Abstract
To investigate the function of N-glycosylation of Cel5A (endoglucanase II) from Hypocrea jecorina, two N-glycosylation site deletion Cel5A mutants (rN124D and rN124H) were expressed in Saccharomyces cerevisiae. The weights of these recombinant mutants were 54 kDa, which were lower than that of rCel5A. This result was expected to be attributed to deglycosylation. The enzyme activity of rN124H was greatly reduced to 60.6% compared with rCel5A, whereas rN124D showed slightly lower activity (10%) than that of rCel5A. rN124D and rN124H showed different thermal stabilities compared with the glycosylated rCel5A, especially at lower pH value. Thermal stabilities were reduced and improved for rN124D and rN124H, respectively. Circular dichroism spectroscopy showed that the modification of secondary structure by mutation may be the reason for the change in enzymatic activity and thermal stability. [BMB Reports 2014; 47(5): 256-261]
Keywords: Cel5A, Circular dichroism spectroscopy, Deglycosylation, Hypocrea jecorina, Saccharomyces cerevisiae
INTRODUCTION
Cellulose is the most abundant polymer on earth. The enzymatic degradation of cellulose is essential for maintaining the global carbon cycle. The cellulase system of the soft rot fungus Hypocrea jecorina (syn. Trichoderma reesei) is the most studied and best understood of all cellulolytic systems (1). H. jecorina produces a suite of cellulolytic enzymes with distinctly different activities. These enzymes are classified into three major families, namely, endoglucanases (EGs; EC 3.2.1.4), cellobiohydrolases (CBHs; EC 3.2.1.91), and β-glucosidases (EC3.2.1.21) (2). According to the classical theory, EGs internally nick the cellulose, thus disrupting its crystallinity and generating new free ends in the polymer. CBHs act processively from these free ends and release soluble cellobiose molecules. β-glucosidases subsequently hydrolyze cellobiose to glucose (3).
Most enzymes secreted by H. jecorina are glycoproteins with both O- and N-linked glycosylation sites (4, 5). N- and O-linked glycosylation could affect the catalytic efficiency, cellulose-binding affinity, and stability of cellulases (6). The extensive glycosylation in cellulytic enzymes in various species (bacteria, fungi, and insects) indicates that they may have a very subtle effect on enzyme catalysis (5, 7, 8). Increased level of N-glycosylation of the Cel7A (CBHI) of H. jecorina resulted in reduced activity and increased non-productive binding on cellulose (9). Deglycosylation of Talaromyces emersonii Cel7A expressed in Saccharomyces cerevisiae resulted in decreased hydrolytic efficiency on crystalline cellulose (10). Different glycoforms of cellobiohydrolase I (CBHI-B) in Penicillium decumbens, which have the same amino acid sequence but different N-glycosylation characteristics, display different biological functions (11). Glycosylation affects not only catalytic efficiency but also enzyme stability. N-glycosylation of EG of Bacillus subtilis protects itself from immobilized-papain attack and accounts for higher thermostability (8).
Cel5A (EGII) is one of the most abundant EGs from H. jecorina (12, 13). The lack of Cel5A production reduces EG activity in the culture supernatant by as high as 55% (14). The presence of both N- and O-linked glycans in Cel5A with only one N-glycosylation site (Asn124) was observed in its catalytic domain (4). The analysis of secretome produced by H. jecorina showed Cel5A heterogeneity. The molecular masses of Cel5A observed on 2D gels were higher than the expected masses calculated from the protein sequences, and the reason may be due to glycosylation (15).
Several mechanisms of glycosylation and deglycosylation effects on Cel7A (CBHI) are well characterized (9, 16-18). However, few data exist on the nature and effect of glycosylation on the effectiveness or stability of Cel5A. According to our previous report, no significant difference is observed in enzymatic activity between the native Cel5A from H. jecorina and the recombinant Ce5A expressed in S. cerevisiae, which has an original 57 kDa molecular mass. Endoglycanase H (Endo H)-treated rCel5A shows a 54 kDa molecular mass (19).
