Abstract
Nuclear hormone receptor coregulator (NRC) is a 2,063-amino-acid coregulator of nuclear hormone receptors and other transcription factors (e.g., c-Fos, c-Jun, and NF-κB). We and others have generated C57BL/6-129S6 hybrid (C57/129) NRC+/− mice that appear outwardly normal and grow and reproduce. In contrast, homozygous deletion of the NRC gene is embryonic lethal. NRC−/− embryos are always smaller than NRC+/+ embryos, and NRC−/− embryos die between 8.5 and 12.5 days postcoitus (dpc), suggesting that NRC has a pleotrophic effect on growth. To study this, we derived mouse embryonic fibroblasts (MEFs) from 12.5-dpc embryos, which revealed that NRC−/− MEFs exhibit a high rate of apoptosis. Furthermore, a small interfering RNA that targets mouse NRC leads to enhanced apoptosis of wild-type MEFs. The finding that C57/129 NRC+/− mice exhibit no apparent phenotype prompted us to develop 129S6 NRC+/− mice, since the phenotype(s) of certain gene deletions may be strain dependent. In contrast with C57/129 NRC+/− females, 20% of 129S6 NRC+/− females are infertile while 80% are hypofertile. The 129S6 NRC+/− males produce offspring when crossed with wild-type 129S6 females, although fertility is reduced. The 129S6 NRC+/− mice tend to be stunted in their growth compared with their wild-type littermates and exhibit increased postnatal mortality. Lastly, both C57/129 NRC+/− and 129S6 NRC+/− mice exhibit a spontaneous wound healing defect, indicating that NRC plays an important role in that process. Our findings reveal that NRC is a coregulator that controls many cellular and physiologic processes ranging from growth and development to reproduction and wound repair.
Nuclear hormone receptors (NRs) are ligand-dependent transcription factors that regulate target genes necessary for cell growth, differentiation, development, and cellular homeostasis. These receptors mediate the effect of thyroid hormone (T3), estrogen (E2), progestins, glucocorticoids, and androgens, as well as lipids, cholesterol metabolites, and bile acids, which act as ligands for the peroxisome proliferator-activated receptors (PPARs), liver X receptors (LXRs), and farnesoid X receptors (FXRs) (4). Over the past 6 to 7 years, a number of nuclear proteins that modulate NR activity have been identified and are collectively referred to as coactivators or coregulators. These coactivators interact with NRs through their LXXLL motifs (12, 22, 41, 44), which bind to a hydrophobic cavity on the surface of the ligand-bound receptor (12, 14, 49). Some of the well-studied coactivators include members of the p160 family, SRC-1/NCoA-1 (30, 50), SRC-2/TIF-2/GRIP-1/NCoA-2 (23, 59, 61), and SRC-3/AIB1/p/CIP/ACTR/RAC3/TRAM-1 (3, 11, 39, 55, 59); members of the CREB-binding protein (CBP)/p300 family (9, 21, 30), RIP140 (8), NRC/ASC-2/PRIP/RAP250/TRBP (7, 31, 34, 41, 70), PGC-1 (52), ARA70 (69), and p/CAF (5, 67); and NRIF3, which exhibits specificity for only the thyroid receptors (TRs) and the retinoid X receptors (RXRs) (36, 37). In addition to mediating effects of NRs, certain coactivators, including NR coregulator (NRC), appear to enhance the activity of other transcription factors such as NF-κB, CREB, c-Fos, and c-Jun (31, 34, 40, 45, 46). CBP/p300 is believed to act as a transcriptional integrator, which is recruited to the NRs and other transcription factors through their association with other coactivators (e.g., the SRCs and NRC) (10, 20, 41).
PBP/TRAP220/DRIP205, a component of the DRIP/TRAP complexes, also interacts with liganded NRs through LXXLL motifs (15, 54, 72). These complexes are related to the yeast SRB/Mediator complex and were identified through biochemical purification using ligand-bound VDR (DRIP) and TR (TRAP). The DRIP/TRAP complex is one of several related complexes that share components and modulate the activities of a variety of transcription factors, including the NRs, Sp1, NF-κB (p65), VP16, and p53 (6, 25, 48, 54).
To further identify the physiologic role of coactivators, mice null for expression of a number of coactivators, including NRC, have been generated by homologous recombination. The following findings summarize these results. CBP−/−, p300−/−, TRAP220−/−, and NRC−/− mice are embryonic lethal (2, 20, 26, 32, 33, 57, 68, 73). TRAP220+/− mice are viable but exhibit pituitary hypothyroidism. Null TRAP220 mouse embryonic fibroblasts (MEFs) in culture also fail to undergo adipocyte differentiation, implicating TRAP220 in regulating PPARγ activity (17). SRC-1−/− mice are viable and reproduce normally, but show partial resistance to E2, progesterone, androgen, and T3 (65). SRC-2 (TIF-2 and GRIP1) mice show a decrease in fertility (18). In male SRC-2−/− mice, this results from a defect in sperm and testicular function. In SRC-2−/− females, hypofertility reflects an SRC-2 requirement for optimal placenta development resulting in the loss of about half of the embryos between 12.5 and 18.5 days postcoitus (dpc). Newborn SRC-2−/− pups are about 30% smaller than the wild-type offspring for the first several weeks of life. After weaning, they grow normally and catch up in size and weight with the wild-type offspring. SRC-3−/− mice are viable but show growth retardation, abnormal mammary epithelial development, a delay in puberty, and decreased female fertility (63). Growth retardation is thought to result from decreased insulin-like growth factor 1 (IGF-1) production and cell resistance to IGF-1. These findings indicate that, although there may be some overlap in the activities of the three SRC isoforms, there are significant differences in their physiological functions.
Although much work has been carried out on the DRIP/TRAPs and the p160/SRC family, other candidate coactivators have been identified that are of biological significance. NRC was cloned in our laboratory (41) and others and is also referred to as ASC-2/PRIP/RAP250/TRBP (7, 31, 34, 70). NRC is a novel 2,063-amino-acid nuclear protein with two LXXLL motifs (LXXLL-1 and LXXLL-2). LXXLL-1 interacts with both retinoid-thyroid and steroid hormone NRs, while the LXXLL-2 region interacts preferentially with estrogen receptor alpha (ERα) and LXRα (35, 41). NRC contains two activation domains, and binding of liganded receptor with LXXLL-1 results in a conformational change in NRC leading to transcriptional activation (41). NRC interacts in vivo with CBP with high affinity, and the activity of NRC is inhibited by the adenoviral E1A(12S) protein, which blocks CBP activity (41). In addition to NRs, NRC also activates a variety of other transcription factors, such as c-Fos and c-Jun (31, 34, 40, 41). Mice null for expression of NRC have been recently reported by three different laboratories (2, 32, 73). Deletion of the NRC gene in mice revealed that null mutants (NRC−/−) die during early stages of embryogenesis (9.75 to 13.5 dpc) while mice heterozygous for the NRC gene (NRC+/−) in a mixed genetic background (C57/129) appear to be normal in growth and development. Intrauterine death of NRC-null embryos has been attributed to placental dysfunction, cardiac hypoplasia, and liver dysfunction, among other changes (2, 32, 73). Null PRIP MEFs in culture also fail to undergo adipocyte differentiation (53). To date, all of the studies on the physiological role of NRC using gene knockout studies have been carried out with NRC−/− mutant embryos and have focused on the etiology of embryonic lethality.
