Abstract
A novel form of ovomacroglobulin/ovostatin (OVOS2) predicted from EST data was previously identified in the chicken ovarian cancer model using a mass spectrometry-based shotgun label-free proteomics strategy. The quantitative label-free data from plasma showed a significant increase over time with the spontaneous onset and progression of ovarian cancer making it a potential protein biomarker for further study. Two other proteins of interest identified from this initial study included vitellogenin-1(Vit-1), a lipid-transport protein tied to egg production, and transthyretin (TTR), a retinol binding transport protein currently used in the clinical management of ovarian cancer. A multiplexed protein cleavage isotope dilution mass spectrometry (PC-IDMS) assay was developed to quantify OVOS2, Vit-1, and TTR by selected reaction monitoring (SRM). A total of 6 stable isotope labeled (SIL) peptide standards were used in the assay with three tryptic peptides from OVOS2, one for Vit-1, and two for TTR. The assay was developed for use with un-depleted raw plasma combined with the filter assisted sample preparation (FASP) method and its use was also demonstrated for matched ovary tissue samples. The PC-IDMS data for the two TTR peptides did not correlate with each other with more than a 10-fold difference in concentration for all 5 timepoints measured. The PC-IDMS data from the longitudinal plasma samples correlated well for OVOS2 and Vit-1 whereas TTR was inconclusive. Interestingly, the absolute amount for one of the OVOS2 SIL peptides was 2-fold less compared with the other two SIL peptides. These data illustrate the successes and challenges of qualifying quantitative levels of proteins from an in-gel digestion sample preparation followed by LC-MS/MS (GeLC) label-free discovery-based approach to a targeted SRM-based quantitative assay in plasma and tissues.
INTRODUCTION
The predictive value of current early-stage detection strategies for epithelial ovarian cancer (EOC) remains insufficient for population-wide screens.1 Identifying more predictive early-stage EOC molecular markers have the potential to significantly improve 5-year survival rates from ~40% to better than 90%. Mass spectrometry (MS)-based proteomics is a commonly used approach to identify candidate protein biomarkers for disease diagnosis. Several candidate biomarkers have been identified using these approaches and their potential as diagnostic markers are being evaluated to varying degrees using targeted assays.2 However, to date none of these markers have exceeded the performance of existing EOC markers such as CA-125, OVX-1, HE-4 either alone, in panels, or in conjunction with imaging techniques (e.g., transvaginal ultrasound).3–7
One approach to improving the predictive value of existing EOC biomarkers is to measure them serially in retrospective studies where longitudinal plasma samples exist. Skates and coworkers have published several papers introducing and evaluating a screening algorithm for CA-125 that takes into account serial values in pre-EOC women.8–11 McIntosh and coworkers have also contributed to developing a method for screening patients for CA-125 as a function of time and reported improvement in the specificity and sensitivity of CA-125 measurements.12 These studies build on earlier concepts put forth by R.J. Williams in the 1950 s13, 14 and quantitatively expressed by E. K. Harris as the “index of individuality” which provided a metric comparing intra-individual to inter-individual biological variability.15 Since many clinically important analytes, including tumor markers CA-125, CEA, CA-19, and TPA, circulate and fluctuate at levels specific to individuals, these studies underscore the potential value of carrying out biomarker discovery and validation with longitudinal rather than single time point samples.16
The chicken is an emerging experimental model for biomarker discovery that develops EOC spontaneously after 2 years of age17 and presents several analogous pathophysiological and molecular characteristics relative to humans18–26 including differential expression of CA-12524, p53 mutation and over-expression of Her-2/neu in EOC tumors23, correlation of EOC prevalence with ovulation rates19 (i.e., incessant ovulation theory27), and similar histopathological features (e.g., serous, clear cell, endometriod, and mucinous).18 Furthermore, multiple milliliters of blood can be drawn from birds over time without sacrificing the animal thereby facilitating longitudinal studies. These aspects combined with the ability to control genetic, environmental, and sample collection variables make the chicken a powerful experimental system for MS-based strategies at the global and targeted level. We recently explored the intra-individual and analytical variability of the plasma proteome28 and N-linked glycome29 from healthy and EOC birds to identify candidate biomarkers. In this study, we developed a protein cleavage (PC) isotope dilution mass spectrometry (IDMS) selected reaction monitoring (SRM) assay to validate observations in the plasma proteomics study.28
PC-IDMS was initially demonstrated by Dass et al.30 in 1991 to quantify beta endorphin and later by Barr et al.31 to quantify apolipoprotein A-1 by liquid chromatography (LC) fast atom bombardment (FAB)-MS. Following enzymatic digestion of the neat protein, the peptides were quantified using stable isotope labeled (SIL) peptides as internal standards. This approach has been adapted to modern LC-MS/MS for quantifying proteins in cell culture32 and in plasma33, 34 (for more examples see reviews by Brun et al.35, Ciccimaro and Blair36, and Elschenbroich and Kislinger37). Recently, multiplexed PC-IDMS-SRM has been frequently employed to quantitatively study the expression and regulation of peptides and proteins in complex biological samples.38–41 Furthermore, the robustness and reproducibility of this method has been investigated by Addona et al..to establish the interlaboratory reproducibility of 11 plasma proteins.42
Herein we describe the development and application of an SRM-based assay for quantifying a predicted form of ovomacroglobulin or ovostatin (OVOS2), vitellogenin-1 (Vit-1), and transthyretin (TTR) in un-depleted raw plasma and tissue samples from chickens. Longitudinal plasma samples and the matched ovary tissue samples were processed using a modified filter assisted sample preparation (FASP)43 method followed by spiking with a multi-peptide SIL cocktail for absolute quantification of the three proteins. The SIL cocktail included three tryptic peptides for OVOS2, one tryptic peptide for Vit-1, and two tryptic peptides for TTR. A comparison of previous label-free quantification in the longitudinal samples28 to the resultant SRM-based quantitative assay will be discussed in detail including discrepancies in absolute abundances for some SIL peptides.
EXPERIMENTAL
Materials
Thiourea, ethylenediaminetetraacetic acid (EDTA), sodium azide, sodium dodecyl sulfate (SDS), dithiothreitol (DTT), urea, iodoacetamide (IAM), ammonium bicarbonate, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO) and used as received. Tris(hydroxymethyl)aminomethane (Tris) for buffer preparation was from BioRad (Hercules, CA). HPLC-grade acetonitrile and water were purchased from Burdick and Jackson (Muskegon, MI) and used as received. The 10 kDa molecular weight cutoff (MWCO) filters were purchased through Sigma-Aldrich from the Pall Corporation (Port Washington, NY).