In the present study the function of N-linked glycosylation in Cel5A was examined using two mutants (rN124D and rN124H) that lack the potential N-linked glycosylation sites. We show that the Asn residue at 124 site not only provides the N-glycosylation site but is also essential in maintaining enzymatic activity. N-linked glycosylation of Cel5A affected the thermal stability of enzyme especially at lower pH value.
RESULTS
Asn residue at 124 site is essential for maintaining enzyme activity
To elucidate the functional role of N-glycosylation sites, two mutants (rN124D and rN124H) were constructed by site-directed mutagenesis. DNA sequencing of the entire gene confirmed the absence of secondary mutations in the mutants. The recombinant enzymes were purified, and SDS-PAGE analysis indicated that both recombinant mutants had lower molecular masses from approximately 57 kDa to 54 kDa (Fig. 1A and B). The reduction in size of the two mutants compared with the rCel5A was expected to be a result of deglycosylation. Zymograms showed three clear activity bands confirming the molecular mass change (Fig. 1B).
Fig. 1. SDS-PAGE, zymograms, and enzymatic activity of recombinant Cel5A, rN124H, and rN124D. (A) SDS-PAGE of purified rCel5A, rN124H, and rN124D. Each mutant had a 54 kDa molecular mass lower than that of 57 kDa for rCel5A. (B) Zymograms of crude fermentation. Each mutant had a 54 kDa activity band, which is lower than that of 57 kDa for rCel5A. (C) Enzymatic activity of purified rCel5A, rN124H, and rN124D.
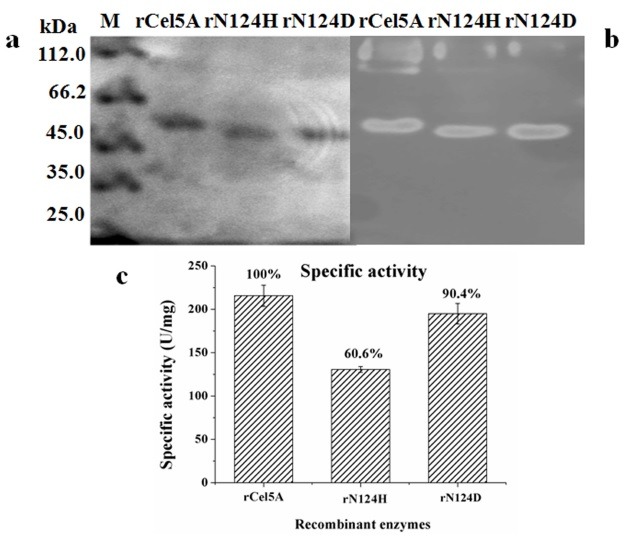
Both recombinant mutants had reduced specific activity. The specific activity of rN124D was 194.9 U/mg, which was slightly lower (10%) than that of rCel5A (215.6 U/mg). However, the mutation rN124H displayed an apparent decrease in its specific activity (130.6 U/mg), approximately 60.6% compared with that of rCel5A (Fig. 1C). For both mutants, the zymograms of the crude fermentation showed the smear migrating above 85 kDa, which were identical to rCel5A. Given the deglycosylation, they might be heterozygous dimers or oligomers of recombinant protein but not hyper-mannosylated glycoproteins as we previously assumed (19) because CMCase activities were detectable regardless of the large molecular masses (Fig. 1B).