In addition to CBP, NRC has been reported to associate with a number of other factors, including CoAA, PIMT, CAPER, and NIF-1 (27, 28, 40, 71). Unlike NIF-1, CAPER, PIMT, and CoAA each contain RNA binding motifs. NIF-1 is a novel zinc finger, leucine zipper-containing nuclear protein that associates with and collaborates with NRC to activate NRs, c-Fos, and c-Jun (40). The similar activation patterns of NIF-1 and NRC and the in vivo association of NIF-1 with NRC in cells suggests that NRC and NIF-1 may be a part of a large coactivator complex involved in transcription. ASC-2 (NRC) is also a component of a 2-MDa complex in HeLa cells (19), which includes the trithorax group proteins ALR-1/2 (MLL2), HALR (MLL3), and ASH2; the retinoblastoma binding protein RBQ-2; and α- and β-tubulin.
In this study, we report a number of novel findings with NRC−/− embryos and NRC+/− mice, which were generated by homologous recombination. Chimeric male mice derived from C57BL/6 blastocysts and NRC+/− embryonic stem (ES) cells (129S6 background) were crossed with C57BL/6 females to obtain NRC+/− mice in a mixed background (indicated as C57/129). As previously reported, the NRC−/− mutation is embryonic lethal and no NRC−/− mice are born while NRC+/− mice in the mixed C57/129 background are viable and able to reproduce (2, 32, 73). We have observed these NRC+/− mice over a 2-year period and have noted that they develop an interesting phenotype associated with chronic wound healing. Unlike wild-type mice, NRC+/− C57/129 mice develop skin lesions in the neck and facial areas as a result of grooming, which persist with little or no healing. We have also generated NRC+/− mice in a pure 129S6 background and have uncovered reproductive phenotypes. NRC+/− 129S6 male mice are hypofertile, while about 20% of 129S6 NRC+/− females are sterile and 80% are hypofertile. In addition, a high rate of neonatal death was also noted among 129S6 NRC+/− mice. We also generated MEFs from C57/129 wild-type, NRC+/− and NRC−/− embryos at 12.5 dpc. In comparison with wild-type MEFs, NRC−/− MEFs exhibit slower growth rates and NRC−/− MEFs display a high level of spontaneous apoptosis, which can be blocked by incubating the cells with zVAD-fmk, an irreversible pan-caspase inhibitor. That the enhanced rate of apoptosis in NRC−/− MEFs reflects a deficiency in NRC was documented by transfecting wild-type MEFs with a small interfering RNA (siRNA) against NRC mRNA. These cells exhibited levels of apoptosis similar to that of the NRC−/− MEFs. Our results collectively identity novel actions of NRC that are important for a number of physiological processes, including cell survival, growth, embryonic development, wound repair, and reproduction.
MATERIALS AND METHODS
Construction of a targeting vector and homologous recombination.
The targeting vector was constructed by using a 15-kb mouse genomic clone containing a region of NRC (Fig. 1). A 1.1-kb DNA fragment containing the LXXLL-1 motif of NRC as a short arm was produced by PCR using primers PP18 and PP15. PP18 is located in the intron, about 2 kb downstream of the exon encoding the LxxLL-1 motif. PP15 is located in the intron 500 bp downstream of the same exon. The short arm was inserted 5′ of the Neo gene cassette by using the HindIII site. The long arm was a 7-kb NsiI genomic fragment that starts 1.5 kb upstream of the same exon. The long arm was cloned into the 3′ end of the Neo gene cassette. This strategy replaces the LXXLL-1 region with an intact Neo gene cloned into the targeting vector derived from pSP72. The targeting vector was linearized by NotI and then transfected into 129S6 (129S6/SvEv) ES cells by electroporation. After selection using G418, surviving colonies were expanded and analyzed for homologous recombination by PCR by using PP21 (CTAAGGACAACCAATCACTATGCC) and F-Neo1 (TGCGAGGCCAGAGGCCACTTGTGTAGC) primers. The correctly targeted ES cells were microinjected into C57BL/6 blastocysts to generate germ line chimeras that showed disruption of one of the NRC alleles. Chimeric males were then mated with C57BL/6 females or 129S6 females to produce progeny in mixed and pure genetic backgrounds, respectively.
FIG. 1.
Targeted disruption of the mouse NRC gene. The structures of the targeting vector (TV) and the wild-type (WT) NRC genomic sequence containing the NR box LXXLL-1 sequence are shown. The targeting vector contains the Neo gene flanked by 5′ and 3′ DNA sequences derived from the wild-type allele. The knockout (KO) allele is shown with a Neo gene replacing the NR box region with the Neo cassette. Bent arrows indicate the directions of transcription. The letters A, N, H, RV, and K represent AvaI, NsiI, HindIII, EcoRV, and KpnI, respectively. Arrowheads indicate the positions of primers used for PCR of genomic DNA.
Genotyping and Southern blotting.
DNA was isolated from tail tips excised from newborn pups and from MEFs derived from embryos and genotyped either by PCR or Southern blotting. DNA was isolated by digesting tails overnight at 55°C in lysis buffer containing 50 mM Tris Cl (pH 8), 0.5% sodium dodecyl sulfate (SDS), 100 mM EDTA, and 500-μg/ml proteinase K. After digestion, NaCl was added to the final concentration of 1.8 M and mixed, and the samples were spun down at room temperature. DNA from the supernatant was precipitated by adding 0.5 volume of isopropanol. Finally DNA was dissolved in 50 to 200 μl of 1× Tris-EDTA (TE) buffer. PCR using primers PP31 and PP21 amplified the wild-type alleles, while F-Neo1 in conjunction with PP21 was used to PCR genotype NRC mutant alleles. DNA was also used for Southern blotting.
Isolation of primary MEFs from wild-type and NRC mutant embryos.
Primary MEFs were isolated from wild-type, NRC+/−, and NRC−/− 12.5-dpc embryos as described previously (58). Briefly the liver, head, and other internal organs were removed from the embryos and were used for genotyping. The remaining embryo tissue was rinsed with phosphate-buffered saline (PBS) and trypsinized at 37°C for 15 to 30 min. MEF cells were cultured and split with Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum on a rigid schedule until the MEF lines spontaneously immortalized.
Transfections, MEF growth studies, and TUNEL assay for apoptosis.
