Abstract
In the present study, we demonstrate that ectopic expression of 56-kDa human selenium binding protein-1 (hSP56) in PC-3 cells that do not normally express hSP56 results in a marked inhibition of cell growth in vitro and in vivo. Down-regulation of hSP56 in LNCaP cells that normally express hSP56 results in enhanced anchorage-independent growth. PC-3 cells expressing hSP56 exhibit a significant reduction of hypoxia inducible protein (HIF)-1α protein levels under hypoxic conditions without altering HIF-1α mRNA (HIF1A) levels. Taken together, our findings strongly suggest that hSP56 plays a critical role in prostate cells by mechanisms including negative regulation of HIF-1α, thus identifying hSP56 as a candidate anti-oncogene product. [BMB Reports 2014; 47(7): 411-416]
Keywords: Hypoxia inducible factor, Prostate cancer, Protein-protein interaction, pVHL-interacting deubiquitinating enzyme, Selenium binding protein
INTRODUCTION
The 56-kDa human selenium binding protein-1 (hSP56) is expressed widely in normal cells and tissues (1-3). Interestingly, hSP56 expression is reduced markedly in several primary cancers and cancer cell lines compared to their normal counterparts (1, 4-13). Reduced hSP56 expression also has been correlated with poor outcome in several cancers (4, 6, 10, 11), implicating hSP56 in the modulation of both the incidence and the aggressiveness of cancer. However, this association has not been studied at the molecular level and it is unknown if reduced hSP56 expression itself results in a more malignant phenotype. We have investigated the effect of hSP56 expression on cancer cell growth properties using two well-studied human prostate cancer cell lines, LNCaP and PC-3. Relatively high levels of hSP56 are expressed in LNCaP cells, while little-to-no expression of hSP56 is detected in PC-3 cells at either the mRNA or protein levels (1, 14). We show that hSP56 influences the cell growth and phenotype significantly both in vitro and in vivo.
We also show that PC-3 cells expressing hSP56 exhibit a significant reduction of HIF-1α protein levels under hypoxic conditions without altering HIF-1α mRNA (HIF1A) levels. HIF-1α is a key transcription factor for the cellular response to oxygen availability (15-17). HIF-1α is rapidly degraded under normoxic conditions through the interactions of its hydroxylated proline residues with pVHL, a component of an E3 ubiquitin ligase complex that mediates ubiquitination-dependent protein degradation of target proteins (16). Under hypoxic conditions, HIF-1α is not hydroxylated, resulting in its protection from the pVHL-mediated protein degradation. The stabilized HIF-1α is translocated into the nucleus where it activates a number of genes, including those important for cell proliferation, angiogenesis, glycolysis and erythropoiesis (17) by binding to hypoxia response elements (HREs). Much has been learned about HIF-1α in recent years, but more detailed mechanisms and factors involved in HIF-1α regulation remain to be understood. Our findings suggest that hSP56 plays an important role in regulating HIF-1α, which may be one of mechanisms of hSP56 expression in suppressing the malignant characteristics of prostate cancer cells.
RESULTS AND DISCUSSION
hSP56 suppresses malignant characteristics of prostate cancer cells
We established PC-3 cells stably expressing hSP56 (PC-3/hSP56) and LNCaP cells with hSP56 stably knocked down (LNCaP/hSP56KD) to be used in this study (Fig. 1A). PC-3 cells or PC-3 cells stably transfected with vector (PC-3/V) did not exhibit detectable hSP56 expression (1, 14). PC-3/hSP56C1 (clone 1) expressed hSP56 at levels similar to LNCaP cells, while PC-3/hSP56C6 expressed approximately 10% of hSP56 compared to that of LNCaP cells. LNCaP/hSP56KDF10 cells exhibited undetectable hSP56 expression compared to LNCaP cells or LNCaP cells stably transfected with another shRNA construct designed for hSP56, which failed to down regulate hSP56 expression (designated LNCaP/C).
Fig. 1. hSP56 expression exhibited profound effects on prostate cancer cell growth. (A) Establishment of stable cell lines, PC-3/hSP56 and LNCaP/hSP56KD cells. (B, C) Cell growth curves of PC-3 cells and derivatives (B), or LNCaP cells and derivatives (C) in anchorage-dependent liquid cultures. (D) Soft agar colony-forming assay. Number of colonies and their size were analyzed using the ImageJ software (NIH). Similar results were obtained in repeated experiments. Scale bar, 500 μm. (E) In vivo tumorigenicity experiment. (F) Pictures of representative mice taken at week 9. The site of injection is marked with dotted circle in one of the pictures.

