Abstract
Histone acetylation/deacetylation has been known to be associated with the transcriptional regulation of various genes. The role of histone deacetylase-3 in the expression regulation of MDR1 was investigated. The expression level of HDAC3 showed an inverse relationship with the expression level of MDR1. Wild-type HDAC3, but not catalytic mutant HDAC3S424A, negatively regulated the expression of MDR1. Wild-type HDAC3, but not catalytic mutant HDAC3S424A, showed binding to the promoter sequences of HDAC3. HDAC3 regulated the expression level, and the binding of Ac-H3K9/14 and Ac-H4K16 around the MDR1 promoter sequences. The nuclear localization signal domain of HDAC3 was necessary, and sufficient for the binding of HDAC3 to the MDR1 promoter sequences and for conferring sensitivity to microtubule-targeting drugs. [BMB Reports 2014; 47(6): 342-347]
Keywords: Anti-cancer drug-resistance, Expression regulation, Histone deacetylase-3, MDR1, Nuclear localization
INTRODUCTION
Histone deactylase-3 (HDAC3) is ubiquitously expressed and conserved in a wide range of species (1) HDAC3 has been shown to form large corepressor complexes containing N-CoR/SMRT (2). HDAC3 regulates the JNK pathway (3), MAPK activation (4), and apoptosis (5). HDAC3 represses CREB3-mediated transcription, and the migration of metastatic breast cancer cells (6).
A phase I trial revealed that albumin-bound paclitaxel shows encouraging activity against advanced metastatic melanomas (7). Resistance to taxol, a microtubule-targeting drug, in hepatoma cells is related to JNK activation (8). The inhibition of MAPK enhances taxol-induced apoptosis (9).
Taxol-resistance is correlated with the expression level of MDR1 (10). Elevated levels of histone acetylation in the MDR1 promoter in drug-resistant cancer cells have been described (11).
In this study, the effect of HDAC3 on the expression of MDR1 and acetylated histones was examined. The necessity of HDAC3 activity for regulating the expression of MDR1 was also examined. The domain of HDAC3 that was necessary for exerting regulation on the expression of MDR1, and for the binding to the promoter sequences of MDR1, was identified.
RESULTS AND DISCUSSION
HDAC3 regulates the expression of MDR1 by affecting histone acetylation
Apicidin, an inhibitor of HDAC, induces anti-cancer drug-resistance via the up-regulation of MDR protein (12). The deletion of HDAC3 increases the acetylation of histone H3 at Lys9/14, and histone H4 at Lys5/12 (13). The expression level of acetylated H3Lys9 is elevated 100 fold in the promoter of the MDR1 gene, in drug-resistant breast cancer cell lines, compared to drug-sensitive cell lines (14). In this study, we found lower expression of HDAC3, among HDAC(s), in various cancer cell lines naturally resistant (HpeG2, Hpe3B and WM266-4), or made resistant to celastrol (SNU387R and Malme3MR), taxol (SNU387R-Taxol and Malme3MR-Taxol) or vinblastine (SNU387R-VBL), than anti-cancer drug-sensitive cancer cell lines such as SNU387 and Malme3M cells (Fig. 1A). In these anti-cancer drug-resistant cancer cell lines, the expression level of HDAC3 showed an inverse relationship with the expression level of MDR1 (Fig. 1A). This led to the hypothesis that HDAC3 would exert a negative regulation on the expression of MDR1. First, the effect of HDAC3 on histone acetylation/deacetylation to regulate the expression of MDR1, was examined. The MDR1 gene promoter contains the binding sites for various transcription factors such as p53, ETS, and AP1 (Fig. 1B). However, the binding of HDAC(s) and modified histones around the MDR1 promoter in hepatoma or melanoma cells remains unknown. The down-regulation of HDAC3 induced the expression of MDR1 (Fig. 1C). Malme3MR/ HDAC3-Flag and SNU387R/HDAC3-Flag cells stably expressing HDAC3 showed lower expression of MDR1 than their counterparts (Fig. 1C). The down-regulation of MDR1 did not affect the expression of HDAC3 (Fig. 1D). The down-regulation of HDAC3 induces the expression of Ac-H3K9/14 and Ac-H4K16 (Fig. 1D). However, the down-regulation of HDAC3 did not affect expression of histone H3, or histone H4 (Fig. 1D). HDAC3 exerted a negative regulation on the expression of Ac-H3K9/14 and Ac-H4K16 (Fig. 1E). However, over-expression of HDAC3 did not affect the expression of histone H3 or histone H4 (Fig. 1E). The down-regulation of HDAC3 induced the binding of Ac-H3K9/14 to a site of the MDR1 promoter sequences and the binding of Ac-H3K16 to site 2 of the MDR1 promoter sequences, in SNU387 cells (Fig. 1F). The down-regulation of HDAC3 also induced the binding of Ac-H3K9/14 and Ac-H3K16 in Malme3M cells (Fig. 1F). Acetyl-H3 and Acetyl-H4 show binding to the MDR1 gene proximal promoter sequences (-116 to -11), in MCF7 cells made resistant to adriamycin (15).