Ten thousand MEF cells were seeded into 9-cm2 multiwell plates and cultured in DMEM containing 10% fetal bovine serum. The medium was replaced daily, and the cells were harvested and counted at different times to assess the growth rates of the wild-type and mutant MEFs. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays, 25,000 MEFs were plated onto 12-mm-diameter coverslips in 24-well tissue culture plates with DMEM containing 10% fetal bovine serum. The next day, the medium was changed and the cells were examined for apoptosis 24 h later. MEFs were transfected with a murine-specific siRNA (UGCAAGCGCAACUUCAGGC) directed against NRC mRNA (synthesized by Dharmacon) by using Oligofectamine (Invitrogen). Cells transfected with an siRNA that targets PSF-TFE3 mRNA (42) (which is only expressed in a subset of human renal carcinomas) as well as cells which only received Oligofectamine served as controls. Two days later, the cells were analyzed for apoptosis by TUNEL assay. For TUNEL assays, cells were washed three times with PBS, fixed in 3.7% formaldehyde, and assayed with the Roche Diagnostics in situ cell death detection kit (TMR red) (Roche Diagnostics GmbH, Penzberg, Germany) according to the manufacturer's protocol. In some cases, cells were also stained with Hoechst dye to visualize nuclei. Cells were then mounted on slides using Dako fluorescent mounting media, (DAKO Corporation, Carpinteria, Calif.) and examined by fluorescent microscopy. All of these expermients were repeated at least twice with duplicate or triplicate samples.
Skin explants, outgrowth, and immunostaining.
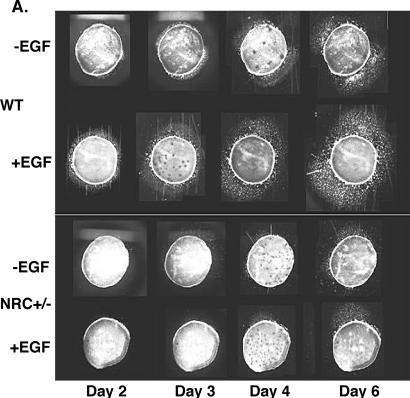
Skin explants were prepared from 2-day-old pups as described previously (43). Briefly, 2-day-old pups were washed with 10% povidone-iodine solution, sterile double-distilled water, 70% ethanol, and then PBS. Tails from each pup were used for genotyping. After an incision along the ventral surface of the body, the skin was peeled away and laid flat, dermis side down. A 4-mm sterile punch was used to remove six explants from each pup, which were placed individually into wells (24-well plate) containing coverslips. After allowing 5 min for the skin to adhere, 200 μl of medium was added to each well followed by the addition of 1.5 ml of medium the next day. Epidermal growth factor (EGF; 20 ng/ml) was added to three wells from each sample for additional stimulation of proliferation/migration. To assess keratinocyte outgrowth as a function of time in culture, phase-contrast micrographs of explants were recorded on days 2, 3, 4, and 6. Micrographs were scanned into a computer with Photoshop 4.0 software. Each experiment was done in triplicate. The extent of outgrowth was quantitated by overlaying the scanned images on a grid and assessing the area of outgrowth relative to the area of the 4-mm explant. Shown in Fig. 6 are individual components of a representative experiment.
FIG. 6.
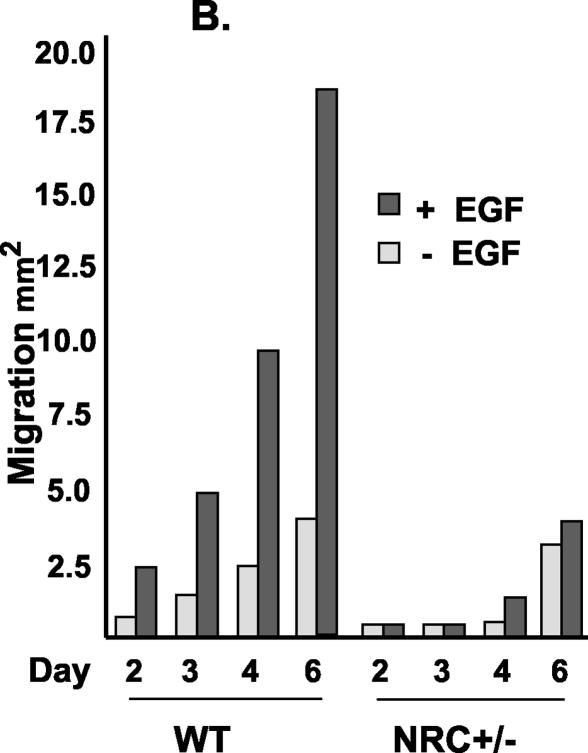
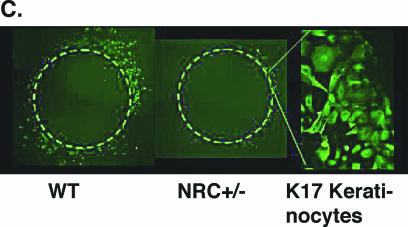
Lack of keratinocyte migration in explants from NRC+/− mice. (A) Keratinocyte outgrowth and migration are shown. Normal keratinocyte migration from explants of wild-type (WT) mice and induction of the migration by EGF are shown at the top. Explants from NRC+/− mice exhibit a delay of initiation and diminished capacity of migration, as well as lack of the response to EGF (bottom). (B) Quantitation of the area of ketatinocyte outgrowth shown in panel A. (C) K17-specific antibody staining of the explant outgrowth identifies keratinocytes as migratory cells.
To assess the expression of keratin 17 (K17) which is expressed in keratinocytes, the coverslips were washed in PBS and fixed in cold (−20°C) acetone-methanol (1:1) for 2 min at room temperature. Following another wash with PBS, a rabbit polyclonal antibody that can detect mouse K17 (a gift from Pierre Coulombe) (24) was applied (1:1,000), diluted in 5% bovine serum albumin in PBS. After overnight incubation with K17 antibody at 4°C, the coverslips were rinsed in PBS and incubated with fluorescein-conjugated anti-rabbit immunoglobulin G (1:600 dilution) (Sigma-Aldrich) for 3 h at room temperature. After a final wash in PBS, the stained coverslips were examined by using a Zeiss fluorescent microscope and digital images were collected with Adobe Photoshop 4.0.
Histological analysis and immunochemistry.
All samples processed for histopathology were stored in buffered 10% formalin, embedded in paraffin blocks, and sectioned with a microtome. The tissue sections were examined after staining with hematoxylin-eosin.
RESULTS
Loss of NRC results in embryonic lethality.
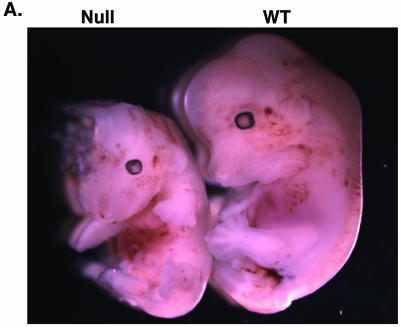
Figure 1 illustrates the targeting vector used to generate NRC+/− 129S6 ES cells for implantation into C57BL/6 blastocysts to generate chimeric offspring from pseudopregnant C57BL/6 females. Male germ line chimeras were crossed with either C57BL/6 or 129S6 female mice to obtain NRC+/− F1 mice either in the C57/129 mixed or 129S6 genetic background. Crosses between C57/129 NRC+/− males and NRC+/− females led to predictable numbers of wild-type and NRC+/− offspring [Table 1; C57/129 (+/−) males × C57/129 (+/−) females]. No NRC−/− null pups were born (Table 1). Thus, we analyzed embryos at various stages of gestation. The number of NRC−/− embryos identified from different NRC+/− females progressively decreased from 8.5 to 12.5 dpc. Partially resorbed NRC−/− embryos were detected at 12.5 dpc, suggesting that the death of null embryos in utero occurred between 8.5 and 12.5 dpc. Null embryos showed an overall decrease in size (about 50%) compared with wild-type or NRC+/− embryos (Fig. 2A). In addition to decreased growth, a number of other defects including decreased vascularization, developmental defects in liver, heart, and brain, and placental dysfunction were identified (data not shown). Some of the embryos lacked one of the digits in the hind limb or separation of the digits did not occur. These findings are similar to that previously been reported in NRC-null embryos (2, 32, 73).