PC-3/hSP56 grew much slower than PC-3 or PC-3/V cells in anchorage-dependent liquid culture in a manner dependent on hSP56 expression level (Fig. 1B). The higher the hSP56 expression level is, the slower the growth becomes, as represented by PC-3/hSP56C1. PC-3/hSP56C6 exhibited an intermediate growth rate between PC-3/V and PC-3/hSP56C1. The slower growth rate of PC-3/hSP56C1 or C6 was not observed at earlier passages after transfection during the clonal selection procedures, therefore implying that hSP56 expression has a long-term effect on cell growth regulation rather than immediate effect. The clones with high levels of hSP56 expression including PC-3/hSP56C1 either stopped growing in later passages or gradually lost hSP56 expression (Supplementary Fig. S1), suggesting that high expression levels of hSP56 may have a pronounced inhibitory action on cell growth. Therefore, we continued our experiments using PC-3/hSP56C6 or using freshly prepared cells with hSP56 expression levels similar to PC-3/hSP56C6 and comprehensively designated as PC-3/hSP56. While PC-3/hSP56 cells exhibited remarkable differences in cell growth properties, LNCaP/hSP56KD F10 or an additional clone A7, expressing also undetectable hSP56, did not appear to have alterations in growth properties in anchorage-dependent liquid culture (Fig. 1C).
hSP56 expression in PC-3 cells had a profound inhibitory effect on anchorage-independent cell growth in soft agar as well (Fig. 1D). PC-3/V cells exhibited robust growth in soft agar, producing 160 colonies per microscopic field with an average size of 3,575 μm2. In marked contrast, PC-3/hSP56 cells exhibited significantly reduced anchorage-independent growth, producing 136 colonies with an average size of 1,509 μm2. Importantly, in the reciprocal (hSP56 knockdown) experiment, LNCaP/hSP56KDF10 cells exhibited significantly enhanced colony formation (49 colonies with an average size of 606 μm2) compared to the virtual absence of colonies formed by LNCaP/C cells (15 colonies, 171 μm2) (additional microscopic fields are provided in Supplementary Fig. S2). To test the effect of hSP56 expression on tumorigenicity in vivo, we transplanted PC-3/V cells or PC-3/hSP56 cells subcutaneously into groups of eight male SCID mice. Starting one month after injection, tumor size was measured weekly and tumor volume was calculated. The growth rate of the PC-3/hSP56 cell tumors was much slower than that of the PC-3/V cell tumors (Fig. 1E and 1F). These results are consistent with a number of findings that reported reduced hSP56 expression in cancers, supporting the conclusion that hSP56 suppresses the malignant characteristics of prostate cancer cells.
hSP56 interacts with both VDU1 and VDU2
We then sought to understand mechanisms by which hSP56 expression suppresses prostate cancer cell growth. We previously identified pVHL-interacting deubiquitinating enzyme 1 (VDU1) as a protein interacting with hSP56 (14), thus extended this finding to examine whether hSP56 interacts also with VDU2, a closely related isoform of VDU1. We incubated soluble extracts of COS-7 cells mock transfected, or transfected with vector alone or with VDU1-HA or VDU2-HA in a 96-well plate coated with BSA or hSP56, and analyzed protein interaction by anti-HA ELISA. The expression levels of VDU1 and VDU2 from the transiently transfected COS-7 cells were similar (Fig. 2A). Interestingly, we observed a stronger interaction between VDU2 and hSP56 than between VDU1 and hSP56 (Fig. 2B). We investigated the interactions of hSP56 with VDU1 and VDU2 further by co-immunoprecipitation. Soluble extracts of LNCaP cells transfected with VDU1-HA or VDU2-HA were immunoprecipitated using anti-HA antibody or normal mouse IgG, and analyzed by anti-hSP56 immunoblotting. The membrane was then stripped and reprobed with anti-HA-biotin/streptavidin-HRP conjugates. hSP56 was co-immunoprecipitated specifically with either VDU1-HA or VDU2-HA using anti-HA antibody, but not using normal mouse IgG (Fig. 2C), confirming that hSP56 interacts with both VDU1 and VDU2. We also attempted co-immunoprecipitation experiments at their endogenous expression levels, however available antibodies against VDU1 and VDU2 have failed to detect the proteins specifically. Similar to our previous demonstration with hSP56 and VDU1 interactions (14), VDU2 (red fluorescence from anti-HA/anti-mouse IgG-Texas Red) and hSP56 (green fluorescence from hSP56-EGFP) overlapped in the cytoplasm (Fig. 2D). These results suggest that the interaction of hSP56 with VDU2 is relevant.