Peptide Selection and Synthesis
Candidate peptides derived from OVOS2, Vit-1, and TTR were initially identified from spectral libraries generated from the previous label-free investigation from healthy and late-stage EOC birds.28 Skyline version 1.1 was used to filter candidate peptides from the spectral library and for developing the SRM-based assay.44 Signature peptide candidates were carefully evaluated in Skyline, and several factors were considered in the selection of peptides including top scoring species from experimental data, unique peptides to chicken proteome IPI version 3.74, 8-25 amino acid residues in length, no missed tryptic cleavages, minimum to no potential post translational modifications (PTMs), feasibility for solid-phase synthesis, and M, W, Q, R, terminal C, and continuous series of hydrophobic residues were avoided.
The peptides for this study (Table 1) were synthesized by the Mayo Clinic Proteomics Research Center (Rochester, MN) incorporating 13C and 15N into the heavy SIL standards. Purity of the peptides was established by direct infusion analysis on a linear trap quadrupole Fourier transform mass spectrometery (LTQ-FT) obtaining both accurate (MS) and tandem (MS/MS) mass spectra for each peptide sequence. Peptide concentration was determined by absorbance at 205 nm on a Thermo Scientific NanoDrop 2000c by the Scopes Method.45
Table 1.
A summary of information for each peptide analyzed by nLC-PC-IDMS-SRM. The corresponding International Protein Index (IPI) database identifier and mass is listed beneath the protein name. The heavy labeled amino acid in the peptides monitored for absolute quantification are in bold and underlined. The number in the peptide label corresponds to the starting amino acid location of the species in the protein. The precursor mass for the natural or endogenous species (NAT) and the SIL internal standard are all 2+ charge state parent ions. The transitions included were used to monitor protein abundance in the plasma and tissue samples with an exception of the y3 transition for OVOS2779 and OVOS2792 in which these transitions are marked with an asterisk.
| Protein Mass (Da) | Peptide | Label | NAT m/z | SIL m/z | Transition Ions | Amount SIL on column | Collision Energy (V) |
|---|---|---|---|---|---|---|---|
| OVOS2 (IPI00587419.3) 184950.5 | AVDQSVFLLQPER | OVOS2779 | 751.4041 | 754.9127 | y4, y3, b9 | 2.5 fmol | 28.0 |
| ELSAESVYYR | OVOS2792 | 608.7959 | 611.8028 | y8, y6, y3 | 2.5fmol | 22.9 | |
| AEVLSPEGNTAR | OVOS21017 | 622.3175 | 625.8261 | y8, b3, b5 | 2.5 fmol | 23.4 | |
| Vitellogenin-1 (IPI00591843) 210630.1 | NIEDLAASK | Vit918 | 480.7535 | 484.2620 | y7, b2, a2 | 50 fmol | 18.3 |
| Transthyretin (IPI00591282.3) 16309.3 | AADGTWQDFATGK | TTR58 | 684.3149 | 689.3286 | y8, y7, y3, y2 | 25 fmol | 25.6 |
| VEFDTSSYWK | TTR93 | 631.2904 | 636.3040 | y7, y5, b2, a2 | 25 fmol | 23.7 |
Plasma and Tissue Sample Preparation
The longitudinal EDTA plasma and matched tissue biorepository has been described in detail elsewhere.28 The non-depleted EDTA plasma was used as stored and the liquid nitrogen (LN2) snap-frozen chicken ovary tissues were prepared using the following protocol. The frozen tissues were immersed in 50 mM Tris-HCl pH 7.8, 8 M urea, 2 M thiourea, 10 mM EDTA, 10 mM DTT, and 0.001% sodium azide to concentration of approximately 200 μg of wet tissue per 1 mL of lysis buffer. The samples were then homogenized using an OMNI Tip Homogenizing Kit from OMNI International (Marietta, GA) for 1 minute followed by the addition of 2% (w/w) and three cycles of vortexing for 1 min followed by incubation on ice for 5 min. The samples were then centrifuged for 30 minutes at 14,000 rpm, and the clarified extract was fractionated and stored at −80°C.
The FASP digestion protocol for the non-depleted EDTA plasma and lysed tissue samples was adapted from Wisniewski et al.46 and described herein. 7 μl aliquots of un-depleted EDTA plasma samples (approximately 600 μg of total protein, Scopes method at A205) and 20 μl of matched tissue lysates were diluted to a final volume of 70 μl with 100 mM Tris-HCl pH 7.6, 100 mM DTT, 4% SDS buffer. The resulting solutions were heated for 5 min at 95°C and transferred to separate 10 kDa MWCO filters and processed in parallel. A 200 μl aliquot of freshly prepared 8 M urea in 100 mM Tris-HCl pH 8.5 was added to each MWCO filter and centrifuged 20°C and 14,000 rpm for 25 minutes. Another 200 μl aliquot of 8M urea in 100 mM Tris-HCl pH 8.5 was added to each MWCO filter centrifuged again for 25 min. Following the sample rinse, 100 μl aliquots of 50 mM IAM were added to each filter, vortexed, and incubated at room temperature in the dark for 20 min to alkylate the samples. The alkylated samples in the MWCO filters were centrifuged for 15 min to remove the alkylating reagents followed by three successive rinse steps involving 100 μl of 8M urea in 100 mM Tris-HCl pH 8.5 and 15 min of centrifugation. Next 100 μl of 50 mM ammonium bicarbonate was added to each filter followed by 15 min of centrifugation and repeated two more times. The filters were transferred to new flow-thru vials to ultimately collect the tryptic peptides. 50 μl of trypsin (1:50, enzyme:sample by weight) in 50 mM ammonium bicarbonate was added to each filter to digest the reduced and alkylated plasma and tissue lysate samples. Following an overnight incubation at 37°C, 50 μl of SIL cocktail was added to the reaction filters for SRM analysis and all the filters were centrifuged for 10 min. The SIL cocktail was prepared such that the quantities listed in Table 1 were the equivalent to what was loaded on column. To complete the FASP digestion protocol, 100 μl of 1% formic acid was added and the reaction filter was centrifuged for 15 min. Each digested sample solution was stored at −20°C until immediately prior to analysis. Samples were removed from the freezer, diluted by a factor of five with 1% formic acid and interrogated by PC-IDMS-SRM.
