Abstract
Yeast Taf1p is an integral component of the multiprotein transcription factor TFIID. By using coimmunoprecipitation assays, coupled with a comprehensive set of deletion mutants encompassing the entire open reading frame of TAF1, we have discovered an essential role of a small portion of yeast Taf1p. This domain of Taf1p, termed region 4, consisting of amino acids 200 to 303, contributes critically to the assembly and stability of the 15-subunit TFIID holocomplex. Region 4 of Taf1p is mutationally sensitive, can assemble several Tafps into a partial TFIID complex, and interacts directly with Taf4p and Taf6p. Mutations in Taf1p-region 4 induce temperature-conditional growth of yeast cells. At the nonpermissive temperature these mutations have drastic effects on both TFIID integrity and mRNA synthesis. These data are consistent with the hypothesis that Taf1p subserves a critical scaffold function within the TFIID complex. The significance of these data with regard to TFIID structure and function is discussed.
TFIID, one of several multiprotein general transcription factors required for RNA polymerase II (PolII)-catalyzed mRNA gene transcription, is comprised of the TATA-binding protein (TBP) and 14 distinct TBP-associated factors (TAFs or Tafps) that range in Mr from 17,000 to 150,000 in Saccharomyces cerevisiae (62, 63, 65, 75). These TFIID subunits share significant sequence conservation with their metazoan orthologs (1, 22, 26). In vitro, PolII needs six general transcription factors, TFIIA, -B, -D, -E, -F, and -H, to form functional preinitiation complexes (PICs) (12, 51, 54); TFIID plays several critical roles in this process. First, TFIID functions during promoter recognition by binding directly to the promoter (67, 76, 77). On promoters containing a TATA box, Tafps help stabilize TBP binding to the TATA element via interaction with TATA-proximal initiator DNA sequences (69). Tafp-DNA interactions may be particularly important for TFIID binding to promoters that lack a canonical TATA element and contain TATA-proximal initiator and distal promoter element sequences (13, 15, 42, 69, 76). Second, Tafps can function as coactivators during transcriptional activation by making direct contacts with specific activators leading to an increase in PolII PIC formation (1, 16). Finally, Taf1p contains several distinct enzymatic activities, those of histone acetyltransferase (HAT) (46), protein kinase (19), and ubiquitin ligase (55). These enzymatic activities presumably modify proteins that stimulate PIC formation and/or function, leading to PolII transcription initiation.
In most contexts, the Tafp subunits of TFIID are essential for survival, as Tafp inactivation or depletion results in cessation of specific mRNA synthesis and loss of cell viability (3, 4, 33, 34, 56, 59, 64, 65, 78). Because of these critical roles, the composition, organization, assembly, structure, and function of the TFIID complex have been topics of great interest. TBP appears to be incorporated into the TFIID complex primarily through its interaction with the bipartite N-terminal domain of Taf1p, the so-called TAND domain (5, 7, 36, 37) comprised of TAND1 and TAND2 elements. Although Taf6p and Taf12p have also been shown to interact with TBP (58, 60), the exact contribution of these TBP-TAF interactions to TFIID integrity remains to be determined. TBP binds to both isolated TAND and intact Taf1p with nanomolar affinity (5, 7), and the structure of the TBP-TAND complex has been solved by nuclear magnetic resonance (41). The binding of TBP to Taf1p remains the best-defined direct interaction of TBP with a subunit of the TFIID complex. However, exactly how this interaction contributes to TFIID structure, organization, and function remains to be determined.
Since a large number of Tafps constitute TFIID, numerous Tafp-Tafp interactions are possible within the TFIID complex. In vitro studies indicate that certain Tafp-Tafp interactions appear to be preferred during TFIID assembly (9, 23, 24, 38, 60, 63, 74). In fact, functional TFIID subcomplexes can be assembled using purified recombinant subunits (16). Based on these reconstitution studies, an obligatory role of metazoan Taf1p in all the functional Tafp subcomplexes was observed. Consequently it was proposed that Taf1p plays a key structural or scaffold protein role during TFIID assembly (16, 79). However, a systematic Taf1p-Tafp interaction analysis of TFIID has yet to be reported. Yeast Taf1p shares extensive structural and functional similarities with its metazoan orthologs, and the TFIIDs isolated from both yeast and metazoan cells display very similar trilobed structures in electron microscopy studies (2, 11, 39). However, despite these many conserved properties, a similar scaffold function for yeast Taf1p has been disputed (1, 60).
A core Tafp-Tafp assembly distinct from Taf1p(TAND)-TBP has also been proposed to contribute importantly to TFIID formation. It was originally hypothesized by Roeder and coworkers (29, 53) that a subset of TFIID subunits, those containing the helix-loop-helix-loop-helix or histone fold domain (HFD), could assemble to form a complex resembling the histone nucleosome octamer (1, 29, 53, 60). This core structure was suggested to depend upon the HFD, a structural motif shared by the core histones (22) and many other proteins (70). Nine of 14 yeast Tafps appear to contain HFDs, and subsets of these can form dimers with specific partner HFD Tafps (3, 6, 9, 23, 24, 29, 45, 47, 80). Indeed, Tan and colleagues (66) have described the formation and characterization of such an octameric complex comprised of four HFD-Tafps, a dimer of Taf6p-Taf9p bound to a dimer of Taf4p-Taf12p. The exact role of this putative octamer in TFIID structure and function remains to be elucidated.
Our previous structure-function analyses of Taf1p indicated that deletion of a region of Taf1p spanning amino acids 200 to 303 (Taf1p-region 4) caused inviability (5). Our preliminary analyses, as scored by the coimmunoprecipitation (co-IP) of a few Tafps (Taf6p, Taf10p, and Taf14p) and TBP, indicated that in this deletion mutant the integrity of the TFIID complex was compromised. These observations prompted us to propose that region 4 of Taf1p played an essential role in the assembly and/or stability of yeast TFIID. In the present study we have rigorously tested this hypothesis by using a combination of biochemical and genetic approaches. We have found that region 4 of Taf1p is essential for assembly of the TFIID holocomplex. We also observed that this region, in isolation, is capable of nucleating the formation of TBP-TAF subcomplexes both in vivo and in vitro. Further, mutations of various residues within region 4 induced temperature-conditional growth, defects in TFIID integrity, and gene-specific defects in mRNA gene transcription. These data lead us to conclude that yeast Taf1p-region 4 serves as a platform for Tafp-Tafp interactions critical for TFIID assembly. The implications of these results are discussed with regard to TFIID organization, structure, and function.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli strains XL-1 Blue and DH5α were used for plasmid cloning and propagation. Ultracompetent E. coli strain XL-2 Blue (Stratagene) was used for high-efficiency transformation of the TAF1-region 4 mutant library (see below). Recombinant proteins were expressed in E. coli BL21(DE3) Gold (Stratagene).
Co-IP and gel filtration chromatography.
For co-IP assays, yeast cells were grown either in YPAD medium or in appropriate synthetic complete (SC) selective medium lacking either histidine (SC-H), leucine (SC-L), or uracil (SC-U) as indicated. Co-IP experiments were performed as described previously (5). Table 1 lists the strains used in our experiments. Gel filtration chromatography of TFIID was conducted essentially as described previously (62) except that the buffer contained 150 mM NaCl. Column fractions were concentrated with trichloroacetic acid and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis to identify the elution position of native TFIID.
TABLE 1.
Deletion variants of Taf1p used in this study (5)
| Taf1p deletion mutant | Deleted amino acids |
|---|---|
| WT | None |
| Δ1 | 2 to 80 |
| Δ2 | 43 to 115 |
| Δ3 | 101 to 208 |
| Δ4 | 200 to 303 |
| Δ5 | 300 to 367 |
| Δ6 | 365 to 435 |
| Δ7 | 430 to 495 |
| Δ8 | 480 to 595 |
| Δ9 | 540 to 600 |
| Δ10 | 600 to 650 |
| Δ11 | 640 to 700 |
| Δ12 | 692 to 774 |
| Δ13 | 749 to 860 |
| Δ14 | 830 to 912 |
| Δ15 | 899 to 1005 |
| Δ16 | 913 to 1037 |
| Δ17 | 1037 to 1066 |
Taf1p-region 4 expression construct.
A DNA fragment corresponding to TAF1-region 4 (bp 598 to bp 909 of the TAF1 open reading frame [ORF]) was amplified with Pfu Turbo polymerase (Stratagene) by using primers containing HindIII sites at the ends and inserted in the HindIII site of the LEU2-marked p425GalL vector (49). Fragments encoding an N-terminal three-hemagglutinin (HA3) tag and nuclear localization signal (NLS) were inserted in frame into HindIII-digested p425GalL, sequenced, and transformed into yeast strain BY4743 (MATa/MATα leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 lys2Δ0/LYS2 met15Δ0/MET15; Research Genetics), and transformants were selected for Leu+ growth (SC-L). For induction of NLS-HA3 or NLS-HA3-region 4 proteins, cells were grown in SC-L medium with raffinose and galactose added to 2% (wt/vol) final concentrations. Ten optical density at 600 nm (OD600) amounts of cells were harvested 6 h after galactose induction and frozen at −80°C for subsequent white cell extract (WCE) preparation and co-IP studies.