Fig. 2. hSP56 interacts with VDU1 and VDU2. (A) Expression of the full-length VDU1-HA and VDU2-HA in COS-7 cells. The interaction of hSP56 with the full-length VDU1 and VDU2 was examined by ELISA-format in vitro binding (B) and co-immunoprecipitation (C). (D) Co-localization of hSP56 with VDU2. 4’,6-diamidino-2-phenylindole (DAPI) was used for nuclear staining. Scale bar, 10 μm. XF, transfection.

hSP56 down-regulates HIF-1α protein
VDU2 stabilizes HIF-1α by its deubiquitinating activity, resulting in the increased expression of hypoxia responsive genes (18). Therefore, we examined the effect of hSP56 expression on HIF-1α stabilization. PC-3 cells were transfected with hSP56 expression plasmid or vector alone and then incubated under the specified conditions for 5 or 24 hr (Fig. 3A). Transient expression of hSP56 resulted in significantly reduced HIF-1α under hypoxic conditions (1% O2) as well as under simulated hypoxic conditions (100 μM CoCl2 treatment). The extent of the HIF-1α protein reduction (to 38-57% relative to vector transfected cells) is remarkable, especially considering that the transfection efficiency was between 48-66% determined by EGFP transfection under similar conditions (Supplementary Fig. S3). hSP56 expression reduced HIF-1α protein stabilized by CoCl2 treatment to 91% at 5 hr and to 66% at 24 hr. The less efficient reduction of HIF-1α by hSP56 expression observed at the early time point with CoCl2 may be due to the robust effect of CoCl2 treatment in stabilization of HIF-1α. These findings suggest that hSP56 may function at a point downstream of already stabilized HIF-1α, i.e., ubiquitination and protein degradation.
Fig. 3. hSP56 regulates HIF-1α protein levels. (A) PC-3 cells were transiently transfected with the indicated plasmids for 24 hr and plated into 6-well plates. After an additional 24 hr, the specified conditions were applied for 5 or 24 hr. (B) PC-3 cells prepared in 6-well plates were transfected with increasing amounts of hSP56 expression plasmid (equal to 1.5 μg total with vector) for 24 hr and treated with CoCl2 for 18 hr. Relative HIF-1α/β-actin levels quantified by the ImageJ software (NIH) are presented as % of that of vector transfected sample in panels A and B. Samples exhibiting significantly reduced HIF-1α levels are indicated in italicized, underlined fonts. (C) Quantitative RT-PCR to determine HIF1A mRNA levels. GAPDH mRNA levels were used as normalization controls. (D) PC-3 cells stably transfected with hSP56 exhibited a reduction of HIF-1α protein level induced by the indicated CoCl2 treatment for 18 hr. Sections within an identical membrane were combined. (E) HIF-1α stabilization was not significantly affected by hSP56 knock-down in LNCaP cells. Increasing concentrations of CoCl2 (0, 12, 25, 50, 100 and 200 μM) were used for the experiment presented on the right panel. (F) LNCaP cells express much lower level of HIF1A mRNA compared to PC-3 cells. (G) HIF-1α stabilization in LNCaP and PC-3 cells. The cells were treated as indicated for 18 hr, and the cell lysates were analyzed by immunoblotting with the indicated antibodies. LNCaP and PC-3 cell samples were blotted to separate membranes, but processed together under the same conditions. Two exposure times (7 sec and 60 sec) are shown for anti-HIF-1α immunoblotting to compare the HIF-1α signals in two cell samples. Note that MG132 robustly increased HIF-1α in PC-3 cells, but not in LNCaP cells.
