Abstract
ZBTB3 belongs to the Zinc finger and BTB/POZ domain containing transcription factor family; however, its biological role has rarely been studied. We demonstrate for the first time, to our knowledge, that ZBTB3 is an essential factor for cancer cell growth via the regulation of the ROS detoxification pathway. Suppression of ZBTB3 using two different short hairpin RNAs in human melanoma, lung carcinoma, and breast carcinoma results in diminished cell growth. In addition, we found that suppression of ZBTB3 activates a caspase cascade, including caspase-9, -3, and PARP leading to cellular apoptosis, resulting from failed ROS detoxification. We identified that ZBTB3 plays an important role in the gene expression of ROS detoxification enzymes. Our results reveal that ZBTB3 may play a critical role in cancer cell growth via the ROS detoxification system. Therefore, therapeutic strategies that target ZBTB3 could be used in selective cancer treatments. [BMB Reports 2014; 47(7): 405-410]
Keywords: Apoptosis, Cancer, ROS, ZBTB3
INTRODUCTION
Zinc finger and BTB/POZ domain containing (ZBTB) proteins are part of an emerging family of transcription factors, which commonly contain a DNA binding zinc finger and a transcription-repressing BTB/POZ domain. These proteins have critical roles in development, differentiation, and tumorigenesis (1). Mechanistically, ZBTB proteins act as transcriptional repressors via BTB/POZ domain-mediated recruitment of transcriptional co-repressors such as NCoR, mSin3A, CtBP, or HDAC to a subset of their target gene’s promoter regions (2, 3).
The human genome encodes over 40 members of the ZBTB protein family, with several closely linked to cancer development and progression (3, 4). ZBTB29, also known as Hyper-methylated in Cancer 1 (HIC1), acts as a repressor of transcriptional gene regulation in cancer growth, angiogenesis, and invasion through the recruitment of C-terminal binding proteins (CtBPs) and Polycomb Repressive Complex 2 (PRC2) to a subset of its target genes (5, 6). In addition, ZBTB7 (also known as LRF) controls cellular transformation (1), ZBTB27 (also known as BCL6) is involved in B-cell lymphoma (7, 8), and ZBTB4 controls cell proliferation and invasion in breast cancer (9). This suggests that the ZBTB family of proteins may have multidimensional roles in cancer development and progression. Although the human genome has been completely explored, the biological functions of many of the other ZBTB proteins are poorly characterized, if at all. Here, we report that ZBTB3 protein and mRNA levels are functionally expressed in several cancer cell lines and their expression is critical for cancer cell survival.
Reactive oxygen species (ROS) can be produced as byproducts of oxidative phosphorylation, metabolic reprogramming, and environmental stresses, such as ionizing radiation, UV, and redox chemicals, and have been recognized to play an important role in many different physiological processes, such as proliferation, metabolism, aging, and cancer development (10-13). Because ROS cause cellular damage by disrupting intracellular macromolecules, such as proteins, lipids, and DNA, cells must have developed a complex antioxidant defense system to be able to adapt to the changing ROS levels and survive (14). Cells have evolved mechanisms to protect themselves from ROS-induced damage by activating various antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), glutathione peroxidases (GPx) and peroxiredoxins (PRDX) (12, 15).
Cancer cells exhibit increased ROS due to unbalanced metabolic activity (16) and activated oncogenic signaling (10, 17). Although they are consistently exposed to high levels of ROS, cancer cells are more resistant to these conditions through an activated antioxidant defense system (15). Several transcription factors such as Forkhead Box M1 (FoXM1) and Nuclear factor erythroid-2 related factor 2 (NRF2) have been shown to be elevated and functionally activated in a variety of cancers, and these activated transcription factors confer resistance to oxidative stress-induced cancer cell apoptosis by increasing the gene expression of various antioxidants (18, 19). Whereas some of the components of the oxidative stress response have been identified in cancer cells, it is likely that key regulators that contribute to cancer growth are still poorly understood. In this study, we report that suppression of ZBTB3 causes the accumulation of intracellular ROS and results in cancer cell apoptosis, suggesting that ZBTB3 is a critical factor for the antioxidant defense system and cancer cell survival.