nLC-PC-IDMS-SRM and Data Analysis
Reversed-phase nLC-MS/MS was performed with a cHiPLC-nanoflex system interfaced with an nLC-2D system from Eksigent, directly coupled to a TSQ Vantage triple quadrupole MS (Thermo Scientific, San Jose, CA). The LC platform was setup in a trap and elute configuration with a 200 μm × 0.5 mm trap column and 75 μm × 15 cm analytical column both packed with ChromXP C18-CL stationary phase (3 μm, 120 Å) by Eksigent Technologies (Dublin, CA). The mobile phase solvents were composed of a HPLC-Grade water/acetonitrile (A: 98/2% and B: 2/98%) with 0.2% formic acid as the ion pairing reagent. For each experiment, a loop overfill (6 μl) was performed on the 5 μl sample loop of the AS1 Autosampler (Eksigent) and approximately 350 ng of protein were loaded. At 2% B, a 1.5 μl/min metered injection of 9000 nl was completed prior to the analytical gradient separation. The gradient started with 2% B maintained for 3 min followed by a ramp to 45% B over 20 min (3–23 min). In 1 min the gradient was ramped to 95% B and this was held for 4 min (24–28 min). Initial conditions were reinstated in 1 min and held for 6 min (29–35 min) at a flow rate of 400 nl/min. A blank injection was made following each plasma injection to minimize carryover.
A TSQ Vantage was operated in the EZ method builder for a scheduled SRM experiment to monitor the transitions of the endogenous and SIL counterpart species of interest. Monitored transitions for each peptide are described in Table 1 with corresponding collision energy values. The allowed cycle time was a maximum of 1 second, Q1 peak width (FWHM) was 0.70, and remaining parameters were default for the EZ method builder. The capillary temperature and spray voltage were 200°C and 1.4 kV, respectively. To maximize signal transmission for detection, Skyline was employed to facilitate collision energy optimization47 and retention time scheduling (2 minute window).44 Resultant.RAW files were uploaded into Skyline where peak areas and peak area ratios were extracted.44 The TSQ Vantage was calibrated externally according to the manufacturer s protocol. JMP software version 9 from SAS (Cary, NC) was employed for statistical analysis.
RESULTS AND DISCUSSION
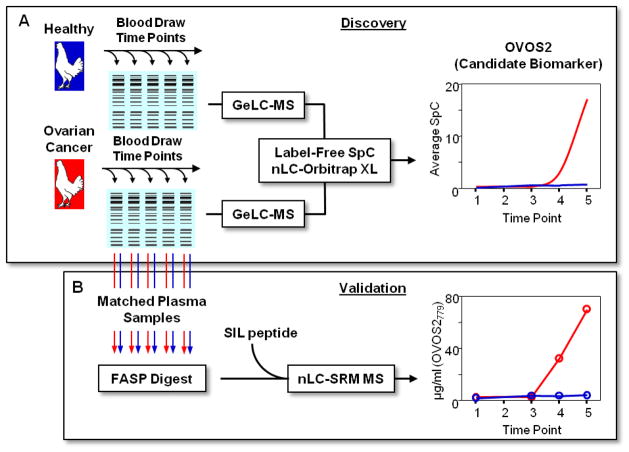
A popular path for MS-based biomarker discovery includes a “Discovery” phase that includes the in-depth analysis of a small numbers of samples to identify candidate markers followed by a series of qualification steps that ultimately result in the final “Validation” phase using targeted assays with high sample throughput.48 In our original study illustrated in Figure 1A, a candidate biomarker was identified using gel electrophoresis prior to LC-MS/MS (GeLC-MS/MS) of longitudinal plasma samples from a healthy and ovarian cancer phenotype. Importantly, the longitudinal analysis of the two phenotypes showed increasing levels of OVOS2 in the late-stage EOC bird suggesting increased tumor burden. Because this represented the first protein-level evidence for OVOS2, we set out to validate the label-free spectral counting (SpC) data using a multi-peptide PC-IDMS-SRM targeted assay applied to the same longitudinal plasma samples (Figure 1B). In addition to OVOS2, Vit-1 and TTR were also quantified as a “biological” and second candidate biomarker, respectively. The peptides used in the PC-IDMS-SRM assay for each of these proteins are given in Table 1 including corresponding labeled amino acid, mass-to-charge (m/z) of endogenous and SIL peptide, monitored transitions, amount of SIL peptide, and the optimized collision energies for each peptide.
Figure 1.
Here illustrates the global, discovery (A) and targeted qualification (B) bottom-up proteomics workflows in which relative and absolute quantification was performed on a set of longitudinal plasma samples of a healthy bird and late-stage EOC bird. The discovery work (A) was previously performed and the SpC trend of a possible EOC candidate marker is representative from Hawkridge et al.28 The targeted approach SRM-based assay (B) rapidly validates selected proteins from the global discovery evaluation and furthers the progression of the potentiality of the candidate biomarker such as demonstrated here.
Vitellogenin-1
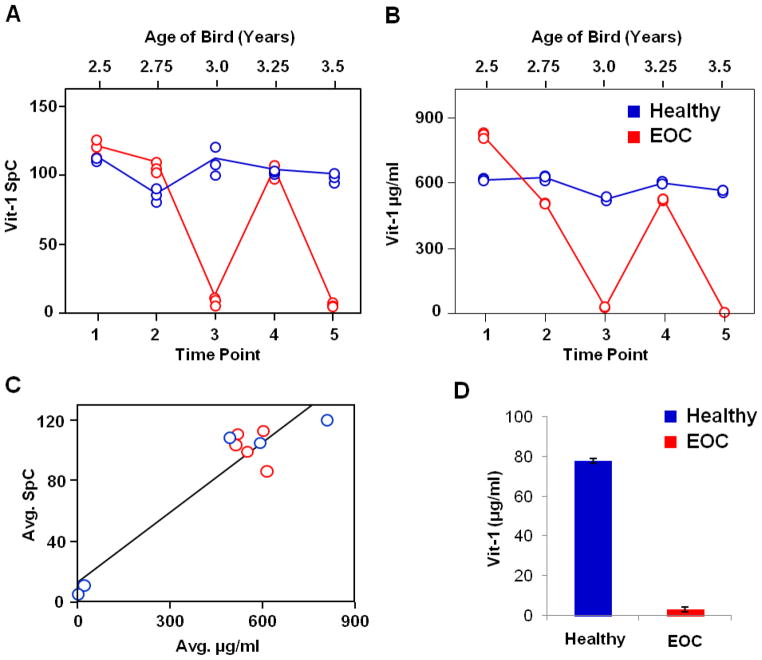
Vitellogenin-1 (Vit-1) is a 210 kDa lipoprotein synthesized in the liver as a yolk precursor that transports lipids and other nutrients to the yolk of the developing oocyte in the ovary.49 Vit-1 plasma levels are highest during egg production in order to maintain the necessary nutrient supply to the developing oocytes. When birds transition out of egg-production, Vit-1 plasma levels drop considerably. Our initial GeLC label-free study quantitatively measured this change for the EOC bird at time points 3 and 5 as shown in Figure 2A. The same longitudinal plasma samples were assayed by PC-IDMS-SRM and the absolute concentrations of Vit-1 as a function of time are shown in Figures 2B. The steady-state levels of Vit-1 in the two egg-producing birds oscillates around 600 μg/mL except for when the EOC bird goes out of egg production at time points 3 and 5. The correlation of label-free SpC with the absolute PC-IDMS-SRM data is shown in Figure 2C. The correlation between the two datasets is excellent (R2 = 0.94), particularly considering the spectral count data is from a GeLC plasma sample preparation with multiple fractions whereas the PC-IDMS-SRM data is from a single fraction of FASP-prepared un-depleted plasma. Further characterization of the bird phenotypes involved the PC-IDMS-SRM assay of the healthy and cancerous ovary tissues shown in Figure 2D. Vit-1 was highly abundant in the fully functional (egg-producing) ovary whereas more than 10-fold less was observed in the EOC tumor tissue with a regressed ovary (non-egg producing).