Construction and isolation of region 4-targeted temperature-sensitive (ts) TAF1 mutants.
A PCR-amplified DNA fragment comprising TAF1 ORF sequences encoding region 4 was cloned into the EcoRI site of pBluescript SK+ plasmid. Random mutations in region 4 coding sequences were generated by error-prone PCR as described previously (43) except that the concentrations of MnCl2 and each deoxynucleoside triphosphate were 250 and 25 μM, respectively. In a 50-μl reaction mixture, 10 pmol of each primer and 2.5 U of Taq DNA polymerase were used with the following amplification cycle: after initial heating at 94°C, 25 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C followed by a 10-min extension at 72°C. The primers used were forward 5′-CCGGAATTCGGCGGCCGCGACTTGATAAACCATGAGAAG-3′ and reverse 5′-CCGGAATTCCTATTAGCGGCCGCCAAGTTCATCAATGGAAACATG-3′, where the underlined sequences represent bp 598 to 618 and bp 889 to 909 of yeast TAF1, upper and lower strands, respectively; the bold letters show an added EcoRI site; and italics show an introduced NotI site. The EcoRI sites were used for cloning the amplified region 4 DNA into EcoRI-digested pBluescript SK+.
The PCR-generated library of region 4 fragments was digested with NotI and ligated into the NotI site of the HIS3-marked pRS313-HA3-TAF1Δ4 plasmid; this plasmid carries an allele of TAF1 that has a NotI site introduced at the site of deletion (5). This construct was modified to remove the NotI site at the multiple cloning site of pRS313 by site-directed mutagenesis prior to reinsertion of region 4 sequences. Ligation reaction mixtures were transformed at high efficiency in E. coli XL-2 Blue ultracompetent cells (Stratagene), transformants were pooled and grown in Luria-Bertani broth for 2 h at 37°C, and plasmids were isolated using the Qiafilter plasmid midi kit (Qiagen). This library of mutations was transformed into yeast strain YBY804 [MATα ura3-52 lys2-801amber ade2-101ochre taf1Δ::TRP1 his3-Δ200 leu2-Δ|pRS316(URA3)TAF1] by the high-efficiency TRAFO method (25). Transformants were selected on SC-H-U plates. In a separate PCR, wild-type (WT) region 4 sequences were amplified using the same primers and Pfu Turbo DNA polymerase. This fragment was separately cloned into the region 4-deletion backbone to generate pRS313-HA3-TAF1Δ4/WT, a construct termed reconstructed WT or WT-r. This plasmid was introduced by transformation into YBY804 as described above. Yeast transformants were subjected to plasmid shuffle on 5-fluoroorotic acid plates (10). More than 100,000 clones were subjected to such screening; the colonies were then screened for temperature sensitivity at 37°C by replica plating. After two rounds of ts mutant screening, HIS3-marked plasmids were recovered directly from these yeast cells as described previously (68), and the TAF1 gene resident on them was sequenced (35). Three mutants displaying ts growth were selected for further study.
Two-hybrid assays.
A yeast two-hybrid bait fragment containing sequences of Taf1p-region 4 (amino acids 231 to 395) was PCR amplified using forward primer 5′-CGCGCGAATTCTCTGTTCCTTATCATTGGCACAGT-3′ (bold letters showing EcoRI site) and reverse primer 5′-GCGCGAGATCTAGCATGTTTAGACTCTTTCAATTT-3′ (bold letters showing BglII site) from a plasmid containing the full-length WT TAF1 ORF. The resulting purified PCR product was digested with EcoRI and BglII and cloned in the EcoRI-BglII sites of the pGBDC1 vector (31) to encode a Gal4-DNA-binding domain (DBD) fusion. For two-hybrid interaction screening, this plasmid construct was introduced into yeast strain PJ69-4A and yeast genomic DNA libraries Y2HL-C1, -C2, and -C3 (31) were transformed into the bait strain. Two-hybrid interactions were screened by activity of three reporter genes: ability to grow without histidine and in the presence of 3-aminotriazole, ability to grow in the absence of adenine, and β-galactosidase activity.
GST pull-down assays.
PCR-amplified DNA fragments corresponding to either WT or mutant region 4 were cloned in the EcoRI site of the pGEX-KG GST fusion vector (27), and the orientation and reading frames of the inserts were confirmed by DNA sequencing. The fusion proteins were expressed in E. coli BL21 cells and purified on glutathione-agarose beads (Sigma) (21). Expression plasmids containing full-length ORF sequences for TAF4 and TAF6 were expressed in E. coli BL21 by IPTG (isopropyl-β-d-thiogalactopyranoside) induction. After induction, bacterial cells were harvested and cell lysates were prepared. Glutathione-agarose beads containing either glutathione S-transferase (GST) or GST fusion proteins were added, and protein binding was allowed to proceed for 4 h with gentle shaking at 4°C, after which the beads were washed by centrifugation three times with 500 μl of BA150. Proteins in the pellets were subjected to SDS-PAGE and analyzed by immunoblotting with antibodies to Taf4p and Taf6p.
Serial chromatin IP (ChIP-ChIP) assays.
Yeast cells were grown to mid-log phase and cross-linked by adding 37% formaldehyde to a final concentration of 1% (vol/vol). ChIP-ChIP was performed as described previously (57). The first IP was performed using monoclonal antibody (MAb) anti-HA (12CA5) whereas the second IP was with anti-Taf4p polyclonal antibodies. Multiple dilutions of input and pellet samples were used as template for real-time PCR amplification to probe for RPS5 gene promoter sequences with the use of the IQ SYBR Green Supermix and iCycler system (both from Bio-Rad Laboratories). Optimal amplification conditions for all the primer pairs were determined empirically. Results were compared based on the threshold cycle (CT) for PCR amplification and normalized to the signals obtained for the POL1 ORF. Serial dilutions of yeast genomic DNA (Research Genetics) were run in parallel for all assays, and melt curve analyses of final products were performed after each amplification to ensure product quality. Previously described PCR primers for the RPS5 promoter (8) and the POL1 ORF (57) were used.
RESULTS
Deletion of region 4 from Taf1p results in decreased steady-state levels of holo-TFIID.
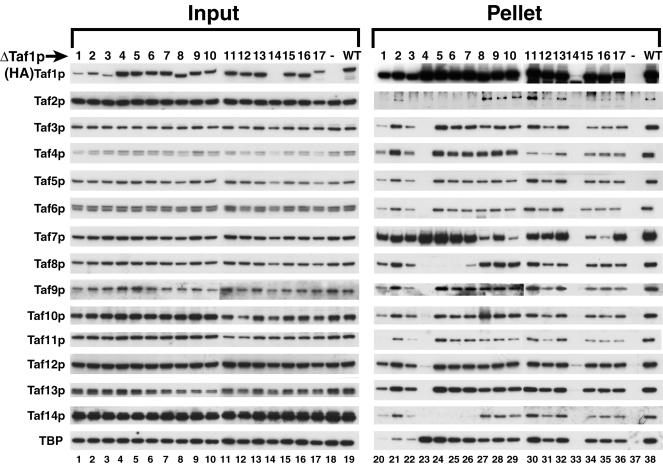
Taf1p is an integral subunit of transcription factor TFIID. In order to elucidate the role that Taf1p plays in TFIID integrity and therefore in TFIID structure and function, we systematically determined which Tafps are associated with different regions of Taf1p by performing co-IP experiments with our collection of 17 deletion mutants that span the entire ORF of TAF1 (5). The strains used for co-IP are pseudodiploid and contain an untagged WT copy of TAF1 in addition to an HA3-tagged deletion variant. WCEs were prepared from the 17 mutant strains, along with two control strains that expressed either an untagged or HA3-tagged WT Taf1p. An aliquot of the inputs was fractionated by SDS-PAGE, blotted, and probed with various anti-Tafp antibodies. The HA-tagged WT and deletion mutant forms of Taf1p were specifically detected [Fig. 1, row marked (HA)Taf1p; compare lanes 1 to 17 and 19, all HA tagged, with lane 18, WT with no tag]. The Δ14 variant of Taf1p (lane 14) is significantly less abundant than other forms of Taf1p. At present we do not know why Δ14Taf1p is 20-fold less abundant (lanes 14 and 33), but this result was reproducibly observed (data not shown). The steady-state levels of the mutant Taf1p proteins were similar to those of WT Taf1p, though mutant Taf1p Δ1, Δ2, and Δ3 were slightly less abundant. All 14 of the other TFIID subunits were present in similar quantities among the strains (Fig. 1, left panel, rows Taf2p to TBP).
FIG. 1.
Deletion of Taf1p-region 4 dramatically reduces steady-state levels of holo-TFIID. WCEs prepared from the indicated yeast strains (Δ1Taf1p to Δ17Taf1p with HA3 tag, WT with no HA3 tag [−], and WT with HA3 tag [WT] [Table 1]) were used for IP with monoclonal anti-HA antibody (MAb 12CA5). One percent of precleared lysate (Input) and 15% of each immunoprecipitate (Pellet) were analyzed by SDS-PAGE and immunoblotting with the antibodies recognizing the TFIID subunits indicated on the left.