N-glycosylation affects the thermal stability of the enzymes especially at lower pH value
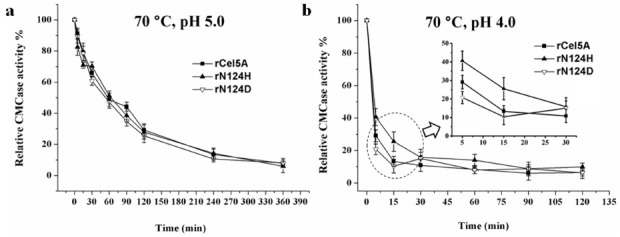
The effect of temperature on enzymatic hydrolysis of rN124D and rN124H at 70℃ was studied at pH 5.0 and 4.0. At pH 5.0, the thermal stability of both mutants was identical to that of rCel5A. Both mutants lost approximately 50% of the initial activity after 1 h incubation (Fig. 2A). At pH 4.0, all three samples showed much less thermal stability than those at pH 5.0. Glycosylated rCel5A and deglycosylated rN124D and rN124H showed different thermal stabilities. Each sample respectively retained approximately 29%, 20%, and 41% of its original activity within 5 min (Fig. 2B).
Fig. 2. Residual enzymatic activity of purified recombinant Cel5A, rN124H, and rN124D. Effect of temperature on enzymatic hydrolysis at 70℃ at (A) pH 5. 0 and (B) pH 4. 0.
Asn residue at site 124 of Cel5A is essential for maintaining enzyme conformation
The mutant rN124H indicated catalytic efficiency loss. Both mutants showed different thermal stabilities at lower pH value compared with that of rCel5A. To investigate whether the catalytic domains of the mutants were correctly folded, circular dichroism (CD) spectroscopy was used to study the secondary structures of rCel5A, rN124D, and rN124H (Fig. 3).
Fig. 3. Far-UV CD spectra of purified rCel5A, rN124H, and rN124D.
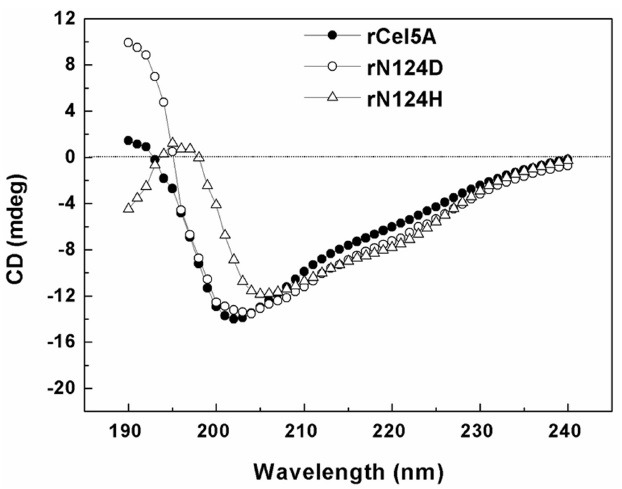
The far-UV CD spectrum of the purified rCel5A indicated a protein rich in β-sheet structure (59.7%) and random coil structure (39%). This result agrees with the high β-sheet content observed in the modeled three-dimensional structure of the enzyme (20). The CD spectra of rN124D indicated a secondary structure similar to that of the wild-type catalytic domain. The slight decrease in negative peak in the spectra range of approximately 202 nm to 205 nm corresponds to an increase in random coil conformation (21). However, the spectra region of rN124H differed greatly with that of rCel5A. Negative peaks of rN124H shifted to the right, with the positive peak in the spectra ranging between 195 and 200 nm. These peaks corresponded to a decrease in random coil but an increase in α- or β-helix conformation (22). The change in secondary structure by mutation was speculated to lead to reduction in hydrolysis activity of rN124H.
DISCUSSION
In Cel5A, there is O-glycosylation and the corresponding number of O-linked glycans ranged from 32 to 42 hexoses residues. However, the O-glycosylation site of Cel5A was not identified yet (4). Endo H is a highly specific endoglycosidase that cleaves asparagine-linked mannose-rich oligosaccharides, generating a truncated sugar molecule with one N-acetylglucosamine residue retained on the asparagines (23). Endo H is assumed to work only on N-glycosylation of Cel5A, not on O-glycosylation, which always occurs on serine, threonine, and hydroxylysine residues. Therefore, rN124D and rN124H mutants were expected to have a lower molecular mass of approximately 54 kDa (Fig. 1A and 1B), consistent with rCel5A treated with endo H (19).