Fig. 1. HDAC3 regulates the expression of MDR1, Ac-H3K9/14, and Ac-H4K16, and the binding of modified histones to MDR1 promoter sequences. (A) Cell lyastes were subjected to Western blot analysis. (B) Human MDR1 gene proximal promoter sequences. Numbers indicate their positions relative to the transcription initiation site (+1). Two sets of primers, designated as P1 and P2, were used for amplification of the proximal MDR1 promoter, respectively. (C) 48 h after transfection with the indicated siRNA, Western blot was performed. Lysates from SNU387R-HDAC3-Flag or Malme3MR-HDAC3-Flag cells were also subjected to Western blot. *denotes exogenous HDAC3. (D) 48 h after transfection, Western blot was performed. (E) 48 h after transfection control vector or Flag-tagged HDAC3 wild-type construct, Western blot was performed. (F) 48 h after transfection, ChIP assays were performed.
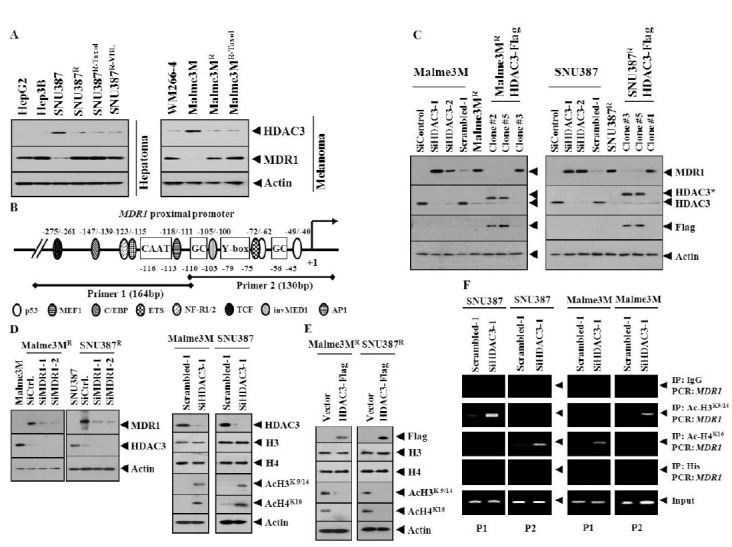
Taken together, these results suggest that HDAC3 regulates the expression of MDR1, by modulating the expression of the modified histones, and the binding of these modified histones to the MDR1 promoter sequences. It would be necessary to identify more modified histones that are regulated by HDAC3.