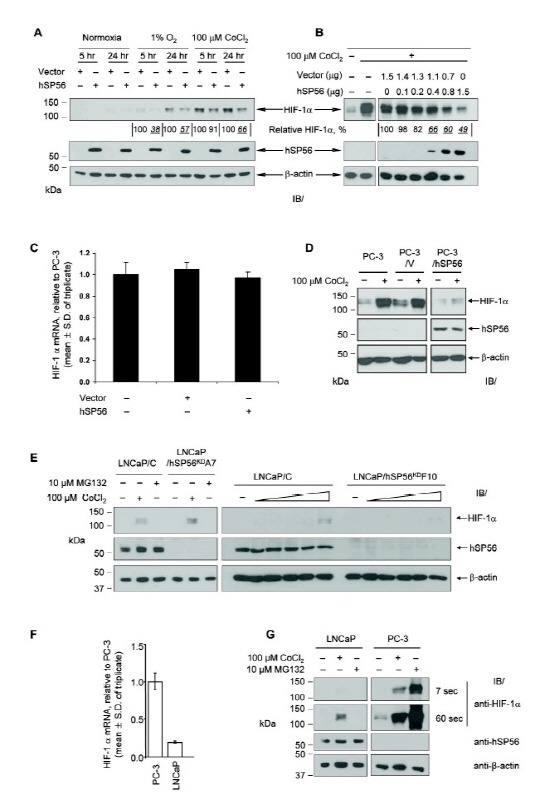
We investigated the negative regulation of HIF-1α by hSP56 further by transfecting PC-3 cells with increasing amounts of hSP56 expression plasmid (Fig. 3B). HIF-1α protein was down regulated up to 49% of vector transfected control, and the reduction correlated with hSP56 expression levels. This hSP56-mediated HIF-1α down regulation did not occur at the gene transcription level, since HIF1A mRNA expression, as measured by quantitative RT-PCR, was unchanged by vector or hSP56 transfection (Fig. 3C). The long-term effect of hSP56 expression on HIF-1α stabilization also was determined using the stable cell lines. PC-3/hSP56 cells exhibited almost no stabilization of HIF-1α in the presence of 100 μM CoCl2 (Fig. 3D), consistent with the immediate early effect of hSP56 expression on HIF-1α (Fig. 3A and 3B).
In contrast, LNCaP/hSP56KD cells did not exhibit an increase of HIF-1α protein with CoCl2 treatment (Fig. 3E). HIF-1α protein level in LNCaP cells appears to be regulated differentially from that in PC-3 cells. Firstly, HIF-1α mRNA expression is much lower in LNCaP cells than in PC-3 cells (Fig. 3F). Secondly, LNCaP cells responded quite distinctively to MG132, a potent proteasome inhibitor. MG132 inhibits the proteasome-mediated protein degradation pathways, consequently preventing HIF-1α degradation (19). However, MG132 did not increase HIF-1α protein level in LNCaP cells, while PC-3 cells exhibited a robust stabilization of HIF-1α by MG132 (Fig. 3G). This finding suggests that the proteasome-mediated protein degradation pathway may not be a major regulatory mechanism for HIF-1α in LNCaP cells. These differences in HIF-1α regulation may be an explanation for the lack of the enhanced HIF-1α stabilization in LNCaP/hSP56KD cells. Further investigation of the differential regulation of HIF-1α in these cells will provide important information regarding the regulation of hypoxic responses in prostate cancer cells.
The function of hSP56 had been difficult to elucidate since its discovery (2, 20, 21). The expression of highly conserved homologs in both animal and plant kingdoms (22) implies a fundamental role for this protein in cell biology. In the present study, we have identified hSP56 as a basic regulator of the cell growth phenotype and as a negative regulator of HIF-1α stabilization in prostate cancer cells. Notably, the mouse homolog of hSP56 has been identified as a novel target of HIF-1α (23). Our current findings, together with this observation, may suggest a feedback regulation between HIF-1α and hSP56. The balance of HIF-1α and hSP56 that is well maintained in normal cellular physiology may be disturbed by yet unidentified mechanisms in human malignancies.