Figure 2.
Relative quantification of Vit-1 in the healthy and EOC birds from the global discovery-based analysis (A) trends rather similarly to the absolute quantification in which the peptide NIEDLAASK was monitored (B). The evaluation of the label-free SpC data from Hawkridge et al.28 and absolute quantification (μg/ml) approximately follow a linear slope (C). As expected in the ovary tissue lysate sample, which correlates to the 5th time point plasma sample, Vit-1 is abundant in the healthy bird which is in egg production (+) as compared to the EOC bird which is out of egg production (−).
Transthyretin
Transthyretin (TTR) is a 14 kDa hormone binding protein synthesized primarily in the liver that circulates in plasma as a homotetramer.50 TTR levels decrease in response to diminished nutrition, inflammation, and liver and kidney disease.51, 52 Point mutations in wild-type TTR are associated with amyloidosis.52, 53 More recently, TTR has been incorporated into a multi-protein biomarker panel OVA1, which can be used to clinically monitor ovarian cancer patients.54 TTR was detected at low abundance levels in the previous study (i.e., 4 SpC average28); however, 4 SpC represents approximately 30% sequence coverage for this low molecular weight protein. While this is a significant coverage level for confident protein identification, the low number of available tryptic peptides renders SpC less sensitive detecting true biological change.55, 56 Furthermore, the quantification accuracy and percent relative standard deviation is known to depend of the number of SpC detected with greater numbers of SpC constituting higher accuracy and lower variation.
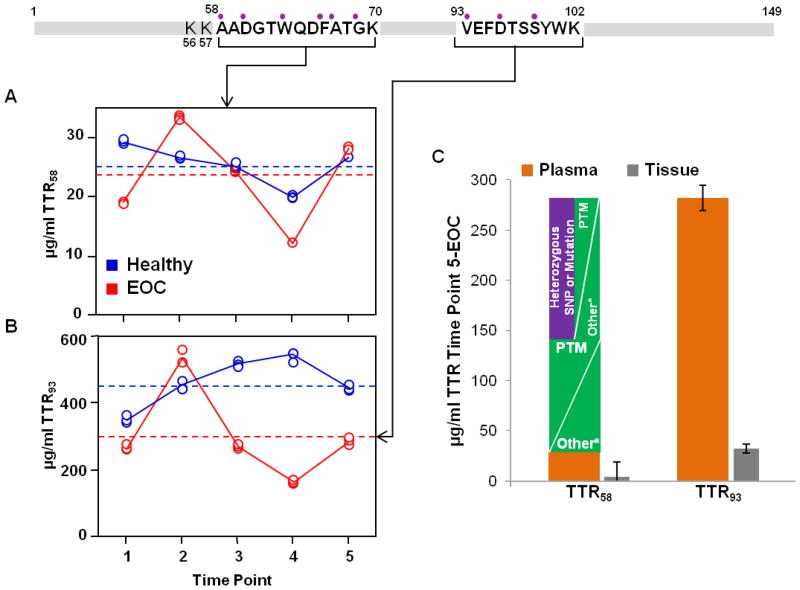
Two tryptic peptides of TTR (TTR58 and TTR93) were selected for the multiplexed PC-IDMS-SRM assay used in this study (Table 1, Figure 3) and the absolute values for each peptide in plasma are plotted as a function of time in Figure 3A and B, respectively. In the EOC bird, TTR concentration measured with either peptide trends similarly, peaking at the 2nd time point and exhibiting the least concentration at the 4th time point. The trends in concentration in the Healthy bird do not correlate as well appearing to be the inverse of each other. To determine if the fluctuation in expression is true biological change for TTR based on the signature peptides in this assay, the coefficient of variance (CV) was considered for the measurements. An estimate of the analytical variability (CVA) was made by measuring each TTR peptide in a pooled plasma tryptic digest using the nLC-PC-IDMS-SRM assay over five days (N=10) and determined to be 9.1% and 21.0% for TTR58 and TTR93, respectively. The measured CV s for TTR58 and TTR93 in the Healthy bird were 12.6% and 15.2%, respectively and in the EOC bird were 32.9% and 42.1%, respectively.
Figure 3.
Absolute quantification for TTR based on two different signature peptides (A) and (B). A clear trend is not realized. The EOC bird follows a similar fluctuation in protein concentration using the two peptides; however, when considering the 5th time point for example, a difference in protein concentration of approximately 10x is exhibited. The dashed lines indicate the average protein concentration derived by each peptide for the Healthy and EOC birds over the longitudinal samples. A plot of the absolute protein concentration for TTR with each peptide for plasma time point 5 (orange) and the ovary tissue (grey) samples (C) includes potential explanations for this concentration discrepancy and comparison to TTR quantified in the tissue. The purple circles above residues in the signature peptides indicate the positions of known variants in human TTR leading to mutation.