TFIID was immunoprecipitated from these WCEs with anti-HA MAb 12CA5, and holocomplex integrity was determined by immunoblotting. The tagged WT and deletion mutant Taf1ps were specifically immunoprecipitated, all with similar efficiencies (compare lanes 20 to 36 and 38, which are tagged, with lane 37, which is not tagged). As expected, the 15 subunits of the holo-TFIID complex specifically coimmunoprecipitated (compare tagged-WT Taf1p, lane 38 immunoprecipitate, with the untagged control in lane 37). Most strikingly, in the deletion mutant Δ4Taf1p IP, only TBP and Taf7p coimmunoprecipitated with Taf1p in normal quantities; essentially no other coprecipitating Tafps were detected (Fig. 1, lane 23), indicating a drastic reduction in steady-state levels of holo-TFIID in this mutant. Such a drastic effect on TFIID integrity was not observed even in the adjoining deletion mutants (compare Δ3-, Δ5-, or Δ6Taf1p with Δ4Taf1p).
The N-terminal Taf1p deletion mutants, Δ1 to Δ3, in contrast to all other deletion mutants had greatly reduced quantities of Taf1p-associated TBP (lanes 20 to 22). These mutant forms of Taf1p also coimmunoprecipitated smaller amounts of certain other Tafps, especially Taf2p, -11p, and -14p. The reduced coprecipitation of TBP with the Δ1Taf1p and Δ2Taf1p deletion mutants is in agreement both with our previous work and with the results of others (5, 7, 37, 52) that showed that TBP avidly binds the N-terminal TAND of Taf1p and that this Tafp is likely the major interaction partner of TBP in the TFIID complex. Moreover, our data indicating that the Taf1p sequences removed by Δ3 are important for Taf1p-TBP interaction (Fig. 1, lane 22 versus lane 38) are entirely consistent with a recent publication by Kokubo and colleagues (73) that mapped a TBP binding domain of Taf1p, termed TAND3 (amino acids 82 to 139). In the co-IPs with Δ5Taf1p, Δ6Taf1p, and Δ7Taf1p, we also observed reduced coprecipitation of Taf2p, Taf8p, and Taf14p (lanes 24 to 26). The concomitant reduction of these three Tafps in the Δ5, Δ6, and Δ7 mutants suggested a possible interaction among Taf2p, -8p, and -14p within the TFIID complex. Although all of the Taf1p variants, ranging from Δ8 to Δ17, were able to coprecipitate all TFIID subunits, the amounts of certain coprecipitated Tafp subunits were reduced (lanes 27 to 36). Taf7p in particular was much reduced in the immunoprecipitates formed with the Δ8, Δ9, Δ10, Δ15, and Δ16 forms of Taf1p (lanes 27 to 29, 34, and 35, respectively).
Based on the results of this co-IP experiment, several important conclusions can be drawn. First and foremost, Taf1p-region 4 plays a key role in the assembly and/or integrity of TFIID, as deletion of region 4 almost completely obliterated the holo-TFIID complex. Second, Taf1p contains separate domains for binding TBP and various Tafps. Third, within the TFIID complex, Taf1p subserves more than a simple structural role because mutants Δ8Taf1p to Δ16Taf1p formed TFIID “holocomplexes” with normal or nearly normal apparent stoichiometry, yet these “TFIID complexes” were unable to support cell growth (5).
Region 4 can nucleate the formation of a partial TFIID complex in vivo.
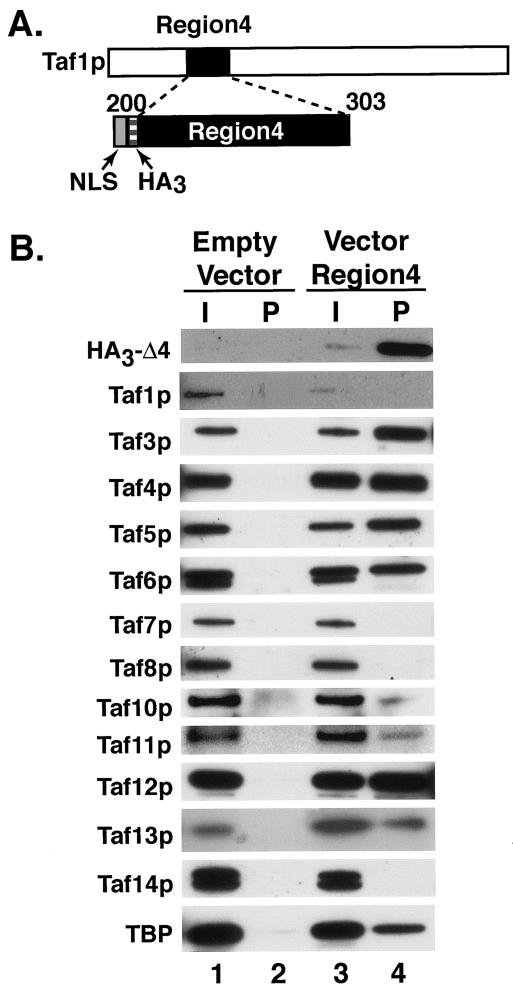
The observed decrease in steady-state levels of holo-TFIID complex in the Δ4Taf1p strain could result either from direct loss of critical TafXp-Taf1p-region 4 sequences or, alternatively, from altered spacing of critical flanking domains that participate in Tafp-Tafp interactions involving these regions (i.e., region 1 and region 3, region 1 and region 5, etc.). To directly assess the role of region 4 sequences in TFIID assembly, we expressed region 4, as an NLS and HA3 fusion protein, in yeast cells under UASGAL control (Fig. 2A) and assayed whether any Tafp could associate with this domain of Taf1p. Empty vector expressing NLS and HA3 tag, but lacking the region 4 ORF, was used as a control. WCEs were prepared from galactose-induced cells, and co-IPs were performed using anti-HA antibody as in Fig. 1. Tagged region 4 polypeptide was specifically and efficiently immunoprecipitated (Fig. 2B, compare lanes 1 and 2 with 3 and 4). Importantly, Taf3p, -4p, -5p, -6p, -10p, -11p, -12p, and -13p and TBP coprecipitated with region 4 (lane 4). It has been hypothesized that human TFIID can dimerize, presumably through direct TBP-TBP interactions (72). To ensure that the Tafp enrichment observed in this co-IP experiment was not due to contaminating TFIID dimerized to coprecipitating TBP, quantitative immunoblot analyses were performed with these two immunoprecipitates (data not shown). The enrichment of Tafp bands in the region 4 pellet (Fig. 2, lane 4) compared to that in the empty vector pellet was as high as 130-fold, whereas TFIID-specific Tafps, Taf1p, Taf2p, Taf7p, and Taf8p, were either absent or consistently near background levels in the region 4 immunoprecipitate. Based upon these data, we concluded that the Tafps associated with Taf1p-region 4 in this experiment did not originate from TFIID contamination but instead represented a subset of TFIID-Tafps that directly associated with region 4 to form a partial TFIID-like complex(es). These results also suggested that the loss of holo-TFIID in the Δ4Taf1p strain was likely due to the direct loss of this TAF-binding domain from Taf1p and was not caused by spacing changes between flanking regions. Taken together, the results of the experiments presented in Fig. 1 and 2 suggest that region 4 of Taf1p is essential for TFIID assembly and integrity.
FIG. 2.
The Taf1p-region 4 polypeptide alone nucleates assembly of a partial TFIID complex in vivo. (A) Schematic representation of Taf1p (top) and region 4 (amino acid residues 200 to 303; bottom). The gray and the hatched boxes represent the NLS (PKKKRKV) and HA3 tag, respectively, of the expression vector fused to region 4 coding sequence and expressed from a GAL enhancer-promoter on a 2μm plasmid. (B) Co-IP analysis of TAF proteins associated with Taf1p-region 4. WCEs were prepared from yeast cells expressing empty vector containing the NLS and HA3 tag only (Empty Vector) or vector-NLS-HA3-region 4 sequence (Vector Region4) and precleared with protein A beads. Immunoprecipitates were formed from these precleared WCEs by using anti-HA immunoglobulin G bound to protein A beads. The resulting immunoprecipitates were fractionated by SDS-PAGE, blotted, and probed with specific antibodies as indicated (left). Input (I) lanes represent 1% of the total WCE used for each IP; pellet (P) lanes represent the total immunoprecipitated material.
Mutations in region 4 confer both temperature-conditional (ts) growth phenotypes and transcription defects.