HDAC3 activity is necessary for the binding of modified histones to the MDR1 promoter sequences
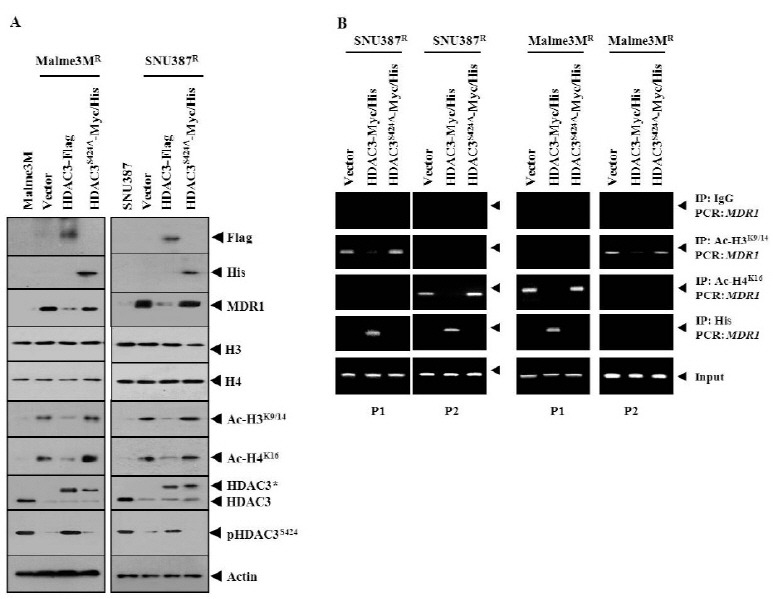
Wild-type HDAC3, but not HDAC3S424A, decreased the expression of MDR1, Ac-H3K9/14, and Ac-H4K16 (Fig. 2A). This suggests that HDAC3 activity is necessary for the expression regulation of Ac-H3K9/14 and Ac-H4K16. HDAC3 did not affect the expression of histone H3 or histone H4 (Fig. 2A). HDAC3Ser424 lacks histone deacetylase activity (16). The protein kinase CK2 phosphoacceptor site in HDAC3 is at Ser424 (16). HDAC3 activity is regulated by interaction with protein serine/threonine phosphatase 4 (PP4c) (16). The expression level of phosphoHDAC3Ser424 was correlated with the expression level of HDAC3 (Fig. 2A). Wild-type HDAC3, but not HDAC3S424A, exerted negative effects on the binding of Ac-H3K9/14 to site 1 of the MDR1 promoter sequences in SNU387R cells, and the binding of Ac-H4K16 to site 2 of the MDR1 promoter sequences in SNU387R cells (Fig. 2B). Wild-type HDAC3, but not HDAC3S424A, exerted negative effects on the binding of Ac-H3K9/14 to site 2 of the MDR1 promoter sequences in Malme3MR cells, and the binding of Ac-H4K16 to site 1 of the MDR1 promoter sequences in Malme3MR cells (Fig. 2B). Wild-type HDAC3, but not HDAC3S424A, showed binding to sites 1 and 2 of the MDR1 promoter sequences in SNU387R cells and binding to site 1 of the MDR1 promoter sequences in Malme3MR cells(Fig. 2B), suggesting an essential role of HDAC3 activity in the expression regulation of MDR1. These results suggest that the expression regulation of MDR1 by HDAC3 involves the regulated binding of modified histones to the MDR1 promoter sequences. For better understanding of the mechanism of the expression regulation of MDR1 by HDAC3, it would be necessary to identify more modified histones that are regulated by HDAC3. MDR1 promoter sequences contain a binding site for p53. The decreased expression of p53 in drug-resistant cancer cell lines was previously reported (17). It would be interesting to examine the possible interaction between HDAC3 and p53, for the expression regulation of MDR1. A target scan analysis predicted the binding of miRNA-326 to the 3' UTR of HDAC3. It would be interesting to examine the effect of miR-326 on the expression of HDAC3 and MDR1. It would also be interesting to identify miRNAs that are regulated by HDAC3, for better understanding of the mechanism of the expression regulation of MDR1.
Fig. 2. HDAC3 activity is necessary for the binding of modified histones to the MDR1 promoter sequences. (A) 48 h after transfection with the indicated construct to SNU387R or Mlame3MR cells, Western blot was performed. Cell lysates isolated from Malme3M or SNU387 cells were also subjected to Western blot analysis. (B) 48 h after transfection, cell lysates were immunoprecipitated with the indicated antibody (2 μg/ml), followed by ChIP assays.