Inspecting Figure 3A and B, there is a significant discrepancy in the absolute concentrations of TTR depending on the peptide monitored in both plasma and matched ovary tissues. TTR measured monitoring TTR58 is computed as approximately 10× less concentrated than that measured with TTR93, 28 and 282 μg/mL, respectively for the EOC bird at the 5th time point, and 3.8 and 32.3 μg/mL, respectively, for TTR58 in the ovary tissue sample. Several factors may contribute to this inconsistency such as the knowledge of over 100 known mutations in human TTR.57 The Chicken TTR sequence identity is 73.3% relative to humans, of which there are overlapping sites at 6 different residues in TTR58 that include 10 known variants (purple circles above sequence in Figure 3). In TTR93, 4 human variants are known in total at 3 different sites. A mutation in TTR58 would activate a two-fold change in protein concentration (see the purple bar in Figure 3C). Unexpected PTMs would alter absolute quantification as well; however, no PTMs were found in the TTR58 and TTR93 sequences after searching the human and chicken TTR protein sequence at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). Other factors that may contribute to this discrepancy in absolute concentration may be the production (i.e., digestion efficiency) or degradation of each tryptic peptide. For example, Shuford et al.58 recently studied the production and degradation kinetics for 24 tryptic peptides in the context of PC-IDMS studies. Irrespective of when the SIL internal standard was added, tryptic peptide degradation led to an overestimation of the absolute amount of the endogenous protein. In some cases, the overestimation was greater than 30-fold. Another potential source of quantitative bias is incomplete digestion which was illustrated for the QconCAT strategy by Brownridge et al.59 The authors noted the inability of trypsin to cleave N-terminal dipeptides. This is a more probable source for the bias observed for TTR58 peptide where the N-terminus contains a di-lysine sequence shown in the TTR sequence (Figure 3). The PeptideCutter tool in ExPASy predicts a 100% and 87.5% cleavage rate at the C-terminus ends of K56 and K57, respectively. We did not however observe the missed cleavage form of TTR58 (i.e., K-TTR58) in our shotgun proteomics datasets. Additional studies including the formation and degradation rates for these two peptides will be necessary to potentially elucidate the source of this concentration bias.
Ovostatin 2 (OVOS2)
Ovostatin (also referred to as ovomacroglobulin) in the chicken is a member of the I39 protease inhibitor family which includes the abundant plasma protein alpha-2 macroglobulin (A2M).50 Ovostatin in chickens is expressed primarily in the oviduct and found in egg white yet not observed in any of the other major organs including the ovary.60–63 Our initial label-free study identified a new protein that was listed as a predicted form of ovomacroglobulin or ovostatin based on partial EST sequence data. This new ~180 kDa protein originated from a neighboring gene on the petite region of Chromosome 1 and has 50% sequence identity to the well-characterized form of ovostatin. To differentiate these two related proteins from different genes, we are referring to the highly abundant form of ovostatin from chicken egg white and oviduct as OVOS1 and the new predicted form of ovostatin identified in our label-free study28 as OVOS2. This terminology also aligns with these two genes in humans which are presently predicted or uncharacterized but have been detected at the transcript level. Interestingly, the human OVOS1, OVOS2, A2M, and several A2M-like proteins are clustered on Chromosome 12 which has significant conserved synteny with the petite region of chicken Chromosome 1.64 Furthermore, a recent report by Lim et al.65 showed mRNA evidence that chicken OVOS1 is up-regulated in some histopathological sub-types of ovarian cancer in chickens. Their data also confirmed earlier reports60, 61 that OVOS1 was expressed primarily in the oviduct of healthy birds. Giles et al.21 showed that ovalbumin, an oviduct and egg-white specific protein, was also present in serous ovarian cancers in chickens. These lines of evidence potentially support emerging data in human ovarian cancer66, 67 that challenge the hypothesis that some EOC histopathological sub-types originate from the ovarian surface epithelium but rather originates in the fallopian tubes, the human analogue of the chicken oviduct. Thus, quantitative measurements of OVOS2 have the potential to provide important diagnostic and pathophysiological information about the onset and progression of ovarian cancer in chickens and humans.
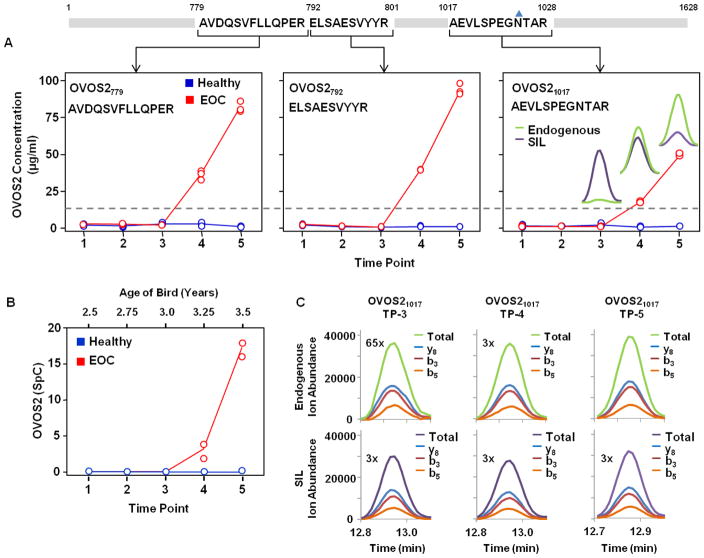
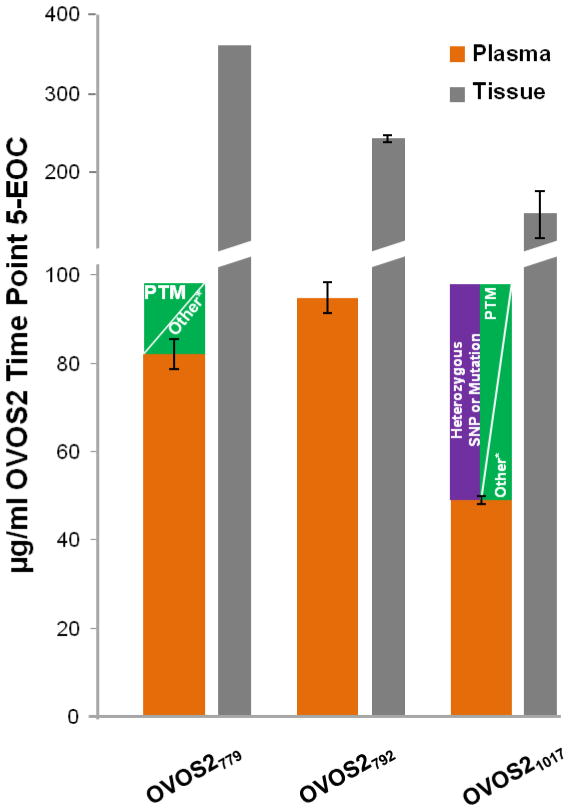
Three SIL peptides were employed for absolute quantification of OVOS2 (Table 1) to confidently qualify the quantitative increase in plasma concentration as a function of time in the late-stage EOC bird. Figure 4 illustrates the resulting concentrations from each OVOS2 peptide, the trend of protein expression from the label-free SpC data, and representative raw data demonstrating the ion abundance of the transitions monitored for three time points with OVOS21017. The trends observed for all three tryptic peptides, OVOS2779, OVOS2792, and OVOS21017, used in the PC-IDMS-SRM assay (Figure 4A) are identical to the original SpC data shown in Figure 4B. No signal from OVOS2 was observed in any of the time points for the Healthy bird. Although the trends correlate, the absolute concentrations derived from OVOS2779 and OVOS2792 are different than OVOS21017 for the EOC bird. For example, the average concentrations for OVOS2779, OVOS2792, and OVOS21017 at the 5th time point are 82, 95, and 49 μg/ml; respectively. The approximately 50% decrease in concentration for the OVOS21017 peptide could be attributable to a number of analytical and/or biological factors including digestion efficiency, proteolytic degradation of the protein, tryptic peptide degradation, PTMs, or single-nucleotide polymorphism (SNP) as illustrated in Figure 5. Dipeptidyl N-terminal lysines or arginines are not present for any of the three tryptic peptides as were seen by Brownridge et al.59 and shown for TTR58 in Figure 3. In-depth manual analyses of our label-free datasets have not detected any of these potential modifications.