rN124D and rN124H showed approximately 10% and 40% reduced activity, respectively (Fig. 1C). Combined with our previous report that endo H-treated rCel5A also showed approximately 10% reduced activity (19), the reduced activity of rN124D can be ascribed to the loss of N-glycosylation. The activity reduction of rN124D is due to the protein structural change, which is the result of N-glycosylation loss by Asp substitution. The CD spectrum of rN124D, which indicates a secondary structure similar to that of rCel5A, suggested that deglycosylation could cause a slight conformational modification, which might be the reason for the reduced activity. The function of glycan chains in protein folding and structure is discussed in a number of reports (5, 24, 25). The long-range effects of local interactions between sugars and amino acid residues are observed in the polypeptide backbone. For example, fucose stabilizes hydrogen bonds on the far side of the β-sheet by making local interactions that determine the equilibrium state of the entire module for Locusta migratoria peptide-C (26). On the contrary, the reduced activity of rN124H should not be completely ascribed to the loss of N-glycosylation completely but also to the substitution of His residue to Asn. According to the homology modeling structure of Cel5A obtained from the SWISS-MODEL (27), similar to other enzymes that belong to clan GH-A, Cel5A shares a (α/β)8 barrel structure, where the two catalytic residues (E218 and E329) are located inside the core of the β barrel (28, 29). The N124 site is located outside of the barrel (20, 30). Therefore, the glycosylation on this site is assumed to not intensively influence the overall structure of the recombinant enzyme.
Although rCel5A, rN124D, and rN124H shared an identical thermal stability at the optimum pH of 5.0, the two deglycosylated samples showed different thermal stabilities compared with the glycosylated rCel5A at pH 4.0 when they were incubated at 70℃. Glycosylated rCel5A showed higher thermal stability than deglycosylated rN124D. The result is expected because many previous studies showed that the increased levels of glycosylation could improve the thermotolerance of glycoproteins (31-33). Although N124 was located outside the (α/β)8 barrel, the covalent binding of glycans to the protein surface may inherently enhance the thermal and kinetic stabilities of proteins. N-linked glycosylation was hypothesized to cause a decrease in dynamic fluctuations throughout the entire molecule, which led to an increase in thermal and structural stabilities (34).
However, the function of glycosylation in improving the thermal stability of glycoprotein is ambiguous. Some reports show that high levels of glycosylation even reduce the thermotolerance capacity of certain proteins (35, 36). However, the decrease of thermal stability for rCel5A compared with rN124H at pH 4.0 was not assumed to be due to its glycosylation. The substitution of His residue to Asn had an essential function in improving the thermal stability for N124H at lower pH value. Although many reports presented that His residues may be responsible for maintaining the pKa of the acid/base catalyst, thereby broadening the optimum pH range of the enzyme, structural analysis indicated that most His residues were located in the catalytic residue vicinity (37, 38). Site 124 of Cel5A was located outside the (α/β)8 barrel, which was far from the two catalytic residues. This phenomenon indicates that the aforementioned site is not likely to have directly interfered with the ionization state of the two catalytic residues. In fact, although the pH-dependent profile of an enzyme is mainly determined by the ionization of the catalytic groups, it can be modulated by various interactions in their microenvironments (39). The CD spectrum difference between rN124H and rCel5A suggests that the substitution of His residue to Asn resulted in a secondary structure change in the protein, which may be due to the increased thermal stability and reduced enzyme activity. However, the molecular basis is not understood yet and should be investigated in the future.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions
S. cerevisiae H158 [GPY55-15B (MATa leu2-3 leu2-112. ura3-52 trp1-289 his4-519 prb1 cir+)] was used as a host for the expression vector pAJ401, which carries a PGK1 gene as promoter and URA3 gene as selective marker (VTT Biotechnology, Finland). The cloning vector pUC18, which is carrying the wild-type Cel5A cDNA (19), was used as a template for PCR. Yeast cells were aerobically cultivated in synthetic medium (SDC: 6.7 g/L yeast nitrogen base without amino acid (Difco Laboratories, MI, USA) with appropriate supplement additions of 20 g/L glucose and necessary amino acids) at 30℃ for 18 h.