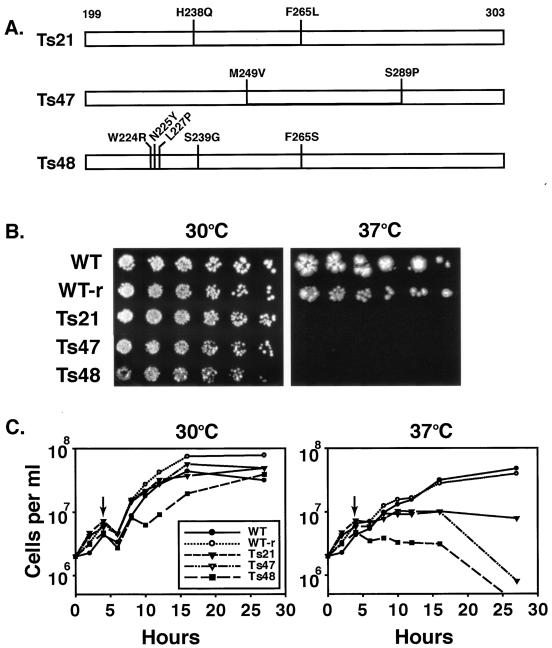
Taf1p-region 4 alone was capable of assembling several Tafps into a partial TFIID complex, presumably through the formation of direct specific Taf1p-region 4-Tafp interactions. We reasoned that, if this were true, then these interactions should be mutationally sensitive. To test this prediction, we generated a library of random mutations in the ORF encoding Taf1p-region 4 via error-prone PCR. After screening of the resulting mutants for ts growth, three ts alleles of TAF1 termed Ts21, Ts47, and Ts48 were selected for further study. DNA sequence analysis of these mutant TAF1 genes revealed that each carried multiple point mutations that mapped throughout the targeted region (Fig. 3A). The mutant strains were viable at a permissive temperature, growing essentially as WT (Fig. 3B), although the Ts48 mutant had a slightly lower growth rate, while all three failed to grow at a nonpermissive temperature. Inviability at 37°C was recessive to WT since ts growth was rescued by introducing a plasmid carrying the WT TAF1 allele (data not shown). Similar results were observed in liquid cultures (Fig. 3C). These data indicated that region 4 of TAF1 represented an independently mutable domain and that mutations in this region induced ts growth at 37°C, results that supported the notion that this domain plays an important role in cellular function, likely through effects upon Tafp-Tafp interactions and thus TFIID integrity.
FIG. 3.
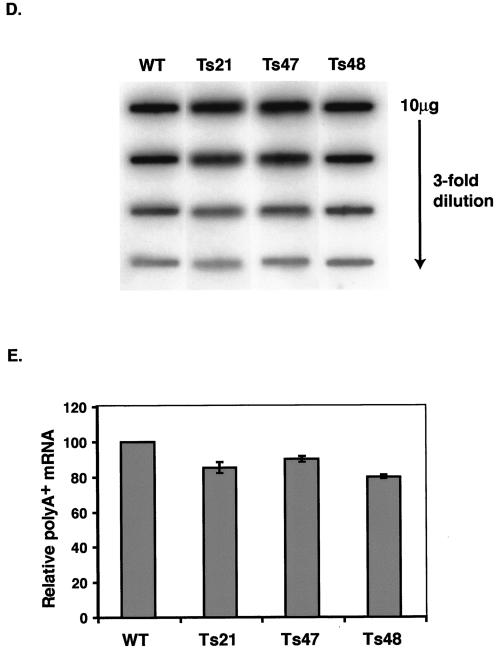
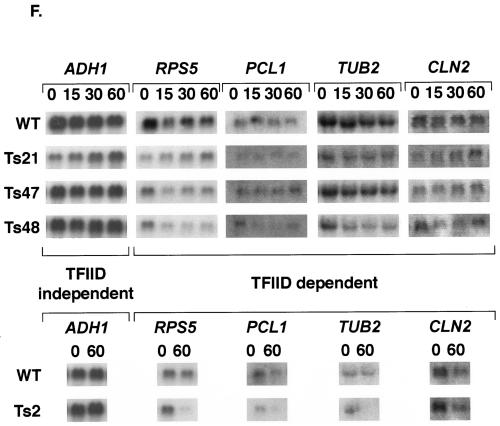
Mutations in Taf1p-region 4 induce both temperature-conditional growth and transcriptional defects. (A) Location of mutations induced by error-prone PCR in three different variants of Taf1p-region 4. The three white bars represent the Taf1p-region 4 coding region corresponding to amino acids 199 to 303, and the location and identities of the WT and mutated amino acids are marked by dark vertical lines where the letters and numbers indicate the positions of mutations (first letter is WT residue and last letter is mutated residue) in Ts21, Ts47, and Ts48. (B) Growth characteristics of WT and mutated strains at permissive (30°C) and nonpermissive (37°C) temperatures. Equal numbers of cells from each of the indicated strains (left) were serially threefold diluted and spotted on minimal medium (increasing dilutions from left to right); plates were incubated at the temperatures indicated for 4 days (30°C) or 7 days (37°C) and photographed. WT, WT cells; WT-r, reconstructed WT strain; Ts21, Ts47, and Ts48, region 4 mutant yeast strains. (C) Effect of shift in temperature on growth of WT and mutant cells in liquid cultures. Equal numbers of cells were grown to log phase at permissive temperature, split into halves, and harvested by centrifugation, and each culture was transferred to either 30 or 37°C as shown at the top; arrows indicate the time point at which cells were pelleted and resuspended at 30 or 37°C. Cell counts were determined at the times indicated using a hemocytometer. (D) Slot blot hybridization of total RNA to measure poly(A)+ mRNA content. Serial threefold dilutions of RNA (10 to 0.33 μg) were blotted and probed with end-labeled 32P-oligo(dT20). Slot blot analyses were done in triplicate, and intensities from 3.3 to 0.33 μg of RNA spots were averaged; only a single analysis for each strain is shown. (E) Normalized amounts of poly(A)+ RNA in yeast strains were obtained by calculating the ratio of the slot blot intensities to the relative amounts of RNA recovered from equal numbers of cells in each strain (WT, 35.5 pg/cell, 100%; Ts21, 110.4%; Ts47, 106.9%; and Ts48, 113.6%). (F) Effect of Taf1p-region 4 ts mutations on TFIID-dependent gene transcription. (Top) Effect of temperature shift on transcription of various mRNAs in cells expressing WT and region 4 mutant alleles. Total RNA was prepared from cells grown for the indicated time (in minutes) at 37°C. Equal microgram amounts of these RNAs were gel fractionated, blotted, and probed with the 32P-labeled DNA probes indicated. Fold changes were normalized to total rRNA scored by staining blots with methylene blue. (Bottom) RNA blot analysis of TAF1 ts2 mutant and its congenic WT strain. RNA samples prepared from cells at time zero and 60 min after temperature shift to 37°C were gel fractionated and probed with radiolabeled cDNA probes as described above.
The ts growth phenotype of the three mutants could be due to the loss of Taf1p function in mRNA synthesis; this was tested in two ways. First, we determined total RNA content and the total poly(A)+ mRNA content of each of these strains. Total RNA was isolated from equal numbers of cells, and mRNA content was measured by slot blot hybridization with 32P-labeled oligo(dT20) as a probe (Fig. 3D). When normalized to the total RNA content of the cells, the mutant cells contained slightly reduced amounts of poly(A)+ mRNA (Fig. 3E). Poly(A)+ mRNA was 80% of the WT levels in Ts48 followed by Ts21 (85% of the WT level) and Ts47 (90% of the WT level). We next used Northern blot analyses to test for the effects of temperature shift on PolII-catalyzed TFIID-dependent mRNA gene transcription (Fig. 3F). Strain Ts21 had greatly reduced transcript levels of PCL1 (eightfold) and TUB2 (sixfold) 60 min after shift to restrictive temperature. In Ts48, transcripts for RPS5, PCL1, TUB2, and CLN2 were reduced approximately 2-, 3-, 2.5-, and 6-fold, respectively, while the Ts47 mutant displayed reduced transcript levels for RPS5 and CLN2. As a control we used the ts2 mutant allele of TAF1, a mutant that has been well characterized elsewhere (67). In ts2 cells, steady-state levels of RPS5 mRNA dropped to undetectable levels after 60 min of growth at restrictive temperature but were unaffected in the congenic WT strain (Fig. 3F). Expression of several other genes was also tested by reverse transcription coupled with real-time quantitative PCR with similar results (data not shown). The results of these experiments showed that, in addition to a general decrease in the poly(A)+ mRNA contents of the mutant yeast strains, TAF1 allele-specific transcription differences also existed. These data were consistent with the idea that the region 4 domain of Taf1p contributes importantly to Taf1p and TFIID function.
The TFIID complex is unstable in the ts mutants.
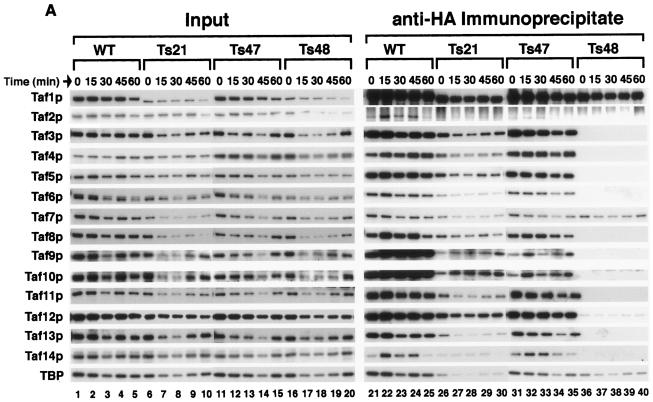
To test whether the integrity of the TFIID complex was affected in the ts mutants, co-IP assays were conducted using WCEs prepared from cells before and after shift to 37°C. The cells used in these experiments were from the same cultures used for the mRNA analyses described above (Fig. 3). Tafp stability was relatively insensitive to shift to 37°C (Fig. 4A, Input, lanes 1 to 20), although Taf1p steady-state levels were lower in Ts21 and Ts48 cells (compare Taf1p signal in lanes 5, 10, 15, and 20). Taf1p was efficiently immunoprecipitated from all strains (lanes 21 to 40). In the WT yeast strain all TFIID subunits coprecipitated with Taf1p (lanes 21 to 25). In contrast, with the mutants, the integrity of the TFIID holocomplex was affected to various degrees. The Ts47 mutant, like WT, showed coprecipitation of all TFIID subunits (lanes 31 to 35), whereas with Ts48, only TBP and Taf7p (and trace amounts of Taf12p) coprecipitated with Taf1p (lanes 36 to 40) even at time zero, indicating a very strong effect of the five mutated residues of region 4 in Ts48 on Tafp-Tafp interactions (Fig. 4A, lane 36). The association of Taf1p with only Taf7p and TBP but not the other Tafps is consistent with the co-IP results seen for Δ4Taf1p in Fig. 1 (lane 23). With Ts21, an intermediate effect was observed; decreased Taf1p association of Taf6p, Taf9p, Taf10p, Taf13p, and Taf14p and TBP was observed (Fig. 4A, lanes 26 to 30). From these co-IP experiments, it appeared that, in the ts mutants, especially the Ts48 mutant, TFIID had either dissociated into “free” Tafps or had split into discrete subcomplexes of Tafps of unique constitution.