TABLE 1.
Genotypes of offspring of the mouse crosses used in this study
| Genotype of offspring | No. of pups at birth | No. with genotype
|
||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| F2 from C57/129 (+/−) males × C57/129 (+/−) females | 94 | 29 | 65 | 0 |
| 129S6 (+/−) males × 129S6 (+/+) females | 85 | 50 | 35 | 0 |
| 129S6 (+/−) males × 129S6 (+/−) females | 85 | 58 | 27 | 0 |
FIG. 2.
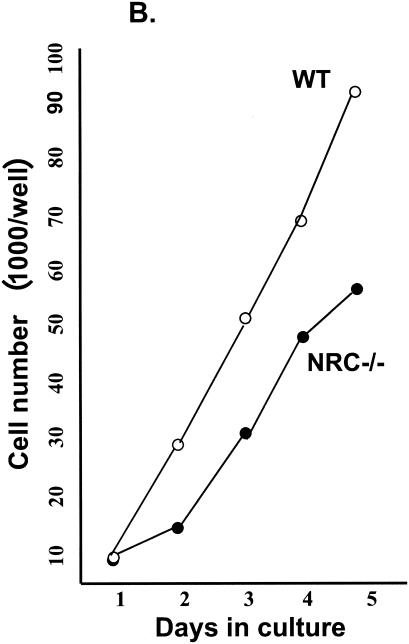
Null mutant embryos are half the size of wild type embryos, and mutant MEFs grow at a slower rate than wild type MEFs. (A) Size difference between wild-type (right) and NRC−/− mutant (left) embryos at 12.5 dpc. The mutant embryo is partly resorbed. (B) Ten thousand wild-type and NRC−/− MEF cells were seeded in 9-cm2 wells, and the cells were harvested and counted each day for 5 days. Shown is a representative experiment.
NRC−/− null MEFs grow at a reduced rate and exhibit apoptosis.
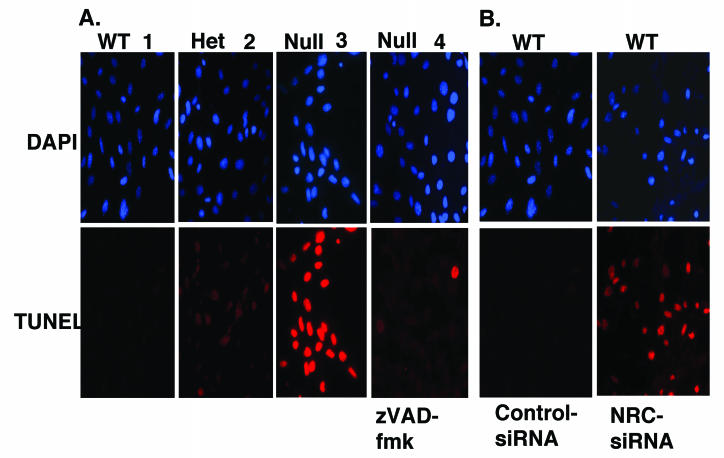
NRC-null embryos that die in utero are about half the size of wild-type embryos. This growth retardation in null embryos suggests that a slower rate of cell proliferation, cell cycle dysfunction, or enhanced apoptosis may lead to embryonic lethality. To examine these possibilities, we developed immortalized MEFs from wild-type, NRC+/−, and NRC−/− 12.5-dpc embryos through repeated passage. Under identical culture conditions, the null MEFs took longer to establish than wild-type or NRC+/− MEFs. After the lines were established, we determined the rate of cell growth of wild-type, NRC+/−, and NRC−/− MEFs. The results suggested that NRC−/− MEFs double at about half the rate of wild-type MEFs (Fig. 2B). We did not perform as extensive growth studies with the NRC+/− MEFs. However, they appeared to exhibit a growth profile intermediate between the wild-type and NRC−/− MEFs (data not shown). During the course of these studies, we observed that NRC−/− null MEFs in late log phase develop a morphology reminiscent of apoptosis. TUNEL assay indicated that a high percentage of the NRC−/− MEFs undergo apoptosis as the cells approach late log phase (Fig. 3A, panels 1 to 3). Apoptosis was not noted in the mid-log phase of NRC−/− MEF growth, suggesting that metabolic changes or environmental signals associated with the late log phase require NRC for cell survival. NRC+/+ cells showed no evidence of apoptosis under the same conditions. With NRC+/− MEFs, the cells undergo apoptosis but at much lower rates than the NRC−/− MEFs as they approach the late log phase of growth. The apoptosis identified in the NRC−/− MEFs was inhibited by incubating cells with zVAD-fmk, an irreversible pan-caspase inhibitor supporting a role for caspases in the apoptosis observed in MEFs lacking NRC (Fig. 3A, panel 4).
FIG. 3.
NRC−/− cells undergo apoptosis and an siRNA that targets NRC mRNA leads to apoptosis of wild-type MEFs. (A) MEF cells as indicated were plated on coverslips and analyzed for apoptosis (red) after 2 days of growth by TUNEL assay as described in Materials and Methods. Cells were also stained with Hoechst dye (DAPI [4′,6′-diamidino-2-phenylindole]) for nuclear staining (blue) (panels 1 to 3). Apoptosis of the NRC−/− MEF cells (panel 4) was inhibited when they were incubated for 2 days with zVAD-fmk, a pan-caspase inhibitor, prior to the TUNEL assay. (B) Cells were transfected with an siRNA specific for NRC mRNA and a control siRNA that targets PSF-TFE3 mRNA, which is not expressed in the MEFs. Two days later, the cells were analyzed for apoptosis by TUNEL assay as described in Materials and Methods.
We used RNA interference (RNAi) to document that a deficiency of NRC is responsible for the apoptosis found in the NRC−/− MEFs. An siRNA designed to target mouse NRC mRNA was transfected into wild-type MEF cells by using Oligofectamine. An siRNA against PSF-TFE3 mRNA, which is only expressed in a subset of human kidney tumors, was used as a control (42). Wild-type MEFs were also treated with Oligofectamine reagent alone as a control for possible cell death due to the transfection reagent. Cells were analyzed for apoptosis by TUNEL assay 2 days after transfection. As shown in Fig. 3B, wild-type MEFs transfected with the NRC siRNA exhibited apoptosis similar to that found with the NRC−/− MEFs. No apoptosis was observed with the control cells, which received the control PSF-TFE3 siRNA (Fig. 3B) or the Oligofectamine reagent alone (not shown). The results strongly suggest that expression of NRC in MEFs is important for cell survival and that NRC may play an important role in regulating antiapoptotic or prosurvival genes.