Site-directed mutagenesis
Overlap extension PCR was used to generate site-directed mutagenesis mutant (40). The N-linked glycosylation site, Asn124, was mutated by creating codons for His (CAC) and Asp (GTC). The following mutant primer combinations were used (underline indicates the mutated site). For the N124H, 5'-TTTACAGATCATCAAGGAAG-3' and 5'-GCCGGTGAAGTGCTTCAACGG-3' were used to generate an upstream fragment (94℃ 1 min; 52℃ 30 s; and 72℃ 1 min), and 5'-GTGTGGAATTGTGAGCGGAT-3' and 5'-CCGTTGAAGCACTTCAC CGGC-3' were used to generate a downstream fragment. For the N124D, 5'-TTTACAGATCATCAAGGAAG-3' and 5'-CCGT TGAAGACTTCACCGGC-3' were used to generate an upstream fragment, and 5'-GTGTGGAATTGTGAGCGGAT-3' and 5'-GCCGGTGAAGTCCTTCAACGG-3' were used to generate a downstream fragment. The two fragments were then purified and fused without primers for 8 cycles. The nested primers 5'-GAAGTTCGGAATTCGGCA-3' and 5'-ACCATGATTACGCCAAGC-3' were added to the reaction system to obtain the mutant PCR product. The mutant PCR products were digested with EcoR I and Xho I and inserted into pAJ401, which was digested previously with the same enzyme to yield expression vectors of pAJ401-cel5A-N124D and pAJ401-cel5A-N124H. These vectors were then transformed into S. cerevisiae H158 according to the LiAc/ssDNA/PEG method previously described (41).
Purification of recombinant enzyme
The recombinant enzymes were purified according to a method described previously (19). Briefly, the concentrated supernatant of yeast culture broth was loaded onto a CM-Sepharose FF column (Amersham, UK), which is balanced with 20 mM acetate buffer at pH 3.8. The bounded proteins were eluted with a linear salt gradient of 0 M to 1 M NaCl. The elution was then desalted and buffer-exchanged (pH 6.0) using a Sephadex G20 column and separated by a Sephadex G75 column according to the manufacturer's instructions (Amersham, UK). Protein concentration was assayed following the method of Lowry (42). The molecular mass and purity of recombinant Cel5A were analyzed using the Image Quant TL software (Amersham Biosciences Corp., USA).
SDS-PAGE and zymograms
SDS-PAGE analysis was performed to detect the purified protein. Zymograms were prepared according to the general procedure described by Medve (43). The 12% separating gel contained 0.15% sodium carboxymethyl cellulose (CMC-Na). The gel was then washed in a solution containing 0.5 M NaAc and 25% isopropanol to remove the sodium dodecyl sulfate. The proteins were renaturated in a 50 mM acetate buffer (pH 5.0) containing 5 mM mercaptoethanol by rocking the gel overnight at 4℃. The gel was transferred to a 50 mM NaAc-HAc buffer (pH 5.0) and incubated at 50℃ for 2 h. The gel was stained in 0.2% Congo Red for 1 h and destained in 1 M NaCl. Clear bands against a red background indicated CMC-Na hydolysis.
Activity measurement and effect of temperature on enzyme activity
The activities of the purified recombinant enzymes were determined by measuring the increase in reducing sugar liberated from 0.5% of CMC-Na in 50 mM acetate (pH 5.0) using the dinitrosalicylic acid reagent method (44). The hydrolysis time of CMC-Na was 30 min at 50℃. Effect of temperature on enzyme activity was tested by incubating the enzymes at 70℃ in 50 mM acetate buffer at pH 5.0 and pH 4.0, followed by measuring the residual activity at different incubation time points.