RESULTS
Inhibition of ZBTB3 decreases cancer cell growth
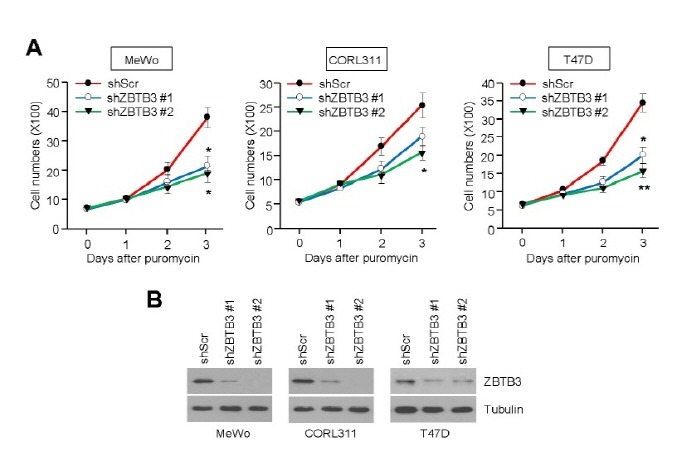
To understand the functional role of ZBTB3 in cancer growth, we initially examined whether depletion of ZBTB3 affects cancer cell survival. We found that two different shRNAs targeting ZBTB3 (shZBTB3 #1 or shZBTB3 #2) strongly decreased cell viability (Fig. 1A) as well ZBTB3 protein levels (Fig. 1B) in human melanoma (MeWo), human lung small cell carcinoma (CORL311), and human breast carcinoma (T47D) cells. Therefore, we speculate that ZBTB3 plays an important role in cancer cell growth.
Fig. 1. ZBTB3 knockdown suppresses cell viability. (A) The effects of ZBTB3 knockdown on cell viability in MeWo (melanoma), CORL311 (lung), T47D (breast) cancer cells infected with lentiviruses expressing shRNA against either control or two different sequence of ZBTB3 (shZBTB3 #1 or shZBTB3 #2). Values represent mean ± SD of independent experiments performed three times; *P < 0.05 and **P < 0.01 vs. control shRNA. (B) ZBTB3 protein levels in the control cells and those with ZBTB3 knocked down.
ZBTB3 depletion causes cellular apoptosis
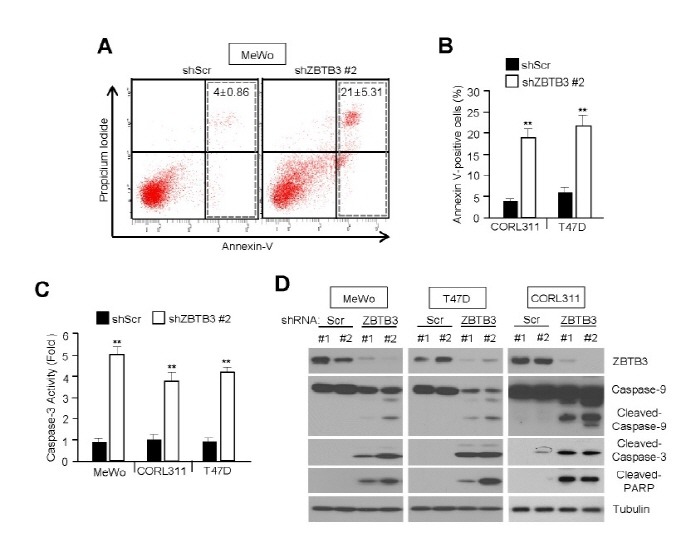
The fact that cancer cell growth was decreased in ZBTB3-depleted cells suggests that it may have an anti-apoptotic function during cancer growth. To determine the anti-apoptotic function of ZBTB3, we analyzed the levels of apoptosis in control and ZBTB3-depleted cells using a FITC-conjugated Annexin-V antibody. As expected, the knockdown of ZBTB3 in MeWo, CORL311, and T47D cells significantly increased apoptosis (Fig. 2A and B). We further investigated the mechanism responsible for apoptotic induction after ZBTB3 knockdown. We found that apoptosis was mediated by the activation of the caspase pathway, indicated by the activation of cleaved caspase-9, -3 and PARP in MeWo, CORL311, and T47D (Fig. 2D). In addition, we found that caspase-3 was highly activated in ZBTB3-depleted cells (Fig. 2C). These results suggest that the inhibition of ZBTB3 strongly induces cancer cell apoptosis mediated by the caspase pathway.
Fig. 2. ZBTB3 knockdown causes apoptosis. (A, B) The effects of ZBTB3 knockdown on cell apoptosis in MeWo, CORL113, and T47D cells. Values (percentage of apoptotic cells) represent mean ± SD of three independent experiments performed three times; **P < 0.01 vs. control shRNA. (C) The effects of ZBTB3 knockdown on caspase-3 activity in several cancer cell lines. Values represent mean ± SD of independent experiments performed three times; **P < 0.01 vs. control shRNA. (D) Expression of apoptotic proteins in cells with ZBTB3 knocked down.