HIF-1α and hypoxia inducible genes, such as VEGF, play important roles in tumor progression (17). As noted earlier, reduced hSP56 expression has been observed in many types of human cancer. Also, decreased expression of hSP56 has been associated with a poor clinical outcome in several human cancers, including lung adenocarcinoma (4), pleural mesothelioma (24) and colorectal carcinomas (6, 7). These associations in several cancer types may be explained by loss of hSP56’s function in destabilization of HIF-1α. The expression of the gene encoding hSP56 (SELENBP1) in colon cancer was shown to be downregulated by hypermethylation in its promoter region (25). It will be important to investigate whether this mechanism is operative in the reduction of hSP56 expression in multiple cancers. Demethylating agents that can reverse the reduced hSP56 expression, consequently recovering the normal balance of HIF-1α and hSP56, may become interesting candidates for future chemopreventive drug development. Understanding the mechanisms by which hSP56 expression is regulated may become useful to understand the functions of hSP56 in other diseases, since SELENBP1 gene expression has been shown to be upregulated in major psychotic disorders such as schizophrenia (26, 27).
Our findings in prostate cancer cells perhaps can be extrapolated into other types of cancer as well, an idea supported by data that we obtained by expressing hSP56 in a human lung carcinoma cell line, A549, where ectopic expression of hSP56 downregulated HIF-1α stabilization (Supplementary Fig. S4). In a hepatocellular carcinoma cell line (SMMC7721), however, downregulation of hSP56 rather decreased HIF-1α protein level (28). Different cell types may have distinct mechanisms maintaining the balance between hSP56 and HIF-1α. It will be further investigated whether the protein-protein interactions of hSP56 with VDU 1 and/or 2 are essential for HIF-1α regulation, or whether the negative regulation of HIF-1α by hSP56 is essential for suppressing the malignant characteristics of cancer cells. Our findings suggest that hSP56 exhibits an anti-cancer action by mechanisms including that involved in negative regulation of HIF-1α stabilization in prostate cancer cells.
MATERIALS AND METHODS
Plasmid construction and in vitro binding experiments
Construction of hSP56 expression plasmids is presented in Supplementary Fig. S5. For down-regulation of hSP56 expression, an shRNA construct containing targeting sequence of 5’-catcacccacactccctattt (Open Biosystems) was used. Plasmids expressing VDU1 and VDU2 with a C-terminal HA-tag (VDU1-HA and VDU2-HA) were constructed as shown in Supplementary Fig. S6. Recombinant (His)6-hSP56 expressed in E. coli was purified using Ni-NTA agarose (Qiagen) and used in the ELISA-format in vitro binding experiment as described previously (14).
Mammalian cell culture and transfection
COS-7, PC-3 and LNCaP cells (American Type Culture Collection) were maintained in RPMI-1640 medium (for COS-7, Mediatech) or DMEM/F-12 medium (for PC-3 and LNCaP, Mediatech) containing 10% FBS (HyClone) and 1× penicillin/streptomycin (Invitrogen) at 37℃ in a humidified atmosphere of 95% air and 5% CO2. Hypoxic conditions were established either in a humidified hypoxia chamber (StemCell Technologies) purged with controlled gas mixtures containing 1% O2 or by 100 μM CoCl2 treatment for simulated hypoxic conditions (29). Soft agar colony-forming assays were performed as described (30). FuGene6 (Roche) was used to transfect cells according to the manufacturer’s instructions. Stable cell lines were established by growing antibiotic-resistant clones from single cell colonies.
Immunological methods
Co-immunoprecipitation was performed using soluble cell lysates prepared with CytoBuster (Novagen), containing protease inhibitor cocktail set III (CalBiochem). The lysates were rotated overnight at 4℃ with 2 μg of anti-HA antibody (Covance) or normal mouse IgG (Jackson ImmunoResearch Laboratory). A 50 μl suspension (1 : 1) of Protein A/G-agarose beads (Pierce) was added and incubated by rotation at 4℃ for 2 h. The immunoprecipitates were pelleted by centrifugation and washed three times with the lysis buffer. Both the immunoprecipitates and total cell lysates were analyzed by anti-hSP56 and anti-HA immunoblotting. Immunoblotting and immunofluorescence analyses were described previously (14).
RNA isolation and quantitative RT-PCR
RNA isolation and cDNA synthesis were described previously (31). Quantitative real-time PCR (qRT-PCR) was performed using the ABI 7300 real-time PCR system (Applied Biosystems) with iTaq SYBR Green Supermix (Bio-Rad). qRT-PCR validated primer sets for HIF1A and GAPDH mRNA detection were purchased from RealTime Primers.
In vivo tumorigenicity
Male SCID-ICR mice (6-week-old) were purchased from Taconic and housed in AALAC accredited animal facility. All procedures were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Mice were randomized into two groups (n=8 each group) and injected subcutaneously, in the dorsal flank, with PC-3/Vector or PC-3/hSP56 cells (5 × 106 cells/animal). Tumor size was measured using a Vernier caliper and tumor volume was calculated using the standard formula, volume = length × (width)2 × 0.52.