Inbred 129S6 NRC+/− heterozygous mice display compromised reproductive phenotypes and increased neonatal mortality.
NRC+/− mice in a 129S6 background were produced from crosses between the chimeric founder male and 129S6 wild-type females. NRC+/− 129S6 males (F1) were found to be hypofertile when crossed with wild-type 129S6 females, with the average litter size comprising three pups. The cause of hypofertility in the NRC+/− 129S6 males is presently unknown. Numerical evaluation as depicted in Table 1 [129S6 (+/−) males × 129S6 (+/+) females] revealed a ratio of 0.7:1 for NRC+/− versus wild-type newborn pups, which deviates from an expected 1:1 Mendelian ratio. In contrast, NRC+/− and wild-type pups in the C57/129 hybrid background are produced at an expected ratio as shown in Table 1 [C57/129 (+/−) males × C57/129 (+/−) females]. This suggests that some NRC+/− 129S6 embryos die in utero, similar to NRC−/− embryos. This is also consistent with the finding that pups from crosses between NRC+/− 129S6 males and wild-type 129S6 females exhibited increased neonatal mortality (about 15% of the NRC+/− neonates died within 1 week after birth). The cause of death among these 129S6 NRC+/− neonates has not yet been determined. The NRC+/− 129S6 neonates in general were also found to be smaller (10 to 15%) than their wild-type littermates. However, the differences in weight were found to diminish with age between 1 and 2 months after birth (data not shown). We have also noted similar growth retardation among NRC+/− C57/129 neonatal mice. However, this occurs with a very low frequency (less than 5%). Interestingly, about 3% of NRC+/− 129S6 newborn mice were extremely small (stunted), weighing 70% less than the wild-type littermates (Fig. 4), and these mice often die before weaning. Our findings indicate that the penetrance of the NRC phenotype is more pronounced in the 129S6 background.
FIG. 4.
NRC+/− 129S6 mice exhibit stunted growth. All NRC+/− 129S6 newborn mice are 10 to 15% smaller than their wild-type littermates. Approximately 3% of postnatal NRC+/− mice are severely stunted and are about 70% of the size of their wild-type littermates. The figure shows the difference in size between a 10-day-old wild-type mouse and a severely stunted NRC+/− mouse.
Unlike C57/129 NRC+/− mice, inbred crosses in the 129S6 background between NRC+/− females and NRC+/− males often failed to yield pups. Thirty-five matings confirmed that the failure to produce pups was due primarily to infertility of 20% of NRC+/− 129S6 females. In contrast, NRC+/− males could produce pups when mated with wild-type 129S6 females [Table 1, 129S6 (+/−) males × 129S6 (+/+) females]. NRC+/− 129S6 females produced small litters when crossed with wild-type males. The litter size was further reduced when such females were mated with NRC+/− 129S6 males (approximately three pups per litter). The ratio of heterozygous NRC+/− to wild-type pups was estimated to be 0.5:1 [Table 1, 129S6 (+/−) males × 129S6 (+/−) females]. The NRC+/− pups also displayed an increase in neonatal mortality. NRC+/− 129S6 females thus fall into two categories: (i) those that are totally sterile (∼20%) and (ii) those that are hypofertile (∼80%), which produce approximately three pups per litter. These NRC+/− hypofertile females were crossed repeatedly, which revealed a progressive decline in the fertility of these females.
We have not yet defined the cause of hypofertility or infertility among NRC+/− 129S6 females. However, given the role of NRC as a coactivator of NRs, hypofertility may involve a wide variety of factors, ranging from hormone resistance to developmental defects in the reproductive system (e.g., ovulation, implantation, and placental function) or behavioral problems. A number of infertile female 129S6 NRC+/− mice were screened for the presence of vaginal plugs when mated with 129S6 NRC+/− or wild-type male mice. Approximately 80% of the females displayed vaginal plugs, suggesting successful copulation or normal sexual activity. However, these mice were found to lack embryos when dissected at 8.5 to 12.5 dpc. The regular estrus cycle in female mice is generally used as a measure for reproductive function. Examination of the estrus cycle was performed for 9 days on 20 age-matched adult (129S6) NRC+/− and wild-type female mice by microscopically analyzing cells from vaginal smears. As with the wild-type females, the majority of NRC+/− 129S females displayed a regular, normal cycling profile, suggesting that they respond to estrogen and that the cause of infertility among these females is likely due to other factors.
NRC heterozygous mice exhibit a chronic wound healing phenotype.
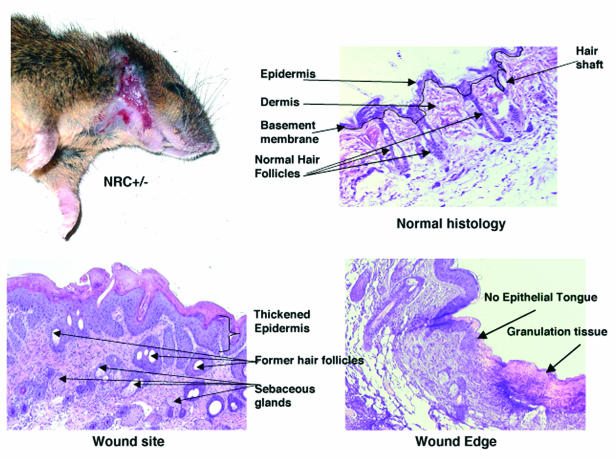
After weaning, NRC+/− mice in a mixed C57/129 background appear normal and grow and reproduce similar to wild-type mice, as also reported by others (2, 32, 73). We have observed the C57/129 NRC+/− mice for over 2 years and have noted an interesting phenotype associated with chronic wound healing as they age. NRC+/− C57/129 mice as early 4 months spontaneously develop wounds or ulcers around the neck, ears, snout, and facial area. These regions correspond to the areas where mice groom and likely scratch themselves. This occurs in approximately 20% of both male and female NRC+/− C57/129 and NRC+/− 129S6 mice (Fig. 5). No skin lesions were noted in wild-type mice. Mice suffering from skin lesions were separated immediately and monitored for wound healing. Approximately 25% of the mice with lesions showed a delay in wound healing (1 to 2 months), while the lesions in the remaining mice did not heal. Owing to the severity of the chronic wounds, these mice were sacrificed and skin biopsies were collected from affected and normal regions of the skin and analyzed for histopathology.
FIG. 5.
NRC+/− mice exhibit a wound healing defect. Shown is a representative NRC+/− mouse with skin lesions in the neck area. The skin from the wound site and the edge of the wound and a normal skin area were processed for histology and stained with hematoxylin-eosin. The skin lesions show an increase in sebaceous glands, a lack of hair follicles, and no keratinocyte migration (epithelial tongue).
Figure 5 illustrates a mouse with such a lesion along with representative histopathology studies that show thickening of the epidermis, increased sebaceous glands, and a reduction or total loss of hair follicles and the lack of a leading edge of keratinocyte migration (epithelial tongue), which is seen with normal wound healing. The lesions were deep, extending through the epidermis into dermis and hypodermis. In such areas, granulation tissue replaced the preexisting structures. The chronicity of the lesion is reflected by the presence of a thickened epidermis due to hyperproliferation of keratinocytes. No ectoparasites were noted in the sections. Specially stained sections of the skin did not reveal bacterial or fungal organisms associated with the lesions. The location of these lesions is consistent with self-inflicted trauma as an inciting cause, as part of normal grooming.