Inhibition of ZBTB3 increases ROS levels leading to cellular apoptosis
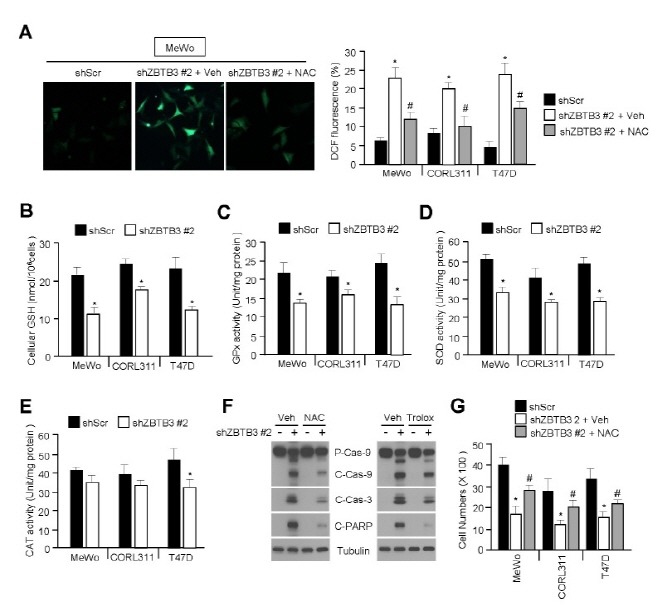
Because increased intracellular ROS levels can lead to cellular apoptosis, we investigated the involvement of ZBTB3 using an oxidation-sensitive fluorescent dye, DCFH-DA. Fig. 3A shows that the intracellular ROS levels were significantly elevated in ZBTB3-depleted cells, but not after pretreatment with N-acetyl-L-cysteine (NAC). Therefore, we hypothesized that the elevated ROS levels in ZBTB3-depleted cells may be associated with a reduction of glutathione (GSH) levels. As expected, intracellular GSH levels were significantly decreased in ZBTB3-depleted cells (Fig. 3B). Given that the intracellular GSH as well ROS levels are largely controlled by a variety of antioxidant enzymes, we measured the activity of these enzymes in the ZBTB3-depleted cells. Consistent with an increase in intracellular ROS levels in these cells, free radical eliminating enzymes, such as GPx, SOD, and CAT, were significantly inactivated by the knockdown of ZBTB3 (Fig. 3C-E). Moreover, two different antioxidants, NAC and trolox, largely prevented caspase and PARP cleavage, thereby suggesting that elevated ROS levels are required to induce caspase cascade-dependent apoptosis in ZBTB3-depleted cells (Fig. 3F). Based on the above presented results that ZBTB3 affects cell survival by regulating antioxidant capacity, we examined whether antioxidants could rescue cell growth by eliminating the increased ROS levels in ZBTB3-depleted cells. ZBTB3 knockdown-mediated reduction of cell viability was significantly rescued by the antioxidant, NAC (Fig. 3G). Collectively, these results indicate that the inhibition of ZBTB3 contributes to the accumulation of abnormal levels of ROS causing apoptosis in cancer cells.
Fig. 3. Oxidative stress induced by ZBTB3 knockdown causes apoptosis. (A) The effects of ZBTB3 knockdown on accumulation of intracellular H2O2 levels in several cancer cells measured using an oxidation-sensitive dye DCFH-DA. Representative images are shown (left) and values represent mean ± SD of independent experiments performed three times; *P < 0.05 and #P < 0.05. (B) The effects of ZBTB3 knockdown on cellular GSH levels in several cancer cell lines. Values represent mean ± SD of independent experiments performed three times; *P < 0.05. (C-E) The effects of ZBTB3 knockdown on antioxidant activities such as GPx, SOD, and CAT in cancer cells. Values represent mean ± SD of independent experiments performed three times; *P < 0.05. (F) The effects of antioxidants on ZBTB3 knockdown-induced apoptosis. (G) The effects of the antioxidant NAC on viability in cells with ZBTB3 knocked down. Values represent mean ± SD of independent experiments performed three times; *P < 0.05 and #P < 0.05.