FIG. 4.
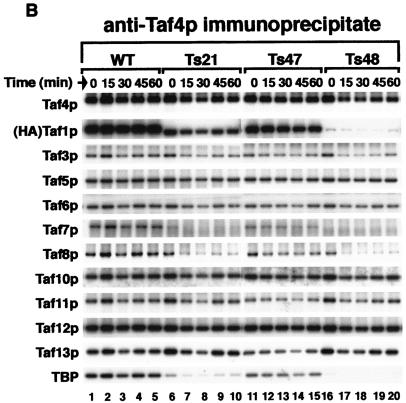
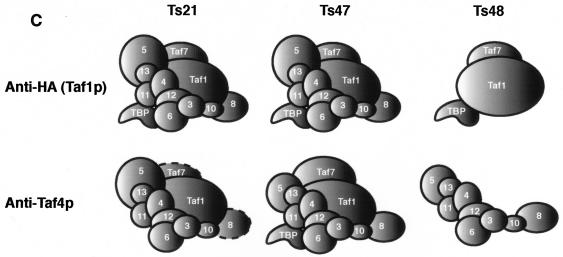
Effect of temperature shift on TFIID integrity in WT and TAF1 region 4 mutant strains. WT and ts mutant cells (as indicated, top) were grown to log phase at 30°C and shifted to 37°C. Cells were collected at 0, 15, 30, 45, and 60 min after the temperature shift (top), WCEs were prepared and normalized to total protein content, and equal amounts of total protein were used for co-IP assays with anti-HA antibody (MAb 12CA5) (A) or anti-Taf4p antibodies (B). Immunoprecipitates were fractionated by SDS-PAGE and blotted, and specific antibodies (left) were used to probe the blots as detailed for Fig. 1 and 2. The same precleared lysate was used for all IPs. Input lanes represent 1.5% of the total precleared lysate; pellet lanes contain approximately 15% of the immunoprecipitate. (C) Summary of the results of the two sets of co-IP experiments; dashed lines denote reduced amounts of association as scored by co-IP.
If the latter hypothesis were correct, then we should be able to detect such subcomplexes by appropriate co-IP studies. Consequently, we conducted co-IP experiments with antibodies to Taf4p, using the same WCEs as used in the Fig. 4A experiment. Taf4p was selected for several reasons: it is TFIID specific, total Taf4p levels did not change over the time course of this experiment, Taf4p was dissociated from Taf1p in the Ts48 mutant, and finally, Taf4p has been shown by others to be able to interact with other Tafps (60, 66, 74). The results of this anti-Taf4p co-IP study are shown in Fig. 4B. Taf4p was efficiently immunoprecipitated from all strains (Fig. 4B, lanes 1 to 20, rows labeled Taf4p). In the WT, all the tested Tafp subunits efficiently coprecipitated with Taf4p (lanes 1 to 5). Similarly, and in agreement with the previous co-IP with Ts47, all the tested Tafps coprecipitated with Taf4p (Fig. 4B, lanes 11 to 15). In the Ts48 samples, neither Taf1p, Taf7p, nor TBP efficiently coprecipitated with Taf4p (Fig. 4B, lanes 16 to 20) while all the other TFIID Tafps did. These results are consistent with the notion that TFIID in Ts48 cells could split into at least two subcomplexes, one containing Taf1p, TBP, and Taf7p and the other containing Taf4p and the remaining Tafps (compare Fig. 4A, lanes 36 to 40, and Fig. 4B, lanes 16 to 20). Slightly less Taf7p, Taf8p, and TBP were Taf4p associated in Ts21 WCE (lanes 6 to 10). These results for Ts21 were somewhat surprising, since this mutant did not display drastic dissociation of Tafps in the Taf1p co-IP experiment presented in Fig. 4A.
A schematic summarizing the results of these co-IP experiments is presented in Fig. 4C. The data are consistent with the idea that mutation of Taf1p-region 4 resulted in an unstable TFIID complex and that Tafps could dissociate from the TFIID complex at a nonpermissive temperature. Indeed, mutations in the Ts48 mutant had the most profound effect on TFIID integrity, apparently “splitting” TFIID into two subcomplexes under the conditions of our co-IP experiments. This effect was observed even when WCEs were prepared from cells grown at a permissive temperature where no large growth deficiencies were observed. Decreased affinities of Tafp-Tafp association could explain the conditional growth and transcriptional defects of our ts-TAF1 strains.
Taf1p and Taf4p co-occupy the RPS5 promoter in Ts48 cells.
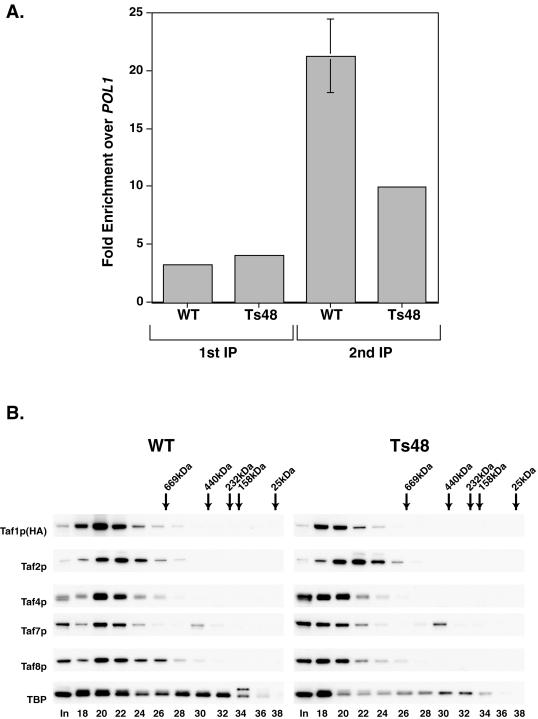
To further explore the hypothesis that region 4 mutation decreased Taf1p-Tafp affinity-stability, we conducted an in vivo serial ChIP-ChIP assay. This ChIP-ChIP study should allow us to test whether both Taf1p and Taf4p (putative cognate subcomplexes [Fig. 4C]) resided upon the same promoter in WT and Ts48 cells. For ChIP-ChIP assays, we first immunoprecipitated Taf1p using anti-HA antibody followed by elution of the immunoprecipitated chromatin with HA peptide and a second IP with anti-Taf4p antibodies. The occupancy of TFIID on the constitutively expressed TFIID (Taf1p)-dependent RPS5 gene promoter was determined by quantitative real-time PCR. We reasoned that, if in vivo TFIID was intact on gene promoters, then both Taf1p and Taf4p should be cross-linked to the promoter in WT and Ts48 cells. Such promoter occupancy in vivo is readily scored by quantitative PCR analysis of the immunoprecipitated DNAs. We observed that comparable amounts of DNA were coprecipitated with Taf1p in the WT and Ts48 mutant samples with anti-HA antibody (Fig. 5A). In the second IP performed with anti-Taf4p antibody, we again observed enrichment of the RPS5 promoter in both strains, indicating that, in a significant fraction of cells, both Taf1p and Taf4p are resident on the RPS5 promoter in strain Ts48. These data are consistent with the idea that Ts48 cells actually contain an intact TFIID holocomplex.
FIG. 5.
TFIID is intact in WT and Ts48 cells. (A) Measurements of Taf1p and Taf4p co-occupancy on the RPS5 gene promoter analyzed by sequential ChIP (ChIP-ChIP). Sheared formaldehyde cross-linked WT and Ts48 chromatin preparations were subjected to a first IP with anti-HA (Taf1p) antibody. Immunoprecipitated Taf1p-DNA complexes were eluted with HA peptide, and a second IP was performed on this eluted material with anti-Taf4p antibodies. Aliquots of both immunoprecipitates were analyzed by real-time quantitative PCR, and results were normalized and plotted as RPS5 gene promoter enrichment over DNA PolI (POL1) ORF. (B) Gel filtration of TFIID extracted from WT and Ts48 cells. Equal aliquots of alternate fractions from gel filtration chromatography of WT and Ts48 WCE were trichloroacetic acid precipitated, fractionated by SDS-PAGE, and blotted, and different TFIID-specific Tafps were detected with specific antibodies (left). The WCE input (I) and column fractions analyzed are indicated (bottom); the elution positions of various size markers run in parallel are shown (top).