The nuclear localization signal domain of HDAC3 is necessary and sufficient for conferring sensitivity to microtubule-targeting drugs
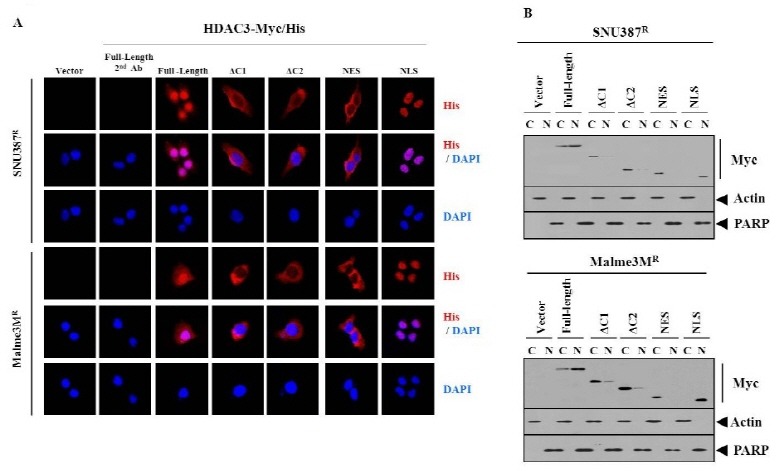
HDAC3 consists of functional domains, such as the oligomerization, nuclear export signal sequence and nuclear localization signal sequence (18). There have been no reports concerning the domain of HDAC3 that confer sensitivity to microtubule-targeting drugs. Therefore, a series of HDAC3 deletion constructs were made, to determine the functional domain of HDAC3 that confers sensitivity to microtubule-targeting drugs. The full-length HDAC3 (HDAC3-FL) decreased the expression of MDR1 in Malme3MR cells (Fig. 3A). HDAC3-△C1 (1-313 of HDAC3) or HDAC3-△C2 (1-180 of HDAC3) did not affect the expression of MDR1 (Fig. 3A). HDAC3-NES (180- 313 of HDAC3), which possesses a nuclear export signal, did not affect the expression of MDR1 (Fig. 3A). HDAC3-NLS (nuclear localization signal sequence, 313-428 of HDAC3), which possesses Ser424, decreased the expression of MDR1 (Fig. 3A). HDAC3-NLS, but not other deletion constructs, decreased expression of Ac-H3K9/14 and Ac-H4K16 (Fig. 3A). The full-length HDAC3 or HDAC3 deletion constructs did not affect the expression of histone H3 or histone H4 (Fig. 3A). HDAC3-FL and HDAC3-NLS, but not other deletion constructs, conferred sensitivity to microtubule-targeting drugs (Fig. 3B). HDAC3-NLS conferred apoptotic effects against celastrol and taxol (Fig. 3C). HDAC3-NLS, among HDAC3 deletion constructs, showed binding to the MDR1 promoter sequences (Fig. 3D). It is reasonable that nuclear localization of HDAC3, is necessary for the expression regulation of MDR1 by HDAC3. Immunofluorescence staining revealed the localization of the full-length HDAC3, in both the nucleus and cytosol (Fig. 4A). HDAC3, unlike HDAC1/2, is present in both the nucleus and cytosol of DT40 chicken B cells (19). The localization of HDAC3-△C1 and HDAC3-△C2 was mostly cytosolic (Fig. 4A). The localization of HDAC3-NES was also mostly cytosolic (Fig. 4A). HDAC3-NLS showed exclusively nuclear localization (Fig. 4A). As for the localization of these HDAC3 constructs, a cellular fractionation study showed the same results as immunofluorescence staining (Fig. 4B). Taken together, these results suggest that the NLS domain of HDAC3 may be necessary and sufficient for conferring sensitivity to microtubule-targeting drugs, and also for direct regulation of MDR1. It would be necessary to identify sequences within the NLS domain that are necessary for the expression regulation of MDR1 and modified histones.