CD spectroscopy
Far-UV CD spectra of the purified recombinant enzymes were measured in 10 mM acetate buffer (pH 5.0) at 25℃ with a Jasco J-810 spectropolarimeter (Hachioji City, Japan) with a 3 mm-path-length cuvette and a protein concentration of 0.1 mg/ml. The protein far-UV spectra were recorded over a wavelength range of 190 nm to 240 nm. For each measurement, a total of six spectra were collected, averaged, and corrected by subtraction of a buffer blank.
Acknowledgments
We thank the grants from National Basic Research Program of China (Grant no. 2011CB707403) and National Natural Science Foundation, China (Grant No. 31030001 and 31370086).
References
- 1.Kubicek C. P. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J. Biotechnol. (2013);163:133–142. doi: 10.1016/j.jbiotec.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynd L. R., Weimer P. J., van Zyl W. H., Pretorius I. S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. (2002);66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Béguin P., Aubert J. P. The biological degradation of cellulose. FEMS Microbiol. Rev. (1994);13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 4.Hui J. P. M., White T. C., Thibault P. Identification of glycan structure and glycosylation sites in cellobiohydrolase II and endoglucanases I and II from Trichoderma reesei. Glycobiology. (2002);12:837–849. doi: 10.1093/glycob/cwf089. [DOI] [PubMed] [Google Scholar]
- 5.Beckham G. T., Dai Z., Matthews J. F., Momany M., Payne C. M., Adney W. S., Baker S. E., Himmel M. E. Harnessing glycosylation to improve cellulase activity. Curr. Opin. Biotechnol. (2012);23:338–345. doi: 10.1016/j.copbio.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 6.Zhou F., Olman V., Xu Y. Large-scale analyses of glycosylation in cellulases. Genomics Proteomics Bioinformatics. (2009);7:194–199. doi: 10.1016/S1672-0229(08)60049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia D., Wei Y., Zhang G., Zhao Q., Zhang Y., Xiang Z., Lu C. cDNA cloning, expression, and enzymatic activity of a novel endogenous cellulase from the beetle Batocera horsfieldi. Gene. (2013);514:62–68. doi: 10.1016/j.gene.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X. H., Wang W, Wang F. Q., Wei D. Z. A comparative study of β-1, 4-endoglucanase (possessing b-1, 4-exoglucanase activity) from Bacillus subtilis LH expressed in Pichia pastoris GS115 and Escherichia coli Rosetta (DE3) Bioresour. Technol. (2012);110:539–545. doi: 10.1016/j.biortech.2011.12.086. [DOI] [PubMed] [Google Scholar]
- 9.Jeoh T., Michener W., Himmel M. E., Decker S. R., Adney W. S. Implications of cellobiohydrolase glycosylation for use in biomass conversion. Biotechnol Biofuels. (2008);1:10. doi: 10.1186/1754-6834-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voutilainen S. P., Murray P. G., Tuohy M. G., Koivula A. Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng. Des. Sel. (2010);23:69–79. doi: 10.1093/protein/gzp072. [DOI] [PubMed] [Google Scholar]
- 11.Gao L., Gao F., Wang L., Geng C., Chi L., Zhao J., Qu Y. N-glycoform diversity of cellobiohydrolase I from Penicillium decumbens and synergism of nonhydrolytic glycoform in cellulose degradation. J. Biol. Chem. (2012);287:15906–15915. doi: 10.1074/jbc.M111.332890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun H., Kieselbach T., Jönsson L. J. Enzyme production by filamentous fungi: analysis of the secretome of Trichoderma reesei grown on unconventional carbon source. Microb. Cell. Fact. (2011);10:68. doi: 10.1186/1475-2859-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medve J., Karlsson J., Lee D., Tjerneld F. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. (1998);59:621–634. [PubMed] [Google Scholar]
- 14.Suominen P. L., Mäntylä A. L., Karhunen T., Hakola S., Nevalainen H. High frequency one-step gene replacement in Trichoderma reesei. II. Effects of deletions of individual cellulase genes. Mol. Gen. Genet. (1993);241:523–30. doi: 10.1007/BF00279894. [DOI] [PubMed] [Google Scholar]