WCE prepared from Ts48 cells contains intact TFIID.
To directly address the question of TFIID holocomplex integrity in WT and Ts48 cells, we performed gel filtration chromatography experiments to size TFIID in WCEs derived from these two cell types. If the TFIID complex was split into two subcomplexes as observed in the co-IP experiments, then Taf1p and Taf4p would not coelute. TFIID elution was monitored by immunoblotting column fractions with antibodies to five TFIID-specific subunits. We found that TFIID eluted in the same fractions in both the WT and the Ts48 strains (Fig. 5B) with elution profiles consistent with our earlier sizing analyses of TFIID (62). Thus, results from two different and independent assays, ChIP-ChIP and gel filtration, indicated that the TFIID complex in the Ts48 mutant was in fact intact even though our co-IP experiments (Fig. 4) showed that there were significant differences in the strength of Tafp-Tafp interactions between the WT and mutated Taf1p-region 4.
Region 4 directly binds to Taf4p and Taf6p.
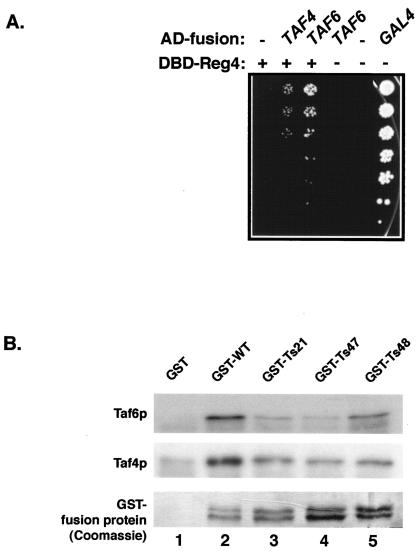
To reveal the identity of the Tafp(s) that might be directly interacting with Taf1p, we performed systematic two-hybrid screens using an overlapping family of 11 gene fragments spanning the entire TAF1 ORF sequentially fused to the DBD of Gal4p as “bait.” Yeast genomic DNA Gal4p-activation domain (AD) fusion libraries (31) were screened against this family of 11 DBD-Taf1p bait molecules. Only those proteins showing interaction with the Taf1p-region 4 DBD bait are discussed here. Roughly 107 individual colonies were screened, and two Tafp-encoding genes, TAF4 and TAF6, were identified. DNA sequencing indicated that both TAF4 and TAF6 AD fusion clones carried essentially full-length genes (data not shown). The interactions of Taf4p and Taf6p with Taf1p-region 4 were specific, as both required bait and prey plasmids (Fig. 6A). The two-hybrid bait used for these screens lacked 31 amino acid residues at the N-terminal end and had an additional 92 amino acid residues at its C-terminal end compared to the deletion mutation in Δ4Taf1p. The fact that Taf1p-region 4 interacts with other Tafps is consistent with the hypothesis that region 4 of Taf1p encodes sequences essential for TFIID integrity and function.
FIG. 6.
Direct interaction of Taf1p-region 4 with Taf4p and Taf6p. (A) Yeast two-hybrid interactions between DBD-Taf1p-region 4 bait and Taf4p- and Taf6p-AD fusion proteins. Yeast strains harboring either no bait (−) or a Taf1p-region 4 (Reg4)-containing DBD bait (amino acids 235 to 395) and either empty AD vector (−), AD-Taf4p, or AD-Taf6p were plated on appropriate selection medium containing 3 mM 3-aminotriazole. Equal numbers of cells for each strain were serially threefold diluted and spotted on plates (dilutions top to bottom). Cells expressing full-length Gal4p served as a positive control. (B) Taf4p and Taf6p interact directly with (GST-)region 4. E. coli carrying either Taf4p or Taf6p expression plasmids was induced with IPTG as detailed in Materials and Methods, and bacterial cell extracts were prepared. Equal amounts of precleared E. coli cell lysates were incubated with equal amounts of GST or region 4-GST fusion proteins (bottom) bound to glutathione-agarose beads. Pellets containing GST-bound proteins were fractionated by SDS-PAGE, blotted, and probed for Taf6p (top) or Taf4p (middle) binding by using antibodies to either Taf6p or Taf4p (left).
To test whether the interactions observed in our two-hybrid assays directly involved Taf1p-region 4 amino acids (i.e., amino acids 200 to 303) identified by our genetic and biochemical assays, we conducted a series of GST pull-down experiments. Recombinant Taf4p and Taf6p were expressed in E. coli, and the resulting cell extracts were incubated with either bead-bound GST alone or bead-bound region 4-GST fusion proteins (WT and Ts21, Ts47, and Ts48) for pull-down assays (Fig. 6B). Tafp-Taf1p-region 4 interactions were scored by SDS-PAGE and immunoblotting of bead-bound proteins. Taf4p specifically interacted with GST-region 4 protein (Fig. 6B, compare lanes 2 to 5 with lane 1). Interestingly, all the mutant region 4-GST-fusion proteins reproducibly bound less Taf4p (12 to 55% of WT level) than did the WT protein, indicating an effect of these mutations on the affinity of Tafp-Tafp interactions. Similar results were obtained with Taf6p; we observed that less Taf6p associated with mutant GST-region 4 than with WT GST-region 4. Taken together, the data presented in Fig. 6 support the hypothesis that at least two integral TFIID Tafps directly and specifically interact with region 4 of Taf1p, results consistent with the notion that this domain of Taf1p contributes critically to TFIID stability and hence function.
DISCUSSION
Structural insights into the organization of the TFIID complex.
Since our first report in 1997 on the structure-function relationships of TAF1 (5), a more complete picture of the TFIID complex has emerged. New TFIID-specific Tafps have been identified (33, 34, 60, 65, 75), novel Tafp-Tafp interactions have been described (22, 23, 63, 66, 81), and low-resolution three-dimensional structures of the TFIID complex have been determined (2, 11, 39, 40, 48). In this study, we have performed a comprehensive analysis of all the TFIID components associated with Taf1p using co-IP assays and our family of Taf1p deletion mutants that encompass the entire TAF1 ORF. As monitored by this assay, full-length WT Taf1p efficiently precipitated along with TBP and the 13 additional Tafp subunits that represent holo-TFIID (Fig. 1). Certain deletion mutants exhibited significantly reduced amounts of holo-TFIID. Based on these co-IP results, a more detailed picture of Taf1p-TBP (5, 7, 73) and Taf1p-Tafp subunit interactions has emerged. Our biochemical and genetic approach for studying the role of Taf1p in TFIID structure, as detailed in this report, has been fruitful, not only confirming known subunit-subunit interactions but also providing new information regarding the physical and structural organization of these subunits within TFIID.
It is clear that in the context of the TFIID complex TBP interacts predominantly with the N-terminal domain of Taf1p. This interaction has been the subject of intense investigation, and results of our systematic Taf1p (WT and mutant)-TBP co-IP analyses are consistent with previous results from genetic and biochemical studies as well as our recent three-dimensional structure of yeast TFIID (40). For example, our co-IP results showing that less TBP is coprecipitated with Δ3Taf1p predicted additional interactions between Taf1p and TBP over and above TAND-TBP binding. This prediction has now been independently confirmed by a report published while the manuscript was being revised. Kokubo and colleagues showed that another Taf1p sequence termed TAND3 (amino acids 82 to 139) interacts with TBP (73). Part of TAND3 is deleted in our Δ3Taf1p mutant (amino acids 101 to 208 deleted). Thus, our co-IP results have strong predictive value for the structural features of TFIID organization.
Similarly, our data have afforded us greater structural insight into Taf1p-Taf7p interactions within the TFIID complex. Taf7p interaction with the C-terminal portion of Taf1p has previously been reported in yeast two-hybrid interaction (81) and recently by electron microscopy structural mapping (40). However, our co-IP experiments on Taf1p further extend these findings by showing that Taf7p makes multiple contacts with the C-terminal end of Taf1p (Fig. 1). In addition, Taf7p's interaction with Taf1p is independent of the remaining TFIID subunit Tafp-Tafp interactions mediated through Taf1p-region 4, since in Δ4Taf1p co-IP experiments normal amounts of Taf7p coprecipitated with Δ4Taf1p (Fig. 1). Moreover, the co-IP results with Δ4-, Δ 5-, and Δ6Taf1p also demonstrated a possible interaction among Taf2p, Taf8p, and Taf14p subunits of TFIID, which to our knowledge has not yet been described. Thus, using our co-IP approach, we have unveiled novel structural features of Tafp-Tafp interactions in the TFIID complex.