Fig. 3. The nuclear localization domain of HDAC3, which encompasses Ser424, is necessary and sufficient for conferring sensitivity to microtubule-targeting drugs. (A) HDAC3-Myc/His6 serial deletion constructs. FL denotes full-length. NES denotes nuclear export signal domain. Numbers in parentheses represent amino acid residues of HDAC3. NLS denotes nuclear localization signal domain. 48 h after transfection, Western blot was performed. (B) Malme3MR cell line was transfected with the indicated construct. The next day, cells were treated with various concentrations of the indicated drug for 24 h, followed by MTT assays. Comparison was made between SNU387R or Malme3MR cells transfected with the control vector, and the same cells transfected with HDAC3-FL. *P < 0.05; **P < 0.005. Comparison was also made between SNU387R or Malme3MR cells transfected with control vector, and the same cells transfected with HDAC3-NLS. #P < 0.05; ##P < 0.005. (C) Malme3MR cell line was transfected with the indicated construct. The next day, cells were treated with celastrol (1 μM) or taxol (1 μM) for 24 h, followed by Western blot. C-PARP denotes cleaved PARP. V denotes control vector. Caspase-3* denotes pro-caspase-3 and caspase-3** denotes active caspase-3. (D) The indicated cell line was transfected with the indicated HDAC3-Myc/His construct. 48 h after transfection, ChIP assays were performed, as described. SNU387R or Malme3MR cells transfected with full-length HDAC3 were also subjected to ChIP assays, employing isotype-matched anti-IgG antibody (2 μg/ml).
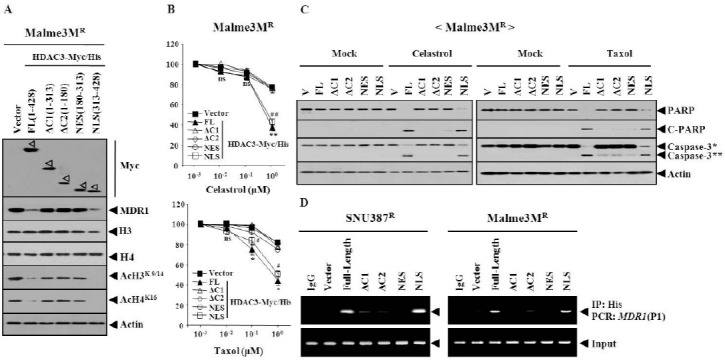
Fig. 4. The localization of HDAC3 deletion constructs. (A) The indicated cell line was transiently transfected with the indicated HDAC3-Myc/His construct. 48 h after transfection, immunofluorescence staining was performed, as described. SNU387R and Malme3MR cell lines transfected with full-length HDAC3 were also subjected to immunofluorescence staining employing only isotype-matched secondary antibody. (B) Cell lysates from each fraction were subjected to Western blot analysis.
MATERIALS AND METHODS
Cell lines and cell culture
The cancer cell lines used in this study were cultured in Dulbecco’s modified minimal essential medium (DMEM; Gibco, Gaithersburg, MD, USA), supplemented with heat-inactivated 10% fetal bovine serum (FBS, Gibco) and antibiotics, at 37℃, in a humidified incubator with a mixture of 95% air and 5% CO2.
SNU387R or Malme3MR cells stably expressing HDAC3 were generated, by co-transfection of HDAC3-Flag, along with pcDNA3.1. Stable transfectants were selected by G418 (400 μg/ml). Cancer cell lines made resistant to microtubule-targeting drugs (celastrol, taxol and vinblastine) were established by stepwise addition of the respective drug. Cells surviving drug treatment (attached fraction) were obtained, and used throughout this study. SNU387/SNU387R or Malme3M/Malme3MR cells that stably express anti-sense HDAC3 cDNA or HDAC3-Flag were selected by G418 (400 μg/ml).
Materials
Anti mouse and anti rabbit IgG-horse radish peroxidase conjugate antibodies were purchased from Pierce Company. Lipofectamin and PlusTM reagent were purchased from Invitrogen (Carlsbad, CA, USA). Bioneer (Daejeon, Korea) synthesized all primers and oligonucleotides used in this study.
Cell viability determination
The cells were assayed for their growth activity, using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT; Sigma). Viable cell number counting was carried out by trypan blue exclusion assays.
HDAC3 constructs
HDAC3S424A-Myc/His(6) expression plasmid (catalytically inactive HDAC3 mutant) was derived from pFlag-HDAC3, with the Quick-change site-directed mutagenesis kit (Stratagene). HDAC3 serial deletion mutant constructs were made, by cloning various PCR-amplified HDAC3 fragments into pcDNA3.1-Myc/His vector.