ZBTB3 suppression results in the reduced expression of antioxidants
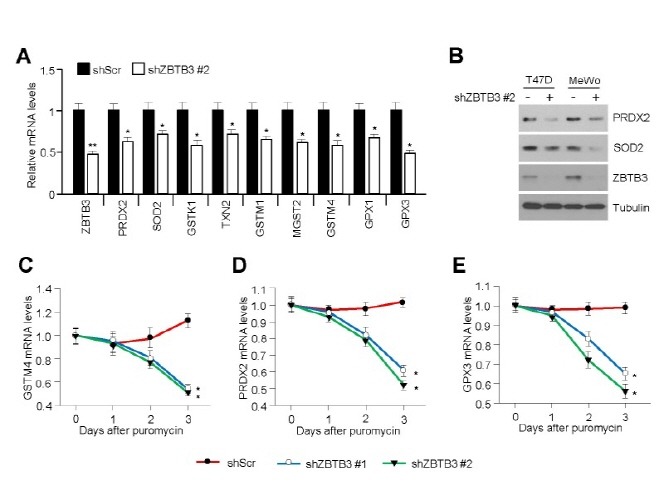
To determine the molecular basis of the oxidative stress induced by ZBTB3 knockdown in cancer cells, we analyzed expression level of mRNAs encoding the antioxidant enzymes affected by depletion of ZBTB3. We found that the mRNA and protein levels of a variety of antioxidant enzymes, including PRDX2 and SOD2, were significantly decreased in ZBTB3-depleted cancer cells (Fig. 4A and B). Quantitative real-time PCR (qRT-PCR) analysis further confirmed the downregulation of GSTM4 (Fig. 4C), PRDX2 (Fig. 4D), and GPX3 (Fig. 4E) in ZBTB3-depleted cells.
Fig. 4. Inhibition of ZBTB3 downregulates antioxidant gene and protein expression. (A) The effects of ZBTB3 knockdown on antioxidant gene expression. Values represent mean ± SD of independent experiments performed three times; *P < 0.05 and **P < 0.01. (B) Antioxidant protein levels in control cells and those with ZBTB3 knockdown. (C-E) Antioxidant mRNA expression levels. Values represent mean ± SD of independent experiments performed three times; *P < 0.05.
DISCUSSION
Cancer cells reprogram their antioxidant defense system in order to survive under oxidative stress induced by metabolic reprogramming and activated oncogenic signaling (10, 16, 17). It is becoming clear, however, that cancer cells are not dependent on one single pathway to accomplish this, as cancers generally choose from a variety of antioxidant pathways for survival. In this study, we provide evidence that the inhibition of ZBTB3 using shRNA in several cancer cell lines results in the elevation of ROS levels causing apoptosis. We did this by showing that antioxidants such as NAC and trolox clearly prevented the apoptosis induced by ZBTB3 inhibition. These results reveal that cancer cells use ZBTB3 as another antioxidant factor for survival and growth.
In pre-clinical settings, ROS-inducing drugs have been considered as an anti-cancer strategy whereby increasing ROS levels above a certain threshold could cause cancer cell apoptosis (15). Indeed, piperlongumine and PEITC have recently been shown as selective anti-cancer drugs acting via the accumulation of ROS in cancer cells (17, 20). Therefore, inhibition of ZBTB3 may sensitize ROS-inducing drug-mediated cancer cell apoptosis and could be useful in selective cancer treatment.
ZBTB proteins exhibit C-terminal zinc fingers, which recognize and interact with specific DNA sequences. They also possess an N-terminal BTB/POZ domain, which mediates the recruitment of transcriptional co-repressors, such as nuclear receptor co-repressor (NCoR), c-terminal binding protein (CtBP), and histone deacetylase (HDAC) to the promoter regions of its target genes (1). For example, ZBTB33, also known as kaiso, transcriptionally represses matrix metalloproteinase (MMP)-7 gene expression through the recruitment of the transcriptional co-repressor, myeloid translocation gene (MTG)-16 to the kaiso binding site in the promoter region (21). Additionally, ZBTB4 has been shown to negatively regulate cell survival oncogene expression such as vascular endothelial growth factor (VEGF), VEGFR1, and survivin in human breast cancer cells by inhibiting specificity protein 1 (Sp1), Sp3, and Sp4 (9). Paradoxically, our results show that inhibition of ZBTB3 strongly downregulates antioxidant mRNA expression suggesting that it may act as a transcriptional activator of antioxidants. This could be explained by the fact that ZBTB3 may downregulate other transcriptional co-repressors or micro RNAs, which in turn inhibits antioxidant gene expression. In addition, transcriptional function of ZBTB3, as either an activator or repressor of gene expression, could be dependent on the cellular context. Indeed, ZBTB19 (also known as PATZ1) has been shown to positively regulate p53 transcriptional activity and enhance apoptosis and senescence in response to DNA damage and oxidative stress in osteosarcoma and endothelial cells (22, 23).