Haploid yeast cells containing Taf1p mutants Δ8-, Δ9-, Δ10-, Δ11-, Δ12-, Δ13-, Δ15-, and Δ16Taf1p are inviable despite the association of all TFIID subunits with the mutant Taf1ps. Therefore, in addition to contributing critically to TFIID assembly and TFIID stability, Taf1p must also play other significant nonstructural roles in TFIID function. The HAT activity of yeast Taf1p has been roughly mapped to the central conserved domain (amino acids 354 to 817) of the protein (46). Portions of the central conserved domain are deleted in mutants Δ5- to Δ13Taf1p, thus suggesting that this HAT function could be one of the nonscaffold functions of Taf1p. The Taf1p domain defined by Δ16 has been implicated in promoter (DNA) binding by others (44). Taken together, these results underscore the modular nature of this protein. Taf1p contains at least five functional domains: the composite tripartite N-terminal TAND that binds TBP; a TFIID assembly domain (localized around region 4); a Taf7p binding domain, distributed through region 8, region 10, region 15, and region 16; a HAT enzyme active center that abuts the TFIID assembly domain (region 5 to region 13); and a promoter recognition domain (region 16). Collectively, these data indicate that Taf1p plays multiple critical roles in TFIID function. The most important structural insight from our experiments has been the discovery that Taf1p-region 4 plays an essential role in TFIID assembly and stability.
Taf1p-region 4 is essential for TFIID assembly and stability.
The assembly-stability or scaffold function of Taf1p critically depends on region 4, since deletion of this domain resulted in the loss of holo-TFIID. This loss of TFIID integrity was not due to nonspecific or trivial effects; the Taf1p protein encoded by taf1-Δ4 was not simply globally unfolded because both Taf7p and TBP retained the ability to associate with Δ4Taf1p. Further supporting a specific scaffold function for region 4 are our data showing that, when expressed in yeast cells, Taf1p-region 4 alone nucleated the assembly of several Tafps into a partial TFIID complex (Fig. 2), a result that supported a direct role for this domain of the protein in the scaffold-assembly function of Taf1p. Comparable detailed protein-protein interaction studies with metazoan Taf1p have not yet been reported; thus, it remains to be determined whether a similar assembly domain exists in higher eukaryotes. A small amount of TBP coprecipitated with the TFIID subcomplex formed on Taf1p-region 4, most likely through interactions with Taf6p and/or Taf12p (58, 60). In this regard, it is notable that C-terminal truncation mutants of Taf1p that contain region 4 sequences have been found to form TFIID-like subcomplexes (18, 44), though in these experiments not all the TFIID-Tafps were probed for. In light of our data, we assume that region 4 most likely contributed critically to the assembly of these TFIID subcomplexes. Surprisingly, in the study by Collart and colleagues (18), TBP was dissociated from the complex despite the presence of the TAND. Further experimentation will be needed to understand whether these partial TFIID complexes are functional in transcription.
The possibility of Tafp contamination through TFIID dimerization (72) in our Taf1p-region 4 co-IP experiments was ruled out through quantitative immunoblotting studies. These experiments showed that not all the TFIID-specific Tafps coprecipitated with Taf1p-region 4: for example, Taf1p, Taf2p, and Taf8p were not detected in these immunoprecipitates. We also note that no such dimer has yet been detected for yeast TFIID by co-IP (14, 62), TFIID molecular sizing (62), or electron microscopy (39, 40).
It is also notable that the expression of Taf1p-region 4 had no dominant-negative effect upon growth even when a Leu2d-based high-copy-number yeast expression vector (61) with a highly inducible Gal promoter was used (data not shown). This result is likely due to the fact that, even after galactose induction, cells carrying this Leu2d-region 4 plasmid did not accumulate large quantities of the region 4 polypeptide. We estimated the expression of region 4 polypeptide in the induced cells by quantitative immunoblotting and found it to be ≤1,000 molecules per cell, only 20 to 30% the level of TFIID. We assume that the low steady-state levels of region 4 peptide reflect the fact that region 4-Tafp complexes are less stable than the holo-TFIID complex because high-salt washes of these immunoprecipitates induce greater complex dissociation compared to that for the full-length Taf1p (i.e., TFIID [data not shown]).
Consistent with the essential role played by Taf1p-region 4 in TFIID assembly, integrity, and cell viability, we were readily able to generate temperature-conditional mutant alleles of TAF1 that had molecular lesions in this region of the ORF. Co-IP assays performed with these mutant strains revealed striking effects of these mutations on the integrity of the TFIID complex, effects that ranged from negligible in the Ts47 mutant to profound in the Ts48 mutant. These data are summarized in Fig. 4C, which depicts the results of co-IP experiments generated by using antibodies to Taf1p (anti-HA) and Taf4p. Although in the co-IP experiments several Tafps, including Taf4p, were found to be dissociated from holo-TFIID in a Ts48 WCE, ChIP-ChIP assays showed that both Taf1p and Taf4p occupied a significantly large proportion of the RPS5 promoters in Ts48 cells, suggesting the presence of an intact TFIID complex in vivo. Gel filtration chromatography of WCE prepared from WT and Ts48 cells further proved that indeed there was intact TFIID holocomplex in these cells and that no partial TFIID complex existed in Ts48 extracts, consistent with our own previous published work (62, 63). We conclude that, in the present study, the disruptive effects of mutations in Taf1p-region 4 were accentuated in our co-IP experiments. While slightly different buffers were used in the co-IP and gel filtration experiments, the differences between the co-IP and gel filtration results seem to originate from the reduced stability of the TFIID complex during the co-IP studies, because only TBP and Taf7p were associated with Taf1p of the Ts48 mutant when either buffer was used in co-IP experiments (data not shown). In this regard, it is important to note that the duration of gel filtration chromatography is only 45 min compared to the approximately 16-h duration of the co-IP. We believe that mutations in Taf1p-region 4 have significantly weakened Tafp-Tafp interactions and that these weakened interactions lead to dissociation of the TFIID complex during the co-IP experiments. We assume that transferring Ts21, Ts47, and Ts48 cells to 37°C exacerbates this decreased strength of Tafp-Tafp interactions.
Given the functional nexus of multiple Tafps with Taf1p-region 4, it is important to understand how this region contributes to the TFIID assembly process. To assess this, we conducted two-hybrid assays with a Taf1p fragment encompassing region 4; we found that this bait interacted with Taf4p and Taf6p (Fig. 6). GST pull-down experiments confirmed the direct interaction of Taf1p-region 4 with both Taf4p and Taf6p. These data indicate that Taf1p-region 4 interacts with at least these two Tafps to help form the TFIID holocomplex. The association of a TFIID-specific Tafp (Taf4p) and a SAGA-TFIID-shared Tafp (Taf6p) could play a critical role in the decision to assemble shared Tafps into either TFIID or SAGA.
Both human and yeast TFIID display a trilobed structure (2, 11, 39), and the HFD-containing Tafps have been mapped to these lobes (39). Based upon our data, TFIID assembly can be considered either as a cooperative process that is nucleated on Taf1p or as a modular process where a subcomplex composed of TBP, Taf1p, and Taf7p associates with another subcomplex(es), perhaps comprised of the remaining Tafps binding either pairwise as dimers (i.e., HFDTafX-HFDTafY) (39) or as a Taf-octamer-like core (66). Our co-IP results with the region 4 mutants appear consistent with the trilobular models of TFIID structure (39, 40). Thus, the apparent “splitting” of TFIID can be viewed as dissociation of both A and B lobes from a Taf1p-Taf7p-TBP “core,” in the case of Ts48 (Fig. 4A), while in the case of Ts21, only one of these lobes dissociates (Fig. 4A). We do not presently know whether the partial TFIID complex observed in the Ts48 mutant is a bona fide in vivo assembly intermediate or simply the result of in vitro dissociation of holo-TFIID. Our future work will address this issue.
The Ts47 mutant strain exhibited ts growth although TFIID integrity in these cells was unaffected by shift to the restrictive temperature. It is possible that critical conformational changes in TFIID, required for proper functioning (17, 20, 22, 28, 30, 32, 50, 53, 71), are affected by the mutations in Taf1p-region 4 in the Ts47 mutant. Alternatively, despite the fact that these mutations did not result in total dissociation of the Tafps, they might have sufficiently weakened Tafp-Tafp associations such that the resulting complex displays functional defects. Obviously, these models are not mutually exclusive. It is important to note that none of the isolated mutants had a single point mutation, suggesting that multiple amino acid residues likely participate in specific Tafp-Tafp interactions important for TFIID assembly, integrity, and/or function. The most severe effect on TFIID integrity was observed in Ts48, a mutant that contains five missense mutations. We observed effects on the steady-state transcript levels of several TFIID-dependent genes when cells carrying region 4 mutations were grown at the restrictive temperature. Mutations in this region of Taf1p may have broader effects on global transcription in yeast. Planned gene array studies will examine whether the various mutants of Taf1p-region 4 differentially affect global mRNA gene transcription.
In summary, our study has systematically documented the scaffold function(s) of yeast Taf1p. Further, our work has contributed important new information regarding the structural organization of TFIID and the critical role played by Taf1p generally and Taf1p-region 4 specifically in this process. Our future studies will be directed at understanding the effects of region 4 mutations on transcription, as well as elucidating in detail the role of region 4 in the structure and organization of TFIID.
Acknowledgments
We thank the members of our lab for valuable suggestions concerning our experiments as well as constructive feedback during the preparation of the manuscript. We thank Michael Green for yeast strains.
This study was supported by NIH grant GM52461.