Figure 4.
Absolute quantification for OVOS2 based on three different signature peptides (A) follow the same trend as the label-free SpC data demonstrated by Hawkridge et al.28 (B). Each peptide exhibits an increase in OVOS2 concentration as a function of time in the EOC bird and no OVOS2 is detected in the Healthy bird. The grey dashed line indicates a signal to noise ratio of 10. The chromatographic peaks inset in (A) OVOS21017 demonstrate the endogenous peptide signal (green) compared to the SIL peptide signal (purple), and correspond to the representative raw data for peptide OVOS21017 time point 3 (TP-3), time point 4 (TP-4), and time point 5 (TP-5) in the EOC bird (C). The ion abundance from each transition contributing to the overall signal used for computing absolute concentration of the protein are set to the same scale and magnified as denoted in (C). The blue triangle above the 1025-arginine represents potential N-linked glycosylation.
Figure 5.
Absolute concentration plot of OVOS2 with each peptide employed for plasma time point 5 (orange) and the ovary tissue (grey) samples. The y-axis is not of the equivalent scale throughout which is indicated by the break in the axis. Potential reasons for underestimation of protein concentration by OVOS2779 and OVOS21017 as compared to OVOS2792 are included for the plasma sample. The asterisk (*) indicates other possible influences of variability in concentration including chemical modifications, degradation, and poor digestion efficiency.
Another possibility is a PTM, particularly N-linked glycosylation which is known to be prevalent in human A2M and predicted on several sites in chicken OVOS1. The 1025-asparagine (blue triangle above sequence in Figure 4) is a predicted amino acid for N-linked glycosylation. Another consideration was that OVOS21017 peptide may fall within the bait region of the protease inhibitor, which inhibits proteases by undergoing conformational changes upon cleavage of the bait region sequence. However, based on alignment of OVOS2 with human A2M, the OVOS21017 peptide is not located in the bait region of A2M.
In addition to the undepleted plasma samples, the PC-IDMS-SRM assay was applied to the matched ovary tissue samples collected immediately after the 5th time point plasma sample. Figure 5 shows the absolute concentration values determined for each of the three peptides for the 5th time point plasma sample (orange) and the ovary tissue lysate (grey). Representative raw data for OVOS2779 transitions for the EOC tissue lysate is included in Supplemental Figure 1. The concentration determined from OVOS2779, OVOS2792, and OVOS21017 for the EOC bird were 361, 221, and 185 μg/ml, respectively. Again, the absolute concentration determined by OVOS21017 is underrepresented compared to that determined by the other two peptides. Viewing this data combined with the plasma samples, and in a different manner, Supplemental Figure 2 provides both the absolute quantification of OVOS2 based on each peptide and the signal ratio of the ion abundance area under the curve (AUC) for NAT to SIL for each measurement.
CONCLUSION
Here we have demonstrated the successful translation of findings in a quantitative label-free proteomic study (Discovery) to a targeted nLC-PCIDMS strategy that qualified quantitative trends and provides a means to investigate large numbers of longitudinal plasma samples in short run times. Multiple peptides were selected for the absolute quantification of Vit-1, TTR, and OVOS2. While selection criteria and previous data are considered in choosing the best signature peptide(s), certain potential biological features and behaviors such as differential peptide/protein degradation and formation rates, unknown SNPs, unexpected PTMs such as N-linked and O-linked glycosylation, and peptide degradation and chemical modification continue to bias absolute concentrations of proteins. Our investigation here underscores the difficulties of selecting signature peptides for quantification, and without multiple peptides, very interesting and dynamic features of proteins in complex biological systems could go undiscovered. With further efforts in developing this SRM-based assay, likely incorporating multiple peptides per protein or expressing a stable isotope labeled form of the intact protein, a large scale study of longitudinal plasma samples from birds with healthy and ovarian cancer phenotypes can be carried out.