To summarize, our findings illustrate that the ZBTB3 transcription factor is closely involved in the antioxidant defense system by mediating antioxidant mRNA expression and this could be used to develop cancer therapy targeting antioxidant defense networks.
MATERIALS AND METHODS
Reagents and antibodies
N-acetyl-L-cysteine, trolox, and 5,6-chloromethyl-2',7'-dichlorodihydrofluorescein diacetate (CM-DCF-DA) were purchased from Sigma Aldrich and Invitrogen. Antibodies against cleaved caspase-3, -9, Poly ADP ribose polymerase (PARP), and anti-ZBTB3 were purchased from Cell Signaling Technology and Bethyl Laboratory. Anti-tubulin, PRDX2 and SOD2 antibodies were purchased from Millipore and Pierce.
Cell culture, lentiviral particle production and infection
Cancer cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and 25 mM glucose. Lentiviruses expressing short hairpin RNA (shRNA) against either control or ZBTB3 (TRCN0000015494 and TRCN0000015495; Sigma Aldrich) were produced from HEK293T cells transfected with pLKO and packaging vectors using PolyFect (Qiagen) as previously described (16). Lentiviral particles from the transfected HEK293T cells were collected and used to infect cancer cells with 8 μg/ml polybrene. Infected cells were selected with 2 μg/ml of puromycin for 4 d prior to further experiments.
Western blotting
Total protein was extracted using a cell lysis buffer containing 0.1% IGEPAL, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl (pH 7.9), 10 mM NaF, 0.1 mM EDTA, and protease inhibitor cocktail. Total protein was electrophoresed on SDS-polyacrylamide gels and transferred onto a PVDF membrane (Millipore). Membranes were incubated with primary antibodies in 5% bovine serum albumin containing 0.05% Tween-20 overnight at 4⅂, and HRP-conjugated secondary antibodies were incubated for 1 h at room temperature. Proteins levels were visualized using an ECL Plus kit (GE healthcare).
Measurement of intracellular H2O2 and GSH levels
Intracellular hydrogen peroxide (H2O2) levels were measured using a CM-DCFH-DA assay. Lentivirus-infected cells were cultured in 6-well culture dishes and incubated for 16 h without serum in phenol red-free DMEM. After serum deprivation, cells were stimulated with 10% FBS for 10 min, washed with Krebs-Ringer solution and incubated for 5 min with CM-DCFHDA. Cells were then rinsed and mounted with mounting medium (Vector Labs) and visualized with a confocal microscope (Zeiss). The relative fluorescence was calculated by mean fluorescence intensities of several fields. To measure intracellular glutathione levels, a glutathione colorimetric detection kit (BioVision Research Products) was used according to the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA was isolated with TRIzol (Invitrogen), and 2 μg was used for cDNA synthesis using a high capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems). Experimental Ct values were normalized to 36B4, and relative mRNA expression was calculated versus 36B4 expression. The sequences of PCR primers are summarized in supplemental material.
Measurement of antioxidant activity
Infected cells were grown for 24 h, and cell extracts were collected for the measurement of human CAT, SOD, and GPx activity. Enzyme activity of each antioxidant was measured according to the manufacturer’s instructions (Cayman Chemical Company).
Apoptosis assay
Apoptotic cells were measured using Annexin-V-FITC and propidium iodide (BD Pharmigen). Briefly, cells were infected with lentivirus particles expressing shRNA against either control or ZBTB3, and infected cells were selected by puromycin for 3 days. After selection, cells were trypsinized and washed with cold PBS, and an equal number of cells were stained with FITC-conjugated to an anti-annexin-V antibody and propidium iodide. Fluorescence was acquired by FACS analysis.
Measurement of caspase-3 activity
The proteolytic activity of caspase-3 was determined using a colorimetric assay kit (Cell Signaling Technology) according to the manufacturer’s instructions. Briefly, cells were infected with lentiviral particles expressing shRNA against ZBTB3 or control in 6-well plates. The cells were selected by puromycin for 3 days. Caspase-3 activity in cell lysates was measured and expressed as fold increase compared with control shRNA.