The histology analysis of the wound edge from NRC+/− mice revealed the lack of a leading edge or epithelial tongue, suggesting a defect in cell migration (Fig. 5). In order to examine keratinocyte migration capacity, ex vivo cultures of skin explants from wild-type and NRC+/− pups were compared over a period of 6 days (Fig. 6A). Keratinocytes derived from wild-type mouse explants migrated outward, as expected, and moved steadily during the 6-day period (Fig. 6A). When stimulated with EGF, these keratinocytes responded, as expected, with enhanced proliferation and migration. Interestingly, keratinocytes from explants derived from NRC+/− pups showed a significant delay in onset of migration as well as a decrease in the migration capacity, as evident by a reduction of the outgrowth area. Furthermore, additional stimulation by EGF did not significantly increase migration or proliferation of keratinocytes derived from NRC+/− pup explants (Fig. 6A). We measured and quantified keratinocyte migration for triplicate explants of all the NRC+/− and wild-type pups from a litter and found significantly reduced migratory capacity as well as response to EGF in the explants from NRC+/− mice when compared with explants from wild-type mice. Quantitation of the keratinocyte outgrowth results of Fig. 6A are shown in Fig. 6B. Lastly, we used K17-specific antibody to identify the origin of the migratory cells. Almost the entire cell population that migrated out of the explant was K17 positive (Fig. 6C), thus confirming that migratory cells are indeed keratinocytes. Taken together, we found that NRC+/− mice exhibit diminished capacity of keratinocyte migration and are unable to respond to EGF, which may contribute to the lack of epithelialization and to impaired wound healing.
DISCUSSION
Over the past several years, a number of coactivators have been characterized that have emerged as key components in the activation of gene expression by NRs. Among well-characterized coactivators, the p160 family and the TRAP220 genes have been knocked out in mice, indicating that these coactivators are necessary for growth, reproduction, and activation by NRs (25, 64). A more recently characterized coregulator referred to as ASC-2, RAP250, PRIP, TRBP, or NRC has been established as a coactivator for NRs and other transcription factors, such as c-Fos, c-Jun, NF-κB, and CREB (31, 34, 40).
The effect of NRC on the activity of a broad spectrum of transcription factors along with the other factors reported to interact with NRC suggests that NRC may play an important role in NR function and in transcriptional activation in general. The finding that NRC (31, 41) and the SRCs (29, 60, 66) both interact with CBP/p300 suggests a possible integrative role of these coactivators in mediating NR function and transcriptional activation. Deletion of both alleles of the SRC-1, TIF-2 (SRC-2), or p/CIP (SRC-3) gene in mice is not embryonic lethal, although these mice show evidence for partial steroid and/or thyroid hormone resistance (SRC-1, SRC-2, and SRC-3) and IGF-1 resistance (SRC-3). Although SRC-1 and SRC-2 may mediate somewhat different functions (51), the lack of embryonic lethality suggests redundancy and/or compensatory expression of the p160 family. In contrast, NRC, TRAP220/DRIP205/PBP, CBP, and p300 are essential coactivators necessary for embryonic development.
Recently RAP250, TRBP, and PRIP mutant mice have been generated by homologous recombination (2, 32, 73). These studies focused on the phenotype and etiology of the NRC-null embryonic lethality and identified placental vascular dysfunction, myocardial developmental defects secondary to decreased cell proliferation, and small livers with cell morphologies suggestive of apoptosis. In our present studies, in addition to examining the role of NRC in embryonic development, we examined the role of NRC in NRC+/− mice and analyzed the developmental and reproductive phenotypes of NRC+/− mice in different genetic backgrounds. Although C57/129 NRC+/− mice are fertile, in the 129S6 genetic background, the fertility of both male and female NRC+/− mice is impaired. We have also uncovered an interesting phenotype of chronic wound healing in both NRC+/− C57/129 and 129S6 mice and provide evidence that deletion of NRC can lead to apoptosis.
In agreement with previous reports, we found that deletion of both NRC alleles leads to embryonic lethality. We also noted that the NRC−/− embryos isolated from 8.5 to 12.5 dpc are always smaller than the NRC+/− and wild-type embryos, suggesting both growth and developmental phenotypes. To study the role of NRC in cell growth, we established spontaneously immortalized MEFs from NRC−/−, NRC+/−, and wild-type embryos. Studies with these MEFs indicated that the NRC−/− MEFs grow at slower rates than wild-type MEFs, while the NRC+/− MEFs displayed an intermediate growth rate. We noted that as the cells approached the late log phase of growth, the NRC−/− MEFs displayed morphological characteristics consistent with apoptosis. Wild-type MEFs were not TUNEL positive, while the NRC+/− cells showed some TUNEL-positive cells but much less than the NRC−/− cells. zVAD-fmk, a pan-caspase inhibitor (except for caspase 2) blocked apoptosis in the NRC−/− MEFs, further supporting the notion that the positive TUNEL response reflects apoptosis involving caspase activation. Lastly, transfection of an siRNA that targets NRC mRNA in wild-type MEFs leads to apoptosis. We attempted to document knockdown of NRC by Western blotting (data not shown). However, NRC is a very-low-abundance protein, and our antibody was not sensitive enough for accurate quantitation of NRC. Nevertheless, our siRNA results strongly suggest that expression of NRC in MEFs is important for cell survival and that NRC may play an important role in regulating antiapoptotic or prosurvival genes. Although we have not defined the mechanism through which NRC reduction leads to apoptosis, the NRC gene is amplified in number of breast cancer cell lines (34) and NRC is known to be important for the action of NF-κB, c-Fos, c-Jun, and CREB (31, 34, 40), all of which are known to mediate antiapoptotic effects in cells.
NRC+/− and wild-type offspring in the C57/129 background are produced at an expected ratio [Table 1, C57/129 (+/−) × C57/129 (+/−) females] and grow normally and are fertile. In contrast, 129S6 NRC+/− mice show several distinct reproductive and growth phenotypes. The NRC+/− 129S6 males and females were found to be hypofertile, with the average litter size comprising three pups. The newborn NRC+/− 129S pups obtained from such crosses are far less than expected from Mendelian distribution. This suggests that like NRC−/− embryos, some NRC+/− 129S6 embryos die in utero. In addition, ∼20% of NRC+/− 129S6 females are sterile, the majority of which displayed a normal cycling profile by vaginal cell analysis, suggesting that the females respond to estrogen and that the cause of infertility may be due to other factors. Thus, the penetrance of the NRC phenotype appears more pronounced in the 129S6 background.
Our studies with NRC+/− mice show some functional correlation with SRC-2 (TIF-2−/−) and SRC-3(pCIP)−/− null mice. Both male and female TIF-2 null mice display a decrease in fertility. Male hypofertility is attributed to defective sperm and an age-dependent testicular degeneration (18). The decrease in offspring in female TIF-2−/− mice results from placental dysfunction due to a requirement for maternal TIF-2 in decidua stromal cells that face the developing placenta (18). SRC-3−/− mice do not respond well to superovulation compared to wild-type females, produce fewer pups, and show a lower pregnancy rate compared to SRC-3+/− and wild-type mice (63).