- 15.Herpoël-Gimbert I., Margeot A., Dolla A., Jan G., Mollé D., Lignon S., Mathis H., Sigoillot J. C, Monot F., Asther M. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol Biofuels. (2008);1:18. doi: 10.1186/1754-6834-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckham G. T., Bomble Y. J., Matthews J. F, Taylor C. B., Resch M. G., Yarbrough J. M., Decker S. R., Bu L., Zhao X., McCabe C., Wohlert J., Bergenstråhle M., Brady J. W., Adney W. S., Himmel M. E., Crowley M. F. The O-glycosylated linker from the Trichoderma reesei Family 7 cellulase is a flexible, disordered protein. Biophys. J. (2010);99:3773–3781. doi: 10.1016/j.bpj.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J. P., Lanthier P, White T. C., McHugh S. G., Yaguchi M., Roy R., Thibault P. Characterization of cellobiohydrolase I (Cel7A) glycoforms from extracts of Trichoderma reesei using capillary isoelectric focusing and electrospray mass spectrometry. J. Chromatogr. B Biomed. Sci. (2001);752:349–368. doi: 10.1016/S0378-4347(00)00373-X. [DOI] [PubMed] [Google Scholar]
- 18.Stals I., Sandra K., Geysens S., Contreras R., Van Beeumen J., Claeyssens M. Factors influencing glycosylation of Trichoderma reesei cellulases. I: Postsecretorial changes of the O- and N-glycosylation pattern of Cel7A. Glycobiology. (2004);14:713–724. doi: 10.1093/glycob/cwh080. [DOI] [PubMed] [Google Scholar]
- 19.Qin Y., Wei X., Liu X., Wang T., Qu Y. Purification and characterization of recombinant endoglucanase of Trichoderma reesei expressed in Saccharomyces cerevisiae with higher glycosylation and stability. Protein Expr. Purif. (2008);58:162–167. doi: 10.1016/j.pep.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Lo-Leggio L., Larsen S. The 1. 62 A structure of Thermoascus aurantiacus endoglucanase: completing the structural picture of subfamilies in glycoside hydrolase family 5. FEBS Lett. (2002);523:103–108. doi: 10.1016/S0014-5793(02)02954-X. [DOI] [PubMed] [Google Scholar]
- 21.Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. (1969);8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 22.Holzwarth G., Doty P. The ultraviolet circular dichroism of polypeptides. J. Am. Chem. Soc. (1965);87:218–228. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- 23.Tarentino A. L, Plummer T. H., Jr., Maley F. The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. J. Biol. Chem. (1974);249:818–824. [PubMed] [Google Scholar]
- 24.Helenius A, Trombetta E. S., Herbert D. N., Simons J. F. Calnexin, calreticulin and the folding of glycoproteins. Trends. Cell Biol. (1997);7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- 25.Mansell T. J., Guarino C., Delisa M. P. Engineered genetic selection links in vivo protein folding and stability with asparagine-linked glycosylation. Biotechnol. J. (2013) doi: 10.1002/biot.201300026.. [DOI] [PubMed] [Google Scholar]
- 26.Kaushik S., Mohanty D., Surolia A. Molecular dynamics simulations on pars intercerebralis major peptide-C (PMP-C) reveal the role of glycosylation and disulfide bonds in its enhanced structural stability and function. J. Biomol. Struct. Dyn. (2012);29:905–920. doi: 10.1080/073911012010525026. [DOI] [PubMed] [Google Scholar]
- 27.Schwede T., Kopp J., Guex N., Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. (2003);31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macarrón R., Acebal C., Castillón M. P., Domínguez J. M., de la Mata I., Pettersson G., Tomme P., Claeyssens M. Mode of action of endoglucanase III from Trichoderma reesei. Biochem. J. (1993);289:867–873. doi: 10.1042/bj2890867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macarron R., van Beeumen J., Henrissat B., de la Mata I., Claeyssens M. Identification of an essential glutamate residue in the active site of endoglucanase III from Trichoderma reesei. FEBS Lett. (1993);316:137–140. doi: 10.1016/0014-5793(93)81202-B. [DOI] [PubMed] [Google Scholar]
