin-like domains typically involved in protein-degradation pathways. At least one of these domains (on Bag6) is involved in recruiting ubiquitination machinery to facilitate degradation of membrane proteins in the cytosol (Hessa et al. 2011). Understanding the ways protein targeting is linked to degradation may well be important not only for quality control but possibly also for cellular regulation of IMP abundance.
At the membrane level, the exact mechanisms of TMD movement from aqueous to hydrophobic environments remain to be defined at high resolution. For the Sec61-dependent pathway, this critical step remains especially poorly understood for polytopic IMPs. Current models are not mutually exclusive and range widely, from sequential insertion to those in which insertion of some TMDs occurs after synthesis. For TA proteins, this issue is totally unresolved, with uncertainty about even the basic question of whether insertion machinery is needed after targeting is achieved. And finally, the question of whether other IMP-insertion pathways remain to be discovered should remain open. Not only do some TA proteins fail to interact with the known machinery, but TA proteins are also able to insert (albeit with reduced fidelity) in yeast devoid of the GET pathway. Furthermore, small IMPs are very poorly understood but need to be inserted in different orientations, possibly necessitating unique machinery. Thus, a wide range of questions from those based on general discovery to ones that are highly mechanistic remains to be explored in the area of IMP insertion into the ER.
SUMMARY POINTS.
Numerous biophysical obstacles must be overcome to target hydrophobic integral membrane proteins through an aqueous cytosol and guide them into the lipid bilayer of the ER.
The insertion and topology of any given TMD are influenced by a complex combination of both local and distal sequence determinants. This substantially complicates highly reliable prediction algorithms for polytopic membrane proteins.
Most integral membrane proteins are cotranslationally targeted by SRP and subsequently inserted via the Sec61 translocon.
The Sec61 translocon consists of an aqueous pore and a lateral gate that allows TMDs to partition into the lipid bilayer. Accessory factors near or associated with the Sec61 channel may facilitate insertion of some proteins.
Insertion of marginally hydrophobic TMDs in complex proteins can be facilitated by strong constraints on the assembly of other parts of the protein. This may permit relatively polar sequences to serve as TMDs in the final 3D structure.
TA proteins are posttranslationally inserted by the TRC40/GET pathway, which consists of machinery completely distinct from the cotranslational pathway.
Transfer of a membrane protein from one complex to the next during targeting and insertion is likely to be highly regulated to minimize exposing TMDs to the cytosol.
The multiple steps involved in TA-protein targeting may serve to increase fidelity of targeting and provide a point of quality control or regulation.
FUTURE ISSUES.
The ability to functionally manipulate insertion machinery in well-defined biochemical systems that efficiently recapitulate membrane protein insertion will be crucial to understanding molecular mechanisms.
Trans-acting factors that contribute to polytopic membrane protein insertion and postin-sertion folding need to be identified and refined.
The posttranslational targeting and insertion of TA proteins needs to be reconstituted with fully recombinant proteins whose structures can guide detailed mechanistic and biophysical analysis.
New pathways of membrane protein insertion may remain to be discovered to account for an increasing number of small membrane proteins whose insertion is poorly studied.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NICHD at the National Institutes of Health.