In addition to hypofertility, NRC+/− 129S6 mice show a growth phenotype similar to that found with TIF2−/− mice. NRC+/− 129S6 mice weighed 10 to 15% less at birth than wild-type littermates. However, the weight deficit difference diminished within 1 to 2 months after weaning, as also reported with TIF2−/− mice (18). Interestingly, a smaller number (∼3%) of newborn NRC+/− 129S6 pups revealed high penetrance of the growth phenotype. These NRC+/− mice were extremely stunted at birth, weighing 70% less than their wild-type littermates and exhibited general growth retardation (Fig. 4). The majority of these pups died before weaning. However, about 30% of these pups survived and by 1 to 2 months weighed almost the same as their other littermates. This stunted growth of NRC+/− neonates and the subsequent “catch-up” growth to approach the size of wild-type littermates resembles the phenotype displayed by TIF-2−/− mice. Although SRC-3−/− mice are also ∼30% smaller, these mice show somatic growth arrest, which continues into adulthood. In contrast with NRC+/− 129S6 mice, NRC+/− C57/129 mice grow and develop normally, although we have very infrequently found that some (<5%) NRC+/− C57/129 newborn mice are somewhat smaller than their wild-type littermates.
Although almost all NRC+/− mice in the C57/129 background have no apparent phenotype and grow and reproduce similar to wild-type C57/129 mice, we found that as these mice grow older they exhibit a wound healing phenotype. They develop skin lesions that either do not heal or show delay in healing. The distribution of these skin lesions suggests that they are self-inflicted as part of the normal grooming process. Wound healing is a complex multicellular process that involves (i) the release of inflammatory cytokines, which leads to paracrine release of growth factors from fibroblasts and keratinocytes; (ii) formation of a fibrin clot and subsequently granulation tissue; (iii) keratinocyte migration at the wound edge to form a leading edge (epithelial tongue) that migrates to cover the wound; and (iv) proliferation and stratification of the layers of epidermis and laying down of the extracellular matrix to form a new area of mature skin.
Interestingly, the wound healing phenotype identified in the NRC+/− mice is similar to that observed in mice, where c-Myc is targeted to the basal layer of mouse epidermis and hair follicles using a K14 promoter expression vector (62). Although mice heterozygous for expression of K14/c-Myc are viable and have no phenotype at birth, like the NRC+/− mice, as they age they develop chronic lesions in the facial, ear, and neck grooming areas. Histological studies showed areas of epidermal hyperplasia, enlarged sebaceous glands, reduced numbers of hair follicles, and lack of keratinocyte migration similar to that which we found in the NRC+/− mice. The mechanism for development of the c-Myc-mediated chronic wounds was suggested to be secondary to repression of integrins (e.g., α6 and β1) (62). These integrins are thought to play an important role in keratinocyte migration and in self-renewal of epidermal stem cells leading to their gradual depletion over time. Other microarray studies (16) indicate that the majority of down-regulated genes identified in the keratinocytes overexpressing c-Myc are involved in cell adhesion and the cytoskeleton, including expression of α4β6 integrin, which may be considered as a marker of epidermal stem cells.
Although the mechanism of c-Myc repression of cell adhesion genes is not known, the similar phenotypes found in the mice heterozygous for expression of K14/c-Myc and in the NRC+/− mice suggest that NRC may play a role in regulating a similar set of genes. For example, NRC is a potent coregulator of c-Jun activity, which plays an important role in regulating genes involved in wound healing (integrins, laminins, and extracellular matrix genes in the epithelium) (1). In addition, c-Jun has recently been shown to stimulate expression of heparin-binding (HB) EGF in the basal layer of wounded epithelium, which in turn stimulates signaling by EGF receptor to regulate motility through regulation of focal adhesion kinase (38). The results obtained with skin explants from wild-type and NRC+/− pups revealed a defect in migration of keratinocytes from NRC+/− explants (Fig. 6) both with and without EGF treatment. The lack of keratinocyte response to EGF is not only important for the appropriate wound healing response but also explains in part the mechanism, because NRC plays an important role in c-Jun signaling. Therefore, the impaired wound healing phenotype of the NRC+/− can be explained, at least in part, by diminished migratory capacity of keratinocytes. Thus, NRC as a potent coregulator of c-Jun activity may also affect various signaling cascades involved during the process of wound healing. Taken together, the decrease in wound healing in NRC+/− mice may reflect a dosage effect of NRC on c-Jun activity in keratinocytes and other cells involved in the wound healing process.
In addition to c-Jun, there are other transcription factors and signaling molecules (such as p53, NF-κB, or PPARs) involved in wound healing that may be regulated by NRC (31, 34, 40, 41). For example, PPARα and PPARβ have been shown to be important in wound healing (47, 56). Although both PPARα and PPARβ are not expressed in adult epithelium (except for hair follicle keratinocytes), both are rapidly expressed after the development of a wound, and PPARβ remains expressed throughout the entire wound healing process (47). PPARα is thought to play an important role in the initial inflammatory process, while PPARβ is involved in keratinocyte proliferation (47). PPARβ+/− mice exhibit a significant delay in wound closure, and in vitro studies with PPARβ+/− keratinocytes indicate a decrease in keratinocyte adhesion and mobility. PPARβ also acts to block apoptosis of keratinocytes by regulation of the Akt1 pathway through stimulation of expression of integrin-linked kinase (ILK) and PDK1 and repression of PTEN (13). Although we have not defined the precise mechanism by which the apparent reduction of NRC in the skin of NRC+/− mice leads to chronic skin wounds, its role as a coactivator for c-Jun and the PPARs and its role as a prosurvival factor suggest that NRC acts through regulation of one or more of the above factors and functional pathways involved in the wound healing process.
In summary, our present studies with NRC knockout mice has uncovered novel phenotypes associated with the loss or reduction of the nuclear receptor coactivator NRC in adult mice and further emphasize the importance of examining the role of a factor in a specific genetic background. Our studies indicate that NRC is a pleiotropic coregulator in the embryo and adult involved in growth, development, apoptosis, reproduction, and wound repair. In addition, the increased response to haploinsufficiency of NRC in 129S6 NRC+/− mice suggests that these mice may also be a useful model for identifying the role(s) of NRC in the action of a variety of NRs as well as NR-mediated metabolic processes.
Acknowledgments
We thank Olivera Stojadinovic and Brian Lee from the Tomic-Canic laboratory for the preparation of tissue samples for histology. We also thank Cindy Loomis for helpful suggestions and Jan Sap (Department of Pharmacology) for many helpful suggestions with the animal studies.
This study was supported by NIH grants DK16636 (H.H.S.) and AR45974 and NR08029 to M.T.-C.