Statistical analysis
All data were analyzed using the unpaired Student’s t-test. Data are represented as means ± standard deviations. P < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by Konkuk University.
References
- 1.Lee S. U., Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol. Rev. (2012);247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly K. F, Daniel J. M. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends. Cell Biol. (2006);16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Costoya J. A. Functional analysis of the role of POK transcriptional repressors. Brief. Funct. Genomic. Proteomic. (2007);6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Torrado R., Yamada D., Defossez P. A. Born to bind: the BTB protein-protein interaction domain. Bioessays. (2006);28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 5.Zheng J., Xiong D., Sun X., Wang J., Hao M., Ding T., Xiao G., Wang X., Mao Y., Fu Y., Shen K., Wang J. Signification of hypermethylated in cancer 1 (HIC1) as tumor suppressor gene in tumor progression. Cancer Microenviron. (2012);5:258–293. doi: 10.1007/s12307-012-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulay G., Dubuissez M., Van Rechem C., Forget A., Helin K., Ayrault O., Leprince D. Hypermethylated in cancer 1 (HIC1) recruits polycomb repressive complex 2 (PRC2) to a subset of its target genes through interaction with human polycomb-like (hPCL)proteins. J. Biol. Chem. (2012);287:10509–10524. doi: 10.1074/jbc.M111.320234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albagli-Curiel O. Ambivalent role of BCL6 in cell survival and transformation. Oncogene. (2003);22:507–516. doi: 10.1038/sj.onc.1206152. [DOI] [PubMed] [Google Scholar]
- 8.Polo J. M., Dell’Oso T., Ranuncolo S. M., Cerchietti L., Beck D., Da silva G. F., Prive G. G., Licht J. D., Melnick A. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. (2004);10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 9.Kim K., Chadalapaka G., Lee S. O., Yamada D., Sastre-Garau X., Defossez P. A., Park Y. Y., Lee J. S., Safe S. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. (2012);31:1034–1044. doi: 10.1038/onc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irani K., Xia Y., Zweier J. L., Sollott S. J., Der C. J., Fearon E. R., Sundaresan M., Finkel T., Goldschmidt-Clermont P. J. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. (1997);275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 11.Wu W. S. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. (2006);25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- 12.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. (2011);194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sena L. A., Chandel N. S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. (2012);48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon S. J., Stockwell B. R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. (2014);10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 15.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach?. Nat. Rev. Drug. Discov. (2009);8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 16.Vazquez F., Lim J. H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C. B., Granter S. R., Widlund H. R., Spiegelman B. M., Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. (2013);23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trachootham D., Zhou Y., Zhang H., Demizu Y., Chen Z., Pelicano H., Chiao P. J., Achanta G., Arlinghaus R. B., Liu J., Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. (2006);10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Singh A., Misra V., Thimmulappa R. K., Lee H., Ames S., Hoque M. O., Herman J. G., Baylin S. B., Sidransky D., Gabrielson E., Brock M. V., Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. (2006);3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H. J., Carr J. R., Wang Z., Nogueira V., Hay N., Tyner A. L., Lau L. F., Costa R. H., Raychaudhuri P. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. (2009);28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj L., Ide T., Gurkar A. U., Foley M., Schenone M., Li X., Tolliday N. J., Golub T. R., Carr S. A., Shamji A. F., Stern A. M., Mandinova A., Schreiber S. L., Lee S. W. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. (2011);475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Barrett C. W., Smith J. J., Lu L. C., Markham N., Stengel K. R., Short S. P., Zhang B., Hunt A. A., Fingleton B. M., Carnahan R. H., Engel M. E., Chen X., Beauchamp R. D., Wilson K. T., Hiebert S. W., Reynolds A. B., Williams C. S. Kaiso directs the transcriptional corepressor MTG16 to the Kaiso binding site in target promoters. PLoS One. (2012);7:e51205. doi: 10.1371/journal.pone.0051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho J. H., Kim M. J., Kim K. J., Kim J. R. POZ/BTB and AT-hook-containing zinc finger protein 1 (PATZ1) inhibits endothelial cell senescence through a p53 dependent pathway. Cell Death Differ. (2012);19:703–712. doi: 10.1038/cdd.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valentino T., Palmieri D., Vitiello M., Pierantoni G. M., Fusco A., Fedele M. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis. (2013);4:e963. doi: 10.1038/cddis.2013.500. [DOI] [PMC free article] [PubMed] [Google Scholar]