Glossary
- IMP
integral membrane protein
- TMD
transmembrane domain
- ER
endoplasmic reticulum
- SRP
signal recognition particle
- SR
SRP receptor
- RNC
ribosome-nascent chain
- TRAM
translocating-chain associating membrane protein
- TA
tail-anchored
- TRC
TMD recognition complex
- GET
guided entry of TA proteins
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Abell BM, Rabu C, Leznicki P, Young JC, High S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 2007;120:1743–1751. doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Walter P, Blobel G. Signal recognition protein is required for the integration of acetyl-choline receptor delta subunit, a transmembrane glycoprotein, into the endoplasmic reticulum membrane. J. Cell Biol. 1982;93:501–506. doi: 10.1083/jcb.93.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P, Rao JKM, Hargrave PA. Structural prediction of membrane-bound proteins. Eur. J. Biochem. 1982;128:565–575. doi: 10.1111/j.1432-1033.1982.tb07002.x. [DOI] [PubMed] [Google Scholar]
- Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5Å resolution. Proc. Natl. Acad. Sci. USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae . Genetics. 2006;174:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Jonikas MC, Walter P, Weissman JS, Koller D. Automated identification of pathways from quantitative genetic interaction data. Mol. Syst. Biol. 2010;6:379. doi: 10.1038/msb.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae . J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins. J. Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta. 2010;1808:937–948. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt G, Wild K, Amlacher S, Hurt E, Dobberstein B, Sinning I. The structure of Get4 reveals an α-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 2010;584:1509–1514. doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Makarow M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, et al. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Kuhn A, Dalbey R. The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 1995;14:866–875. doi: 10.1002/j.1460-2075.1995.tb07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-W, Chuang Y-C, Ho Y-C, Cheng M-Y, Sun Y-J, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Suloway CJM, Zaslaver MÄ, Clemons WM. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. USA. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Gilmore R. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat. Struct. Mol. Biol. 2006;13:930–936. doi: 10.1038/nsmb1146. [DOI] [PubMed] [Google Scholar]
- Chi JH, Roos J, Dean N. The OST4 gene of Saccharomyces cerevisiae encodes an unusually small protein required for normal levels of oligosaccharyltransferase activity. J. Biol. Chem. 1996;271:3132–3140. doi: 10.1074/jbc.271.6.3132. [DOI] [PubMed] [Google Scholar]
- Cöıc Y-M, Vincent M, Gallay J, Baleux F, Mousson F, et al. Single-spanning membrane protein insertion in membrane mimetic systems: role and localization of aromatic residues. Eur. Biophys. J. 2005;35:27–39. doi: 10.1007/s00249-005-0002-1. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Denzer AJ, Nabholz CE, Spiess M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 1995;14:6311–6317. doi: 10.1002/j.1460-2075.1995.tb00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman A, Puziss J, Bassford P, Jr., Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli . EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu. Rev. Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Enquist K, Fransson M, Boekel C, Bengtsson I, Geiger K, et al. Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J. Mol. Biol. 2009;387:1153–1164. doi: 10.1016/j.jmb.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V, Vilardi F, Schlecht R, Mayer MP, Dobberstein B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 2010;123:1522–1530. doi: 10.1242/jcs.055970. [DOI] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, et al. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Kida Y, Hagiwara M, Morimoto F, Sakaguchi M. Positive charges of translocating polypeptide chain retrieve an upstream marginal hydrophobic segment from the endoplasmic reticulum lumen to the translocon. Mol. Biol. Cell. 2010;21:2045–2056. doi: 10.1091/mbc.E09-12-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Junne T, Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992a;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies K-U, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992b;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Granseth E, Seppaälä S, Rapp M, Daley DO, von Heijne G. Membrane protein structural biology—how far can the bugs take us? Mol. Membr. Biol. 2007;24:329–332. doi: 10.1080/09687680701413882. [DOI] [PubMed] [Google Scholar]
- Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol. Chem. 2009;390:775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, et al. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 2010;22:437–446. doi: 10.1016/j.ceb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Rapoport TA. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 2003;22:3654–3663. doi: 10.1093/emboj/cdg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005a;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011 doi: 10.1038/nature10181. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005b;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- Higy M, Gander S, Spiess M. Probing the environment of signal-anchor sequences during topogenesis in the endoplasmic reticulum. Biochemistry. 2005;44:2039–2047. doi: 10.1021/bi047976z. [DOI] [PubMed] [Google Scholar]
- Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J. Cell Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Nilsson I, von Heijne G. Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol. Gen. Genet. MGG. 1993;239:251–256. doi: 10.1007/BF00281625. [DOI] [PubMed] [Google Scholar]
- Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA, Hartmann E. Protein translocation: common themes from bacteria to man. FEBS Lett. 1994;346:73–77. doi: 10.1016/0014-5793(94)00367-x. [DOI] [PubMed] [Google Scholar]
- Junne T, Kocik L, Spiess M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol. Biol. Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J. Biol. Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch T, Cambon A, Wattenberg BW. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8:1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- Kauko A, Hedin LE, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane α-helices during membrane protein folding. J. Mol. Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Kaye FJ, Modi S, Ivanovska I, Koonin EV, Thress K, et al. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 2000;467:348–355. doi: 10.1016/s0014-5793(00)01135-2. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Kida Y, Mihara K, Sakaguchi M. Translocation of a long amino-terminal domain through ER membrane by following signal-anchor sequence. EMBO J. 2005;24:3202–3213. doi: 10.1038/sj.emboj.7600788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y, Sakaguchi M, Fukuda M, Mikoshiba K, Mihara K. Amino acid residues before the hydrophobic region which are critical for membrane translocation of the N-terminal domain of synaptotagmin II. FEBS Lett. 2001;507:341–345. doi: 10.1016/s0014-5793(01)03000-9. [DOI] [PubMed] [Google Scholar]
- Kim PK, Hollerbach C, Trimble WS, Leber B, Andrews DW. Identification of the endoplasmic reticulum targeting signal in vesicle-associated membrane proteins. J. Biol. Chem. 1999;274:36876–36882. doi: 10.1074/jbc.274.52.36876. [DOI] [PubMed] [Google Scholar]
- Kim PK, Janiak-Spens F, Trimble WS, Leber B, Andrews DW. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry. 1997;36:8873–8882. doi: 10.1021/bi970090t. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synapto-brevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lakkaraju AKK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lingappa VR, Katz FN, Lodish HF, Blobel G. A signal sequence for the insertion of a transmembrane glycoprotein. Similarities to the signals of secretory proteins in primary structure and function. J. Biol. Chem. 1978;253:8667–8670. [PubMed] [Google Scholar]
- Lipp J, Dobberstein B, Haeuptle MT. Signal recognition particle arrests elongation of nascent secretory and membrane proteins at multiple sites in a transient manner. J. Biol. Chem. 1987;262:1680–1684. [PubMed] [Google Scholar]
- Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Kim H, Nilsson I, White SH, von Heijne G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for Nin–Cout and Nout–Cin transmembrane helices. Proc. Natl. Acad. Sci. USA. 2008;105:15702–15707. doi: 10.1073/pnas.0804842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Masaki R, Kameyama K, Yamamoto A. Post-translational targeting of a tail-anchored green fluorescent protein to the endolpasmic reticulum. J. Biochem. 2003;134:415–426. doi: 10.1093/jb/mvg159. [DOI] [PubMed] [Google Scholar]
- Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick PJ, Miao Y, Shao Y, Lin J, Johnson AE. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- Meindl-Beinker NM, Lundin C, Nilsson I, White SH, von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–1116. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Me´ne´tret J-F, Neuhof A, Morgan DG, Plath K, Radermacher M, et al. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Meyer H-A, Grau H, Kraft R, Kostka S, Prehn S, et al. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Ménétret J-F, Neuhof A, Rapoport TA, Akey CW. Structure of the mammalian ribosome-channel complex at 17Å resolution. J. Mol. Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von Heijne G, et al. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Ito K, Isaksson LA. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- Navarre C, Catty P, Leterme S, Dietrich F, Goffeau A. Two distinct genes encode small isoproteolipids affecting plasma membrane H+-ATPase activity of Saccharomyces cerevisiae . J. Biol. Chem. 1994;269:21262–21268. [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kD heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Nilsson I, Witt S, Kiefer H, Mingarro I, von Heijne G. Distant downstream sequence determinants can control N-tail translocation during protein insertion into the endoplasmic reticulum membrane. J. Biol. Chem. 2000;275:6207–6213. doi: 10.1074/jbc.275.9.6207. [DOI] [PubMed] [Google Scholar]
- Oliver J, Jungnickel B, Görlich D, Rapoport T, High S. The Sec61 complex is essential for the insertion of proteins into the membrane of the endoplasmic reticulum. FEBS Lett. 1995;362:126–130. doi: 10.1016/0014-5793(95)00223-v. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Ott CM, Lingappa VR. Integral membrane protein biosynthesis: why topology is hard to predict. J. Cell Sci. 2002;115:2003–2009. doi: 10.1242/jcs.115.10.2003. [DOI] [PubMed] [Google Scholar]