Immunofluorescence staining
SNU387R or Malme3MR cells were seeded onto glass cover slips in 24-well plates. Cells were fixed with 4% paraformaldehyde (v/v), for 10 min and then permeabilized with 0.4% triton X-100 for 10 min. Nonspecific antibody binding sites were blocked by incubation with 1% BSA in TBST, for 30 min. Cells were then incubated with primary antibody specific to His (Santa Cruz, 1:100) for 24 h, followed by washing with TBS-T, three times. Anti-rabbit IgG-Alexa Fluor 546 (for detection of His) secondary antibody (Molecular Probes) was added to cells and incubated for 1 h. Cover slips were then washed, and mounted by applying Mount solution (Biomeda, Foster City, CA, USA). Fluorescence images were acquired using a confocal laser-scanning microscope and software (Fluoview ver. 2.0), with a 40X objective (Olympus FV300, Tokyo, Japan). DAPI staining was also performed, for nuclear staining.
Transfection
All transfections were performed according to the standard procedures (20). The construction of siRNA was carried out according to the instruction manual provided by the manufacturer (Ambion, Austin, TX).
ChIP assays
Assays were performed according to the standard procedures (21). PCR was done on the phenol-chloroform-extracted DNA with specific primers of MDR1 promoter-1 [5’-AATCCAAGGC ATCAATTTCACCCTGTCTC-3’ (sense)] and [5’-AAGTGAAAT TGATGCCTTGGACCTGTCTC-3’ (antisense)]; and MDR1 promoter-2 [5’-AATTGCATACGCTAAGAGTTCCCTGTCTC-3’ (sense)] and [5’-AAGAACTCTTAGCGTATGCAACCTGTCTC-3’ (antisense)] sequences were used.
Preparation of SiRNA duplexes
The construction of SiRNA was carried out according to the instruction manual provided by the manufacturer (Ambion, Austin, TX).
Statistical analysis
All data were expressed as a mean value ± standard deviation (S.D.), and differences between groups were analyzed using Student’s t-test. Mean values were considered significantly different, when P < 0.05.
Acknowledgments
This work was supported by a grant from the National Research Foundation (2012H1B8A2025495, 2014R1A2A2A01002448, C1008749-01-01). This study was supported by a grant from BK21 plus program. This study was also supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1320160). This work was also supported by a grant from Kangwon National University (120140305).
References
- 1.Mahlknecht U., Emiliani S., Najfeld V., Young S., E.Verdin E. Genomic organization and chromosomal localization of the human histone deacetylase 3 gene. Genomics. (1999);56:197–202. doi: 10.1006/geno.1998.5645. [DOI] [PubMed] [Google Scholar]
- 2.Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. (2000);19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Kalkum M., Chait B. T., Roeder R. G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell. (2002);9:611–623. doi: 10.1016/S1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 4.Mahlknecht U., Will J., Varin A., Hoelzer D., Herbein G. Histone deacetylase 3, a class I histone deacetylase, suppresses MAPK11-mediated activating transcription factor-2 activation and represses TNF gene expression. J. Immunol. (2004);173:3979–3990. doi: 10.4049/jimmunol.173.6.3979. [DOI] [PubMed] [Google Scholar]
- 5.Bardai F. H., D S. R. Selective toxicity by HDAC3 in neurons: regulation by Akt and GSK3beta. J. Neurosci. (2011);31:1746–1751. doi: 10.1523/JNEUROSCI.5704-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H. C., Choi K. C., Choi H. K., Kang H. B., Kim M. J., Lee Y. H., Lee O. H., Lee J., Kim Y. J., Jun W., Jeong J. W., Yoon H. G. HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells. Cell. Mol. Life Sci. (2010);67:3499–3510. doi: 10.1007/s00018-010-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott P. A., Chang J., Madden K., Kannan R., Muren C., Escano C., Cheng X., Shao Y., Mendoza S., Gandhi A., Liebes L., Pavlick A. C. Oblimersen in combination with temozolomide and albumin-bound paclitaxel in patients with advanced melanoma: a phase I trial. Cancer Chemother. Pharmacol. (2013);71:183–191. doi: 10.1007/s00280-012-1995-7. [DOI] [PubMed] [Google Scholar]