Supplementary Material
Acknowledgments
This work was financially supported by NIH-NCI through award K25CA128666 to A.M.H. The authors would like to thank Chris Shuford for providing assistance and technical advice on absolute quantification method development and Angelito Nepomuceno for preparation of the ovary tissue lysate samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bast RC, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nature Reviews Cancer. 2009;9(6):415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams TI, et al. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. Journal of Proteome Research. 2007;6(8):2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 3.Bast RC, et al. Reactivity of a Monoclonal-Antibody with Human Ovarian-Carcinoma. Journal of Clinical Investigation. 1981;68(5):1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bast RC, et al. A Radioimmunoassay Using a Monoclonal-Antibody to Monitor the Course of Epithelial Ovarian-Cancer. New England Journal of Medicine. 1983;309(15):883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- 5.Xu FJ, et al. Ovx1 Radioimmunoassay Complements Ca-125 for Predicting the Presence of Residual Ovarian-Carcinoma at Second-Look Surgical Surveillance Procedures. Journal of Clinical Oncology. 1993;11(8):1506–1510. doi: 10.1200/JCO.1993.11.8.1506. [DOI] [PubMed] [Google Scholar]
- 6.Menon U, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncology. 2009;10(4):327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 7.Molina R, et al. HE4 a novel tumour marker for ovarian cancer: comparison with CA 125 and ROMA algorithm in patients with gynaecological diseases. Tumor Biology. 2011;32(6):1087–1095. doi: 10.1007/s13277-011-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skates S, Knapp RC. Early detection of ovarian cancer with tumor markers as first-line test. Current Problems in Obstetrics, Gynecology, and Fertility. 1996;19(3):85–98. [Google Scholar]
- 9.Skates SJ. Optimal summary of information in longitudinal CA125 measurements for efficient ovarian cancer screening. Journal of Clinical Ligan Assay. 2000;23(2):150–154. [Google Scholar]
- 10.Skates SJ, et al. Calculation of the risk of ovarian cancer from serial CA-125 values for preclinical detection in postmenopausal women. Journal of Clinical Oncology. 2003;21(10 Suppl):206s–210s. doi: 10.1200/JCO.2003.02.955. [DOI] [PubMed] [Google Scholar]
- 11.Skates SJ, et al. Toward an Optimal Algorithm for Ovarian-Cancer Screening with Longitudinal Tumor-Markers. Cancer. 1995;76(10):2004–2010. doi: 10.1002/1097-0142(19951115)76:10+<2004::aid-cncr2820761317>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.McIntosh MW, Urban N. A parametric empirical Bayes method for cancer screening using longitudinal observations of a biomarker. Biostatistics. 2003;4(1):27–40. doi: 10.1093/biostatistics/4.1.27. [DOI] [PubMed] [Google Scholar]
- 13.Williams RJ. Biochemical Individuality; The Basis for the Genetotrophic Concept. John Wiley & Sons, Inc; 1956. p. 267. [Google Scholar]
- 14.Williams RJ, Brown WD, Shideler RW. Metabolic Peculiarities in Normal Young Men as Revealed by Repeated Blood Analyses. Proceeding of the National Academy of Science of the United States of America. 1955;41(9):615–620. doi: 10.1073/pnas.41.9.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris EK. Effects of intraindividual and interindividual variation on appropriate use of normal ranges. Clinical Chemistry. 1974;20(12):1535–1542. [PubMed] [Google Scholar]
- 16.Hawkridge AM, Muddiman DC. Mass Spectrometry Based Biomarker Discovery: Toward a Global Proteome Index of Individuality. Annual Review of Analytical Chemistry. 2009;2(1):265–277. doi: 10.1146/annurev.anchem.1.031207.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredrickson TN. Ovarian Tumors of the Hen. Environmental Health Perspectives. 1987;73:35–51. doi: 10.1289/ehp.877335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barua A, et al. Histopathology of ovarian tumors in laying hens: a preclinical model of human ovarian cancer. International Journal of Gynecological Cancer. 2009;19(4):531–539. doi: 10.1111/IGC.0b013e3181a41613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver DK, et al. Reduction of ovarian and oviductal cancers in calorie-restricted laying chickens. Cancer Prevention Research. 2011;4(4):562–567. doi: 10.1158/1940-6207.CAPR-10-0294. [DOI] [PubMed] [Google Scholar]
- 20.Giles JR, Olson LM, Johnson PA. Characterization of ovarian surface epithelial cells from the hen: a unique model for ovarian cancer. Experimental Biology and Medicine. 2006;231(11):1718–1725. doi: 10.1177/153537020623101108. [DOI] [PubMed] [Google Scholar]
- 21.Giles JR, Shivaprasad HL, Johnson PA. Ovarian tumor expression of an oviductal protein in the hen: a model for human serous ovarian adenocarcinoma. Gynecological Oncology. 2004;95(3):530–533. doi: 10.1016/j.ygyno.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez Bosquet J, et al. Comparison of gene expression patterns between avian and human ovarian cancers. Gynecologic Oncology. 2011;120(2):256–264. doi: 10.1016/j.ygyno.2010.10.030. [DOI] [PubMed] [Google Scholar]
- 23.Hakim AA, et al. Ovarian adenocarcinomas in the laying hen and women share similar alterations in p53, ras, and HER-2/neu. Cancer Prevention Research. 2009;2(2):114–121. doi: 10.1158/1940-6207.CAPR-08-0065. [DOI] [PubMed] [Google Scholar]
- 24.Jackson E, et al. CA125 expression in spontaneous ovarian adenocarcinomas from laying hens. Gynecological Oncology. 2007;104(1):192–198. doi: 10.1016/j.ygyno.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Urick ME, Giles JR, Johnson PA. Dietary aspirin decreases the stage of ovarian cancer in the hen. Gynecological Oncology. 2009;112(1):166–170. doi: 10.1016/j.ygyno.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 26.Urick ME, Johnson PA. Cyclooxygenase 1 and 2 mRNA and protein expression in the Gallus domesticus model of ovarian cancer. Gynecological Oncology. 2006;103(2):673–678. doi: 10.1016/j.ygyno.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Fathalla MF. Incessant ovulation--a factor in ovarian neoplasia? Lancet. 1971;2(7716):163. doi: 10.1016/s0140-6736(71)92335-x. [DOI] [PubMed] [Google Scholar]
- 28.Hawkridge AM, et al. Measuring the intra-individual variability of the plasma proteome in the chicken model of spontaneous ovarian adenocarcinoma. Analytical and Bioanalytical Chemistry. 2010;398(2):737–749. doi: 10.1007/s00216-010-3979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon RB, et al. One-Year Plasma N-linked Glycome Intra-individual and Inter-individual Variability in the Chicken Model of Spontaneous Ovarian Adenocarcinoma. International Journal of Mass Spectrometry. 2011;305(2–3):79–86. doi: 10.1016/j.ijms.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dass C, Kusmierz JJ, Desiderio DM. Mass spectrometric quantification of endogenous beta-endorphin. Biological Mass Spectrometry. 1991;20(3):130–138. doi: 10.1002/bms.1200200306. [DOI] [PubMed] [Google Scholar]