- Pina FJ, O’Donnell AF, Pagant S, Piao HL, Miller JP, et al. Hph1 and Hph2 are novel components of the Sec63/Sec62 posttranslational translocation complex that aid in vacuolar proton ATPase biogenesis. Eukaryot. Cell. 2011;10:63–71. doi: 10.1128/EC.00241-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Wilkinson BM, Stirling CJ, Rapoport TA. Interactions between Sec complex and prepro-α-factor during posttranslational protein transport into the endoplasmic reticulum. Mol. Biol. Cell. 2004;15:1–10. doi: 10.1091/mbc.E03-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J. Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies K-U. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu. Rev. Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Goder V, Heinrich SU, Matlack KES. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61-α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Sato M, Hresko R, Mueckler M. Testing the charge difference hypothesis for the assembly of a eukaryotic multispanning membrane protein. J. Biol. Chem. 1998;273:25203–25208. doi: 10.1074/jbc.273.39.25203. [DOI] [PubMed] [Google Scholar]
- Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Hsu C-M, Kang B-K, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. BioMetals. 2003;16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur. J. Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR, Lingappa VR. Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem. 1993;268:23552–23561. [PubMed] [Google Scholar]
- Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61αin the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- Steel GJ, Brownsword J, Stirling CJ. Tail-anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry. 2002;41:11914–11920. doi: 10.1021/bi026105r. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway CJM, Chartron JW, Zaslaver Ma, Clemons WM. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. USA. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K, Song J, Morimoto RI, Kornbluth S. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 2001;20:1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper DJ, Harley CA. Yeast genes controlling responses to topogenic signals in a model transmembrane protein. Mol. Biol. Cell. 2002;13:1158–1174. doi: 10.1091/mbc.01-10-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, et al. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA. 2000;97:7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider MB, Sansom MSP, Di Nola A. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins: Struct. Funct. Bioinforma. 2005;59:252–265. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]
- Urban S, Freeman M. Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Net N-C charge imbalance may be important for signal sequence function in bacteria. J. Mol. Biol. 1986a;192:287–290. doi: 10.1016/0022-2836(86)90365-7. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J. 1986b;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Spiess M. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J. Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Properties of N-terminal tails in G-protein coupled receptors: a statistical study. Protein Eng. 1995;8:693–698. doi: 10.1093/protein/8.7.693. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miller A, Rusch SL, Kendall DA. Demonstration of a specific Escherichia coli SecY-signal peptide interaction. Biochemistry. 2004;43:13185–13192. doi: 10.1021/bi049485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenberg B, Lithgow T. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic. 2001;2:66–71. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Theibert JL, Murphy C, Jona I, Martonosi A, Collins JH. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch. Biochem. Biophys. 1992;298:620–623. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- Wessels HP, Spiess M. Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell. 1988;55:61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- White SH, von Heijne G. How translocons select transmembrane helices. Annu. Rev. Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- Wickner WT, Lodish HF. Multiple mechanisms of protein insertion into and across membranes. Science. 1985;230:400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nat. Struct. Mol. Biol. 2004;11:1049–1053. doi: 10.1038/nsmb853. [DOI] [PubMed] [Google Scholar]
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Wimley WC, White SH. Designing transmembrane α-helices that insert spontaneously. Biochemistry. 2000;39:4432–4442. doi: 10.1021/bi992746j. [DOI] [PubMed] [Google Scholar]
- Winnefeld M, Grewenig A, Schnölzer M, Spring H, Knoch TA, et al. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp. Cell Res. 312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J. Biol. Chem. 2003;278:3489–3496. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Mimura H, Sato Y, Yamashita M, Yoshikawa A, Fukai S. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Fujita H, Morimoto F, Kida Y, Sakaguchi M. A sugar chain at a specific position in the nascent polypeptide chain induces forward movement during translocation through the translocon. J. Biochem. 2011;149:591–600. doi: 10.1093/jb/mvr011. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Hori O, Stern DM, Hartmann E, Ogawa S, Tohyama M. Stress-associated endoplasmic reticulum protein 1 (Serp1)/ribosome-associated membrane protein 4 (Ramp4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 1999;147:1195–1204. doi: 10.1083/jcb.147.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]