- 8.Chae S., Kim Y. B., Lee J. S., Cho H. Resistance to paclitaxel in hepatoma cells is related to static JNK activation and prohibition into entry of mitosis. Am. J. Physiol. Gastrointest. Liver Physiol. (2012);302:1016–1024. doi: 10.1152/ajpgi.00449.2011. [DOI] [PubMed] [Google Scholar]
- 9.Xu R., Sato N., Yanai K., Akiyoshi T., Nagai S., Wada J., Koga K., Mibu R., Nakamura M., Katano M. Enhancement of paclitaxel-induced apoptosis by inhibition of mitogen-activated protein kinase pathway in colon cancer cells. Anticancer Res. (2009);29:261–270. [PubMed] [Google Scholar]
- 10.Mechetner E., Kyshtoobayeva A., Zonis S., Kim H., Stroup R., Garcia R., Parker R. J., Fruehauf J. P. Levels of multidrug resistance (MDR1) P-glycoprotein expression by human breast cancer correlate with in vitro resistance to taxol and doxorubicin. Clin. Cancer Res. (1998);4:389–398. [PubMed] [Google Scholar]
- 11.Yatouji S., El-Khoury V., Trentesaux C., Trussardi-Regnier A., Benabid R., Bontems F., Dufer J. Differential modulation of nuclear texture, histone acetylation, and MDR1 gene expression in human drug-sensitive and -resistant OV1 cell lines. Int. J. Oncol. (2007);30:1003–1009. [PubMed] [Google Scholar]
- 12.Kim Y. K., Kim N. H., Hwang J. W., Song Y. J., Park Y. S., Seo D. W., Lee H. Y., Choi W. S., Han J. W., Kim S. N. Histone deacetylase inhibitor apicidin-mediated drug resistance: involvement of P-glycoprotein. Biochem. Biophys. Res. Commun. (2008);368:959–964. doi: 10.1016/j.bbrc.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 13.To K. K., Polgar O., Huff L. M., Morisaki K., Bates S. E. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol. Cancer Res. (2008);6:151–164. doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaskara S., Knutson S. K., Jiang G., Chandrasekharan M. B., Wilson A. J., Zheng S., Yenamandra A., Locke K., Yuan J. L., Bonine-Summers A. R., Wells C. E., Kaiser J. F., Washington M. K., Zhao Z., Wagner F. F., Sun Z. W., Xia F., Holson E. B., Khabele D., Hiebert S. W. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. (2010);18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toth M., Boros L. M., Balint M. Elevated level of lysine 9-acetylated histone H3 at the MDR1 promoter in multi-drug resistant cells. Cancer Science. (2012);103:659–669. doi: 10.1111/j.1349-7006.2012.02215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Ozawa Y., Lee H., Wen Y. D., Tan T. H., Wadzinski B. E., Seto E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. (2005);19:827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y., Park H., Park D., Lee Y. S., Choe J., Hahn J. H., Lee H., Kim Y. M., Jeoung D. Cancer/testis antigen CAGE exerts negative regulation on p53 expression through HDAC2 and confers resistance to anti-cancer drugs. J. Biol. Chem. (2010);285:25957–25968. doi: 10.1074/jbc.M109.095950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W. M., Tsai S. C., Wen Y. D., Fejer G., Seto E. Functional domains of histone deacetylase-3. J. Biol. Chem. (2002);277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 19.Takami Y., Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J. Biol. Chem. (2000);275:16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- 20.Zhou C., Shen Q., Xue J., Ji C., Chen J. Overexpression of TTRAP inhibits cell growth and induces apoptosis in osteosarcoma cells. BMB Rep. (2013);46:113–118. doi: 10.5483/BMBRep.2013.46.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Back S. S., Kim J., Choi D., Lee E. S., Choi S. Y., Han K. Cooperative transcriptional activation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 genes by nuclear receptors including Liver-X-Receptor. BMB Rep. (2013);46:322–327. doi: 10.5483/BMBRep.2013.46.6.246. [DOI] [PMC free article] [PubMed] [Google Scholar]