Acknowledgments
This work was supported in part by DOD Grant DAMD17-02-1-0014 and DOD Grant DAMD17-03-1-0233 to AJS, and DOD Grant W81XWH-07-1-0321, an intramural grant from Kosin University College of Medicine and KNRF grant 2012R1A1A2042188 to JYJ.
References
- 1.Yang M., Sytkowski A. J. Differential expression and androgen regulation of the human selenium-binding protein gene hSP56 in prostate cancer cells. Cancer Res. (1998);58:3150–3153. [PubMed] [Google Scholar]
- 2.Chang P. W., Tsui S. K., Liew C., Lee C. C., Waye M. M., Fung K. P. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. J. Cell Biochem. (1997);64:217–224. doi: 10.1002/(SICI)1097-4644(199702)64:2<217::AID-JCB5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Wu C. L., Zhang W. B., Mai K. S., Liang X. F., Xu W., Wang J., Ma H. M. Molecular cloning, characterization and mRNA expression of selenium-binding protein in abalone (Haliotis discus hannai Ino): Response to dietary selenium, iron and zinc. Fish Shellfish Immunol. (2010);29:117–125. doi: 10.1016/j.fsi.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Chen G., Wang H., Miller C. T., Thomas D. G., Gharib T. G., Misek D. E., Giordano T. J., Orringer M. B., Hanash S. M., Beer D. G. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. (2004);202:321–329. doi: 10.1002/path.1524. [DOI] [PubMed] [Google Scholar]
- 5.Brown L. M., Helmke S. M., Hunsucker S. W., Netea-Maier R. T., Chiang S. A., Heinz D. E., Shroyer K. R., Duncan M. W., Haugen B. R. Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Mol. Carcinog. (2006);45:613–626. doi: 10.1002/mc.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H., Kang H. J., You K. T., Kim S. H., Lee K. Y., Kim T. I., Kim C., Song S. Y., Kim H. J., Lee C. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics. (2006);6:3466–3476. doi: 10.1002/pmic.200500629. [DOI] [PubMed] [Google Scholar]
- 7.Li T., Yang W., Li M., Byun D. S., Tong C., Nasser S., Zhuang M., Arango D., Mariadason J. M., Augenlicht L. H. Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Mol. Nutr. Food Res. (2008);52:1289–1299. doi: 10.1002/mnfr.200700331. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P., Zhang C., Wang X., Liu F., Sung C. J., Quddus M. R., Lawrence W. D. The expression of selenium-binding protein 1 is decreased in uterine leiomyoma. Diagn. Pathol. (2011);5:80. doi: 10.1186/1746-1596-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Zhan N., Dong W. G. Altered expression of selenium-binding protein 1 in gastric carcinoma and precursor lesions. Med. Oncol. (2010);28:951–957. doi: 10.1007/s12032-010-9564-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Dong W. G., Lin J. Reduced selenium-binding protein 1 is associated with poor survival rate in gastric carcinoma. Med. Oncol. (2011);28:481–487. doi: 10.1007/s12032-010-9482-7. [DOI] [PubMed] [Google Scholar]
- 11.Xia Y. J., Ma Y. Y., He X. J., Wang H. J., Ye Z. Y., Tao H. Q. Suppression of selenium-binding protein 1 in gastric cancer is associated with poor survival. Hum. Pathol. (2011);42:1620–1628. doi: 10.1016/j.humpath.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Silvers A. L., Lin L., Bass A. J., Chen G., Wang Z., Thomas D. G., Lin J., Giordano T. J., Orringer M. B., Beer D. G., Chang A. C. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clin. Cancer Res. (2010);16:2009–2021. doi: 10.1158/1078-0432.CCR-09-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Wang Y. E., Zhang P., Liu F., Sung C. J., Steinhoff M. M., Quddus M. R., Lawrence W. D. Progressive loss of selenium-binding protein 1 expression correlates with increasing epithelial proliferation and papillary complexity in ovarian serous borderline tumor and low-grade serous carcinoma. Hum. Pathol. (2010);41:255–261. doi: 10.1016/j.humpath.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Jeong J. Y., Wang Y., Sytkowski A. J. Human selenium binding protein-1 (hSP56) interacts with VDU1 in a selenium-dependent manner. Biochem. Biophys. Res. Commun. (2009);379:583–588. doi: 10.1016/j.bbrc.2008.12.110. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin W. G., Jr., Ratcliffe P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. (2008);30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 17.Semenza G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. (2003);3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Wang D., Messing E. M., Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. (2005);6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagg M., Wennstrom S. Activation of hypoxia-induced transcription in normoxia. Exp. Cell Res. (2005);306:180–191. doi: 10.1016/j.yexcr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Jamba L., Nehru B., Medina D., Bansal M. P., Sinha R. Isolation and identification of selenium-labeled proteins in the mouse kidney. Anticancer Res. (1996);16:1651–1657. [PubMed] [Google Scholar]
- 21.Sinha R., Bansal M. P., Ganther H., Medina D. Significance of selenium-labeled proteins for selenium's chemopreventive functions. Carcinogenesis. (1993);14:1895–1900. doi: 10.1093/carcin/14.9.1895. [DOI] [PubMed] [Google Scholar]
- 22.Flemetakis E., Agalou A., Kavroulakis N., Dimou M., Martsikovskaya A., Slater A., Spaink H. P., Roussis A., Katinakis P. Lotus japonicus gene Ljsbp is highly conserved among plants and animals and encodes a homologue to the mammalian selenium-binding proteins. Mol. Plant. Microbe. Interact. (2002);15:313–322. doi: 10.1094/MPMI.2002.15.4.313. [DOI] [PubMed] [Google Scholar]
- 23.Scortegagna M., Martin R. J., Kladney R. D., Neumann R. G., Arbeit J. M. Hypoxia-inducible factor-1alpha suppresses squamous carcinogenic progression and epithelial-mesenchymal transition. Cancer Res. (2009);69:2638–2646. doi: 10.1158/0008-5472.CAN-08-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pass H. I., Liu Z., Wali A., Bueno R., Land S., Lott D., Siddiq F., Lonardo F., Carbone M., Draghici S. Gene Expression Profiles Predict Survival and Progression of Pleural Mesothelioma. Clin. Cancer Res. (2004);10:849–859. doi: 10.1158/1078-0432.CCR-0607-3. [DOI] [PubMed] [Google Scholar]
- 25.Pohl N. M., Tong C., Fang W., Bi X., Li T., Yang W. Transcriptional regulation and biological functions of selenium-binding protein 1 in colorectal cancer in vitro and in nude mouse xenografts. PLoS One. (2009);4:e7774. doi: 10.1371/journal.pone.0007774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glatt S. J., Everall I. P., Kremen W. S., Corbeil J., Sasik R., Khanlou N., Han M., Liew C. C., Tsuang M. T. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc. Natl. Acad. Sci. U. S. A. (2005);102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanazawa T., Chana G., Glatt S. J., Mizuno H., Masliah E., Yoneda H., Tsuang M. T., Everall I. P. The utility of SELENBP1 gene expression as a biomarker for major psychotic disorders: replication in schizophrenia and extension to bipolar disorder with psychosis. Am J. Med. Genet. B. Neuropsychiatr. Genet. (2008);147B:686–689. doi: 10.1002/ajmg.b.30664. [DOI] [PubMed] [Google Scholar]
- 28.Huang C., Ding G., Gu C., Zhou J., Kuang M., Ji Y., He Y., Kondo T., Fan J. Decreased selenium-binding protein 1 enhances glutathione peroxidase 1 activity and downregulates HIF-1alpha to promote hepatocellular carcinoma invasiveness. Clin. Cancer Res. (2012);18:3042–3053. doi: 10.1158/1078-0432.CCR-12-0183. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg M. A., Schneider T. J. Similarities between the oxygen-sensing mechanisms regulating the expression of vascular endothelial growth factor and erythropoietin. J. Biol. Chem. (1994);269:4355–4359. [PubMed] [Google Scholar]
- 30.Freedman V. H., Shin S. I. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. (1974);3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 31.Jeong J. Y., Feldman L., Solar P., Szenajch J., Sytkowski A. J. Characterization of erythropoietin receptor and erythropoietin expression and function in human ovarian cancer cells. Int. J. Cancer. (2008);122:274–280. doi: 10.1002/ijc.23068. [DOI] [PubMed] [Google Scholar]