REFERENCES
- 1.Angel, P., A. Szabowski, and M. Schorpp-Kistner. 2001. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 20:413-423. [DOI] [PubMed] [Google Scholar]
- 2.Antonson, P., G. U. Schuster, L. Wang, B. Rozell, E. Holter, P. Flodby, E. Treuter, L. Holmgren, and J.-Å. Gustafsson. 2003. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol. Cell. Biol. 23:1260-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 4.Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1269-1304. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, J. C. G., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, V. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 7.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. A. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 8.Cavailles, N., S. Dauvois, F. L'Horset, S. Lopez, S. Hoare, P. J. Kushner, and M. G. Parker. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti, D., V. J. LaMorte, M. C. Nelson, T. Nakajima, I. G. Schulman, H. Juguilon, M. Montminy, and R. M. Evans. 1996. Role of CBP/P300 in nuclear receptor signalling. Nature 383:99-103. [DOI] [PubMed] [Google Scholar]
- 10.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 12.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di-Poi, N., N. S. Tan, L. Michalik, W. Wahli, and B. Desvergne. 2002. Antiapoptotic role of PPARb in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol. Cell 10:721-733. [DOI] [PubMed] [Google Scholar]
- 14.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747-1749. [DOI] [PubMed] [Google Scholar]
- 15.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frye, M., C. Gardner, E. R. Li, I. Arnold, and F. M. Watt. 2003. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development 130:2793-2808. [DOI] [PubMed] [Google Scholar]
- 17.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 18.Gehin, M., M. Mark, C. Dennefeld, A. Dierich, H. Gronemeyer, and P. Chambon. 2002. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol. Cell. Biol. 22:5923-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goo, Y.-H., Y. C. Sohn, D.-H. Kim, S.-W. Kim, M.-J. Kang, D.-J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, V. T. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P.-G. Suh, E. J. Song, K.-J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 21.Hanstein, B., R. Eckner, J. DiRenzo, S. Halachmi, H. Liu, B. Searcy, R. Kurokawa, and M. Brown. 1996. p300 is a component of an estrogen receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:11540-11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 23.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutton, E., R. D. Paladini, Q. C. Yu, M. Yen, P. A. Coulombe, and E. Fuchs. 1998. Functional differences between keratins of stratified and simple epithelia. J. Cell Biol. 142:487-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, M., and R. G. Roeder. 2001. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12:127-134. [DOI] [PubMed] [Google Scholar]
- 26.Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 27.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375-33383. [DOI] [PubMed] [Google Scholar]
- 28.Jung, D. J., S. Y. Na, D. S. Na, and J. W. Lee. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229-1234. [DOI] [PubMed] [Google Scholar]
- 29.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17:232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S.-C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 31.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuang, S. Q., L. Liao, H. Zhang, F. A. Pereira, Y. Yuan, F. J. DeMayo, L. Ko, and J. Xu. 2002. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J. Biol. Chem. 277:45356-45360. [DOI] [PubMed] [Google Scholar]
- 33.Landles, C., S. Chalk, J. H. Steel, I. Rosewell, B. Spencer-Dene, N. Lalani el, and M. G. Parker. 2003. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol. Endocrinol. 17:2418-2435. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, L. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, is a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. K., S. Y. Jung, Y. S. Kim, S. Y. Na, Y. C. Lee, and J. W. Lee. 2001. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol. 15:241-254. [DOI] [PubMed] [Google Scholar]
- 36.Li, D., V. Desai-Yajnik, E. Lo, M. Schapira, R. Abagyan, and H. H. Samuels. 1999. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol. Cell. Biol. 19:7191-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, D., F. Wang, and H. H. Samuels. 2001. Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation. Mol. Cell. Biol. 21:8371-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, G., C. Gustafson-Brown, S. K. Hanks, K. Nason, J. M. Arbeit, K. Pogliano, R. M. Wisdom, and R. S. Johnson. 2003. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell 4:865-877. [DOI] [PubMed] [Google Scholar]
- 39.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan, M. A., A. Murray, and H. H. Samuels. 2002. NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC. Mol. Cell. Biol. 22:6883-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathur, M., S. Das, and H. H. Samuels. 2003. PSF-TFE3 oncoprotein in papillary renal cell carcinoma inactivates TFE3 and p53 through cytoplasmic sequestration. Oncogene 22:5031-5044. [DOI] [PubMed] [Google Scholar]
- 43.Mazzalupo, S., M. J. Wawersik, and P. A. Coulombe. 2002. An ex vivo assay to assess the potential of skin keratinocytes for wound epithelialization. J. Investig. Dermatol. 118:866-870. [DOI] [PubMed] [Google Scholar]
- 44.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 46.McKenna, N. J., and B. W. O'Malley. 2002. Minireview: nuclear receptor coactivators—an update. Endocrinology 143:2461-2465. [DOI] [PubMed] [Google Scholar]
- 47.Michalik, L., B. Desvergne, N. S. Tan, S. Basu-Modak, P. Escher, J. Rieusset, J. M. Peters, G. Kaya, F. J. Gonzalez, J. Zakany, D. Metzger, P. Chambon, D. Duboule, and W. Wahli. 2001. Impaired skin wound healing in peroxisome proliferator-activated receptor (PPAR)α and PPARβ mutant mice. J. Cell Biol. 154:799-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 49.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-g. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 50.Onate, S. A., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 51.Picard, F., M. Gehin, J. Annicotte, S. Rocchi, M. F. Champy, B. W. O'Malley, P. Chambon, and J. Auwerx. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931-941. [DOI] [PubMed] [Google Scholar]
- 52.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 53.Qi, C., S. Surapureddi, Y. J. Zhu, S. Yu, P. Kashireddy, M. S. Rao, and J. K. Reddy. 2003. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor γ (PPARγ)-interacting protein, is required for PPARgamma-mediated adipogenesis. J. Biol. Chem. 278:25281-25284. [DOI] [PubMed] [Google Scholar]
- 54.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 55.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 56.Tan, N. S., L. Michalik, N. Noy, R. Yasmin, C. Pacot, M. Heim, B. Fluhmann, B. Desvergne, and W. Wahli. 2001. Critical roles of PPAR β/δ in keratinocyte response to inflammation. Genes Dev. 15:3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi, and S. Ishii. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94:10215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 60.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voegel, J. J., M. J. S. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 62.Waikel, R. L., Y. Kawachi, P. A. Waikel, X.-J. Wang, and D. R. Roop. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28:165-168. [DOI] [PubMed] [Google Scholar]
- 63.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, J., and B. W. O'Malley. 2002. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Rev. Endocr. Metab. Disord. 3:185-192. [DOI] [PubMed] [Google Scholar]
- 65.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 66.Xu, Y., L. Klein-Hitpass, and M. K. Bagchi. 2000. E1A-mediated repression of progesterone receptor-dependent transactivation involves inhibition of the assembly of a multisubunit coactivation complex. Mol. Cell. Biol. 20:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 68.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 69.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. USA 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]
- 71.Zhu, Y., C. Qi, W. Q. Cao, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2001. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl. Acad. Sci. USA 98:10380-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, Y. J., S. E. Crawford, V. Stellmach, R. S. Dwivedi, M. S. Rao, F. J. Gonzalez, C. Qi, and J. K. Reddy. 2003. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J. Biol. Chem. 278:1986-1990. [DOI] [PubMed] [Google Scholar]