- 31.Barr JR, et al. Isotope dilution mass spectrometric quantification of specific proteins: Model application with apolipoprotein A-I. Clinical Chemistry. 1996;42(10):1676–1682. [PubMed] [Google Scholar]
- 32.Gerber SA, et al. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proceeding of the National Academy of Science of the United States of America. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnidge DR, et al. Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC-MS/MS. Journal of Proteome Research. 2004;3(3):658–661. doi: 10.1021/pr034124x. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn E, et al. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and C-13-labeled peptide standards. Proteomics. 2004;4(4):1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 35.Brun V, et al. Isotope dilution strategies for absolute quantitative proteomics. Journal of Proteome Research. 2009;72(5):740–749. doi: 10.1016/j.jprot.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Ciccimaro E, I, Blair A. Stable-isotope dilution LC-MS for quantitative biomarker analysis. Bioanalysis. 2010;2(2):311–341. doi: 10.4155/bio.09.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elschenbroich S, Kislinger T. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Molecular Biosystems. 2011;7(2):292–303. doi: 10.1039/c0mb00159g. [DOI] [PubMed] [Google Scholar]
- 38.Borchers CH, et al. Multiple Reaction Monitoring-based, Multiplexed, Absolute Quantitation of 45 Proteins in Human Plasma. Molecular & Cellular Proteomics. 2009;8(8):1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carr SA, et al. Quantification of Cardiovascular Biomarkers in Patient Plasma by Targeted Mass Spectrometry and Stable Isotope Dilution. Molecular & Cellular Proteomics. 2009;8(10):2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carr SA, et al. Developing Multiplexed Assays for Troponin I and Interleukin-33 in Plasma by Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry. Clinical Chemistry. 2009;55(6):1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kislinger T, et al. In-Depth Proteomics of Ovarian Cancer Ascites: Combining Shotgun Proteomics and Selected Reaction Monitoring Mass Spectrometry. Journal of Proteome Research. 2011;10(5):2286–2299. doi: 10.1021/pr1011087. [DOI] [PubMed] [Google Scholar]
- 42.Addona TA, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature Biotechnology. 2009;27(7):633–U85. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisniewski JR, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–U60. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 44.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scopes RK. Measurement of Protein by Spectrophotometry at 205-Nm. Analytical Biochemistry. 1974;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 46.Wisniewski JR, et al. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 47.MacLean B, et al. Effect of Collision Energy Optimization on the Measurement of Peptides by Selected Reaction Monitoring (SRM) Mass Spectrometry. Analytical Chemistry. 2010;82(24):10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergen HR, et al. Discovery of ovarian cancer biomarkers in serum using NanoLC electrospray ionization TOF and FT-ICR mass spectrometry. Disease Markers. 2003;19(4–5):239–249. doi: 10.1155/2004/797204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salvante KG, Williams TD. Vitellogenin dynamics during egg-laying: daily variation, repeatability and relationship with egg size. Journal of Avian Biology. 2002;33(4):391–398. [Google Scholar]
- 50.Schaller J, et al. Human Blood Plasma Proteins: Structure and Function. West Sussex: John Wiley & Sons, Ltd; 2008. p. 525. [Google Scholar]
- 51.Jacobs D, DeMott W, Oxley D. Jacobs & DeMott Laboratory Test Handbook. 5. Hudson: Lexi-Comp, Inc; 1994. p. 1030. [Google Scholar]
- 52.Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cellular and Molecular Life Sciences. 2009;66(19):3095–3101. doi: 10.1007/s00018-009-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergen HR, et al. Identification of transthyretin variants by sequential proteomic and genomic analysis. Clinical Chemistry. 2004;50(9):1544–1552. doi: 10.1373/clinchem.2004.033266. [DOI] [PubMed] [Google Scholar]
- 54.Fung ET. A recipe for proteomics diagnostic test development: the OVA1 test, from biomarker discovery to FDA clearance. Clinical Chemistry. 2010;56(2):327–329. doi: 10.1373/clinchem.2009.140855. [DOI] [PubMed] [Google Scholar]
- 55.Collier TS, et al. Direct Comparison of Stable Isotope Labeling by Amino Acids in Cell Culture and Spectral Counting for Quantitative Proteomics. Analytical Chemistry. 2010;82(20):8696–8702. doi: 10.1021/ac101978b. [DOI] [PubMed] [Google Scholar]
- 56.Old WM, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Fleming CE, Nunes AF, Sousa MM. Transthyretin: more than meets the eye. Progress in Neurobiology. 2009;89(3):266–276. doi: 10.1016/j.pneurobio.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Shuford CM, et al. Peptide production and decay rates affect the quantitative accuracy of protein cleavage isotope dilution mass spectrometry (PC-IDMS) Molecular & Cellular Proteomics. 2012;11(9):814–23. doi: 10.1074/mcp.O112.017145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brownridge P, Beynon RJ. The importance of the digest: proteolysis and absolute quantification in proteomics. Methods. 2011;54(4):351–60. doi: 10.1016/j.ymeth.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 60.Nagase H, et al. Ovostatin - a Novel Proteinase-Inhibitor from Chicken Egg-White 1. Purification, Physicochemical Properties, and Tissue Distribution of Ovostatin. Journal of Biological Chemistry. 1983;258(12):7481–7489. [PubMed] [Google Scholar]
- 61.Nagase H, Harris ED. Ovostatin - a Novel Proteinase-Inhibitor from Chicken Egg-White.2. Mechanism of Inhibition Studied with Collagenase and Thermolysin. Journal of Biological Chemistry. 1983;258(12):7490–7498. [PubMed] [Google Scholar]
- 62.Saxena I, Tayyab S. Protein proteinase inhibitors from avian egg whites. Cell Mol Life Sci. 1997;53(1):13–23. doi: 10.1007/PL00000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mann K. The chicken egg white proteome. Proteomics. 2007;7(19):3558–3568. doi: 10.1002/pmic.200700397. [DOI] [PubMed] [Google Scholar]
- 64.Scherer SE, et al. The finished DNA sequence of human chromosome 12. Nature. 2006;440(7082):346–51. doi: 10.1038/nature04569. [DOI] [PubMed] [Google Scholar]
- 65.Lim W, et al. Differential expression of alpha 2 macroglobulin in response to dietylstilbestrol and in ovarian carcinomas in chickens. Reprod Biol Endocrinol. 2011:9. doi: 10.1186/1477-7827-9-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proceeding of the National Academy of Science of the United States of America. 2011;108(18):7547–7552. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, et al. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proceeding of the National Academy of Science of the United States of America. 2012;109(10):3921–3926. doi: 10.1073/pnas.1117135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.