REFERENCES
- 1.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Andel, F., III, A. G. Ladurner, C. Inouye, R. Tjian, and E. Nogales. 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153-2156. [DOI] [PubMed] [Google Scholar]
- 3.Apone, L. M., C. A. Virbasius, F. C. Holstege, J. Wang, R. A. Young, and M. R. Green. 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2:653-661. [DOI] [PubMed] [Google Scholar]
- 4.Apone, L. M., C. M. Virbasius, J. C. Reese, and M. R. Green. 1996. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10:2368-2380. [DOI] [PubMed] [Google Scholar]
- 5.Bai, Y., G. M. Perez, J. M. Beechem, and P. A. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bando, M., S. Ijuin, S. Hasegawa, and M. Horikoshi. 1997. The involvement of the histone fold motifs in the mutual interaction between human TAFII80 and TAFII22. J. Biochem. (Tokyo) 121:591-597. [DOI] [PubMed] [Google Scholar]
- 7.Banik, U., J. M. Beechem, E. Klebanow, S. Schroeder, and P. A. Weil. 2001. Fluorescence-based analyses of the effects of full-length recombinant TAF130p on the interaction of TATA box-binding protein with TATA box DNA. J. Biol. Chem. 276:49100-49109. [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus, A. C. Lavigne, I. Davidson, and D. Moras. 1998. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94:239-249. [DOI] [PubMed] [Google Scholar]
- 10.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 11.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 12.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 13.Burke, T. W., P. J. Willy, A. K. Kutach, J. E. Butler, and J. T. Kadonaga. 1998. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harbor Symp. Quant. Biol. 63:75-82. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, K. M., R. T. Ranallo, L. A. Stargell, and K. J. Lumb. 2000. Reevaluation of transcriptional regulation by TATA-binding protein oligomerization: predominance of monomers. Biochemistry 39:2633-2638. [DOI] [PubMed] [Google Scholar]
- 15.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 17.Chi, T., and M. Carey. 1996. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 10:2540-2550. [DOI] [PubMed] [Google Scholar]
- 18.Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler, M. Lemaire, N. Paquet, and M. A. Collart. 2002. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 20.Ellwood, K., W. Huang, R. Johnson, and M. Carey. 1999. Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol. Cell. Biol. 19:2613-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179-187. [DOI] [PubMed] [Google Scholar]
- 22.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 23.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gangloff, Y. G., S. Werten, C. Romier, L. Carre, O. Poch, D. Moras, and I. Davidson. 2000. The human TFIID components TAFII135 and TAFII20 and the yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 26.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 27.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 28.Hai, T. W., M. Horikoshi, R. G. Roeder, and M. R. Green. 1988. Analysis of the role of the transcription factor ATF in the assembly of a functional preinitiation complex. Cell 54:1043-1051. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, A., C. M. Chiang, T. Oelgeschlager, X. Xie, S. K. Burley, Y. Nakatani, and R. G. Roeder. 1996. A histone octamer-like structure within TFIID. Nature 380:356-359. [DOI] [PubMed] [Google Scholar]
- 30.Horikoshi, M., T. Hai, Y. S. Lin, M. R. Green, and R. G. Roeder. 1988. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell 54:1033-1042. [DOI] [PubMed] [Google Scholar]
- 31.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, K. M., J. Wang, A. Smallwood, C. Arayata, and M. Carey. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16:1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1997. Cloning and characterization of an essential Saccharomyces cerevisiae gene, TAF40, which encodes yTAFII40, an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 272:9436-9442. [DOI] [PubMed] [Google Scholar]
- 34.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1996. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 271:13706-13715. [DOI] [PubMed] [Google Scholar]
- 35.Klebanow, E. R., and P. A. Weil. 1999. A rapid technique for the determination of unknown plasmid library insert DNA sequence directly from intact yeast cells. Yeast 15:527-531. [DOI] [PubMed] [Google Scholar]
- 36.Kokubo, T., M. J. Swanson, J. I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotani, T., T. Miyake, Y. Tsukihashi, A. G. Hinnebusch, Y. Nakatani, M. Kawaichi, and T. Kokubo. 1998. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem. 273:32254-32264. [DOI] [PubMed] [Google Scholar]
- 38.Lavigne, A. C., G. Mengus, M. May, V. Dubrovskaya, L. Tora, P. Chambon, and I. Davidson. 1996. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J. Biol. Chem. 271:19774-19780. [DOI] [PubMed] [Google Scholar]
- 39.Leurent, C., S. Sanders, C. Ruhlmann, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21:3424-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leurent, C., S. L. Sanders, M. A. Demeny, K. A. Garbett, C. Ruhlmann, P. A. Weil, L. Tora, and P. Schultz. 2004. Mapping key functional sites within yeast TFIID. EMBO J. 23:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, D., R. Ishima, K. I. Tong, S. Bagby, T. Kokubo, D. R. Muhandiram, L. E. Kay, Y. Nakatani, and M. Ikura. 1998. Solution structure of a TBP-TAFII230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94:573-583. [DOI] [PubMed] [Google Scholar]
- 42.Martinez, E., C. M. Chiang, H. Ge, and R. G. Roeder. 1994. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 13:3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnikov, A., and P. J. Youngman. 1999. Random mutagenesis by recombinational capture of PCR products in Bacillus subtilis and Acinetobacter calcoaceticus. Nucleic Acids Res. 27:1056-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mencia, M., and K. Struhl. 2001. Region of yeast TAF 130 required for TFIID to associate with promoters. Mol. Cell. Biol. 21:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michel, B., P. Komarnitsky, and S. Buratowski. 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2:663-673. [DOI] [PubMed] [Google Scholar]
- 46.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 47.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 48.Muller, F., and L. Tora. 2004. The multicoloured world of promoter recognition complexes. EMBO J. 23:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naar, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakajima, N., M. Horikoshi, and R. G. Roeder. 1988. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol. Cell. Biol. 8:4028-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishikawa, J., T. Kokubo, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1997. Drosophila TAFII230 and the transcriptional activator VP16 bind competitively to the TATA box-binding domain of the TATA box-binding protein. Proc. Natl. Acad. Sci. USA 94:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oelgeschlager, T., C. M. Chiang, and R. G. Roeder. 1996. Topology and reorganization of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 54.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 55.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 56.Poon, D., Y. Bai, A. M. Campbell, S. Bjorklund, Y. J. Kim, S. Zhou, R. D. Kornberg, and P. A. Weil. 1995. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 58.Ranallo, R. T., K. Struhl, and L. A. Stargell. 1999. A TATA-binding protein mutant defective for TFIID complex formation in vivo. Mol. Cell. Biol. 19:3951-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reese, J. C., L. Apone, S. S. Walker, L. A. Griffin, and M. R. Green. 1994. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature 371:523-527. [DOI] [PubMed] [Google Scholar]
- 60.Reese, J. C., Z. Zhang, and H. Kurpad. 2000. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem. 275:17391-17398. [DOI] [PubMed] [Google Scholar]
- 61.Runge, K. W., and V. A. Zakian. 1989. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol. Cell. Biol. 9:1488-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanders, S. L., K. A. Garbett, and P. A. Weil. 2002. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22:6000-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 65.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 66.Selleck, W., R. Howley, Q. Fang, V. Podolny, M. G. Fried, S. Buratowski, and S. Tan. 2001. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol. 8:695-700. [DOI] [PubMed] [Google Scholar]
- 67.Shen, W. C., and M. R. Green. 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 68.Singh, M. V., and P. A. Weil. 2002. A method for plasmid purification directly from yeast. Anal. Biochem. 307:13-17. [DOI] [PubMed] [Google Scholar]
- 69.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 70.Sullivan, S., D. W. Sink, K. L. Trout, I. Makalowska, P. M. Taylor, A. D. Baxevanis, and D. Landsman. 2002. The histone database. Nucleic Acids Res. 30:341-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 72.Taggart, A. K., and B. F. Pugh. 1996. Dimerization of TFIID when not bound to DNA. Science 272:1331-1333. [DOI] [PubMed] [Google Scholar]
- 73.Takahata, S., H. Ryu, K. Ohtsuki, K. Kasahara, M. Kawaichi, and T. Kokubo. 2003. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J. Biol. Chem. 278:45888-45902. [DOI] [PubMed] [Google Scholar]
- 74.Thuault, S., Y. G. Gangloff, J. Kirchner, S. Sanders, S. Werten, C. Romier, P. A. Weil, and I. Davidson. 2002. Functional analysis of the TFIID-specific yeast TAF4 (yTAFII48) reveals an unexpected organization of its histone-fold domain. J. Biol. Chem. 277:45510-45517. [DOI] [PubMed] [Google Scholar]
- 75.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 76.Verrijzer, C. P., J. L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115-1125. [DOI] [PubMed] [Google Scholar]
- 77.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 78.Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 79.Weinzierl, R. O., S. Ruppert, B. D. Dynlacht, N. Tanese, and R. Tjian. 1993. Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J. 12:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie, X., T. Kokubo, S. L. Cohen, U. A. Mirza, A. Hoffmann, B. T. Chait, R. G. Roeder, Y. Nakatani, and S. K. Burley. 1996. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380:316-322. [DOI] [PubMed] [Google Scholar]
- 81.Yatherajam, G., L. Zhang, S. M. Kraemer, and L. A. Stargell. 2003. Protein-protein interaction map for yeast TFIID. Nucleic Acids Res. 31:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]