- 30.Qin Y., Wei X., Song X., Qu Y. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. J. Biotechnol. (2008);135:190–195. doi: 10.1016/j.jbiotec.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Benoit I., Asther M., Sulzenbacher G., Record E., Marmuse L., Parsiegla G., Gimbert I., Asther M, Bignon C. Respective importance of protein folding and glycosylation in the thermal stability of recombinant feruloyl esterase A. FEBS Lett. (2006);580:5815–5821. doi: 10.1016/j.febslet.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 32.Garg N., Bieler N., Kenzom T., Chhabra M., Ansorge-Schumacher M., Mishra S. Cloning, sequence analysis, expression of Cyathus bulleri laccase in Pichia pastoris and characterization of recombinant laccase. BMC Biotechnol. (2012);12:75. doi: 10.1186/1472-6750-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price J. L., Powers D. L., Powers E. T., Kelly J. W. Glycosylation of the enhanced aromatic sequon is similarly stabilizing in three distinct reverse turn contexts. Proc. Natl. Acad. Sci. U. S. A. (2011);108:14127–14132. doi: 10.1073/pnas.1105880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shental-Bechor D., Levy Y. Effect of glycosylation on protein folding: A close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U. S. A. (2008);105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tull D., Gottschalk T. E., Svendsen I., Kramhøft B., Phillipson B. A, Bisgård-Frantzen H., Olsen O., Svensson B. Extensive N-glycosylation reduces the thermal stability of a recombinant alkalophilic bacillus alpha- amylase produced in Pichia pastoris. Protein Expr. Purif. (2001);21:13–23. doi: 10.1006/prep.2000.1348. [DOI] [PubMed] [Google Scholar]
- 36.Chen R., Zhou Z., Cao Y., Bai Y., Yao B. High yield expression of an AHL-lactonase from Bacillus sp. B546 in Pichia pastoris and its application to reduce Aeromonas hydrophila mortality in aquaculture. Microb. Cell. Fact. (2010);9:39. doi: 10.1186/1475-2859-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiraki K., Sakiyama F. Histidine 210 mutant of a trypsin-type Achromobacter protease I shows broad optimum pH range. J. Biosci. Bioeng. (2002);93:331–333. doi: 10.1016/S1389-1723(02)80038-X. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz A., Brecker L., Nidetzky B. Probing the active site of Corynebacterium callunae starch phosphorylase through the characterization of wild-type and His334-->Gly mutant enzymes. FEBS J. (2007);274:5105–5115. doi: 10.1111/j.1742-4658.2007.06030.x. [DOI] [PubMed] [Google Scholar]
- 39.Fang T. Y., Ford C. Protein engineering of Aspergillus awamori glucoamylase to increase its pH optimum. Protein Eng. (1998);11:383–388. doi: 10.1093/protein/11.5.383. [DOI] [PubMed] [Google Scholar]
- 40.Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. (1988);16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gietz R. D., Woods R. A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. (2006);313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 42.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. (1951);193:265–275. [PubMed] [Google Scholar]
- 43.Medve J., Lee D., Tjerneld F. Ion-exchange chromatographic purification and quantitative analysis of Trichoderma reesei cellulase cellobiohydrolase I, II and endoglucanase by fast protein liquid chromatography. J. Chromatogr. A. (1998);808:153–156. doi: 10.1016/S0021-9673(98)00132-0. [DOI] [Google Scholar]
- 44.Meinke A., Damude H. G., Tomme P., Kwan E., Kilburn D. G., Miller R. C., Jr, Warren R. A., Gilkes N. R. Enhancement of the endo-beta-1,4-glucanase activity of an exocellobiohydrolase by deletion of a surface loop. J. Biol. Chem. (1995);270:4383–4386. doi: 10.1074/jbc.270.9.4383. [DOI] [PubMed] [Google Scholar]


