Abstract
Bloom's syndrome (BS) is a genomic instability disorder characterized by cancer susceptibility. The protein defective in BS, BLM, belongs to the RecQ family of DNA helicases. In this study, we found that BLM interacts with hp150, the largest subunit of chromatin assembly factor 1 (CAF-1), in vitro and in vivo. Colocalization of a proportion of the cellular complement of these two proteins is found at specific nuclear foci coinciding with sites of DNA synthesis in the S phase. This colocalization increases in the presence of agents that damage DNA or inhibit DNA replication. In support of a functional interaction between BLM and CAF-1, we show that BLM inhibits CAF-1-mediated chromatin assembly during DNA repair in vitro. Although CAF-1 activity is not altered in BLM-deficient cells, the absence of BLM does impair the ability of CAF-1 to be mobilized within the nucleus in response to hydroxyurea treatment. Our results provide the first link between BLM and chromatin assembly coupled to DNA repair and suggest that BLM and CAF-1 function in a coordinated way to promote survival in response to DNA damage and/or replication blockade.
Bloom's syndrome (BS) is a rare recessive genetic disorder associated with cancer predisposition, immunodeficiency, proportional dwarfism, and a sunlight-induced facial erythema (16). Cells from BS patients display genomic instability, including an excessive number of locus-specific mutations as well as a high frequency of microscopically visible chromatid gaps, rearrangements, breaks, and sister chromatid exchanges (40). BS cells are not defective in the known DNA repair pathways (14, 26, 38) but show a retarded rate of nascent DNA chain elongation (18) and accumulate abnormal replication intermediates (29). The gene mutated in BS encodes a DNA helicase (BLM) belonging to the RecQ family (12). Recombinant BLM exhibits ATP-dependent 3′-5′ DNA helicase activity (22). Immunolocalization studies have shown that BLM is found mainly in the nucleus at discrete foci (shown to coincide with promyelocytic leukemia protein (PML) bodies) (21, 35) and in nucleoli during S phase (55).
Recently, much attention has been paid to the characterization of proteins that associate with the RecQ family of DNA helicases in order to understand their exact roles in the maintenance of genome stability in human cells (19, 34). BLM has been reported to interact with several proteins, including replication protein A (RPA), a DNA binding protein that plays essential roles during DNA replication, repair, and recombination (8); topoisomerase IIIα, a protein that, together with BLM, can resolve intermediates that arise during homologous recombinational repair (53, 54); BRCA1, a protein required for efficient homologous recombination and which acts as a signal processor for DNA damage responses (50); RAD51 and RAD51L3, proteins required for DNA strand invasion and exchange during homologous recombination (6, 7, 52); p53, a tumor suppressor implicated in S-phase processes, DNA repair, and homologous recombination (11, 41, 49); TRF2, a telomere binding protein (36, 44); WRN, Werner syndrome protein, another member of the RecQ helicase family (48); ATM, one of the essential transducers of DNA damage checkpoint responses (5); MLH1 and MSH6, proteins involved in mismatch repair and recombination (26, 37, 38); and five Fanconi anemia proteins: FANCA, FANCG, FANCC, FANCE, and FANCF, a multiprotein complex suggested to function in a DNA damage response pathway (31). The phenotypes exhibited by BS cells, as well as the proteins that can associate with BLM protein, imply that BLM may be involved in DNA recombination and repair processes linked to DNA replication and in telomere maintenance. There are several lines of evidence to show the importance of BLM during DNA replication. First, in Xenopus egg extracts, depletion of the Xenopus BLM homolog (xBLM) leads to an inhibition of DNA replication that can be rescued by addition of recombinant xBLM (27). Second, BLM−/− cells possess replication defects that include the accumulation of abnormal replication fork intermediates, retarded replication fork progression, and the delayed maturation of Okazaki fragments (10). Third, BLM is localized to replication foci, particularly during late S phase (51). However, it remains unclear whether the involvement of BLM in DNA replication is direct or is a secondary effect of lacking efficient repair of stalled replication forks. It is therefore of considerable interest to study the mechanism of how BLM influences DNA synthesis.
In this paper, we report that BLM physically and functionally associates with hp150, the largest subunit of chromatin assembly factor 1 (CAF-1). CAF-1 is a three-subunit complex that promotes the incorporation of histones H3 and H4 into newly synthesized DNA during replication (47) or during nucleotide excision repair (15). Moreover, we show that BLM and hp150 colocalize to sites of DNA synthesis when cells are exposed to DNA replication inhibitors or DNA-damaging agents. In pursuit of a biological function of the BLM-hp150 interaction, we found that BLM specifically attenuates CAF-1-mediated chromatin assembly during DNA repair in vitro. Furthermore, we show that in BLM−/− cells, CAF-1 activity is not altered but that the absence of BLM affects the localization of CAF-1 following hydroxyurea (HU) treatment. Collectively, our data indicate that BLM and CAF-1 may function together in vivo during DNA metabolic processes and provide a novel insight into the role of BLM and CAF-1 in the maintenance of genome stability.
MATERIALS AND METHODS
Constructs.
Construction of part of the BLM deletion mutant series (see Fig. 2) has been described previously (38). PCR products of BLM131-333 and BLM782-952 from plasmid pJK1 (22) were cloned into the pBTM116 vector. The presence of the correct clones was verified by sequencing and by checking expression in yeast cells by Western blotting. hp150 and its deletion mutants were cloned into pBTM116 and pACT2 vectors for yeast two-hybrid (YTH) testing and into the pCITE4 vector (Novagen) for in vitro transcription-translation analyses. Precise details of sequences and construction schemes are available on request.
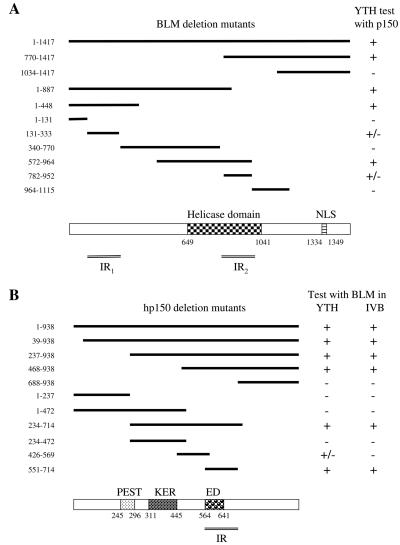
FIG. 2.
Interaction region mapping of BLM and hp150. (A) Two hp150-interacting regions (IR1 and IR2) were identified on BLM. Full-length BLM protein and its deletion mutants used in the YTH assay are shown in the upper panel. The numbers on the left represent the starting and ending amino acids of the protein fragments. In the column on the right, headed YTH test with p150, + indicates a strong interaction, +/− indicates a weak interaction, and − indicates no interaction in the YTH assay (as judged by the intensity of blue color in the test). The lower panel depicts the helicase domain and the nuclear localization signals (NLS) in the full-length BLM (on the same scale as in the upper panel), as well as the two minimal regions in BLM (IR1 and IR2) that mediate the interaction with hp150. (B) The BLM-interacting region on hp150 is located between amino acids 551 and 714. In the upper panel, the sequence boundaries of the deletion mutants tested in the YTH and in vitro binding assays (IVB) are shown with the corresponding amino acid positions indicated on the left. The results are summarized on the right. The lower panel depicts the domain diagram of hp150 showing the PEST, KER, and ED domains to scale with the upper panel. The region (IR) of hp150 that is necessary and sufficient for the interaction with BLM is shown.
Yeast two-hybrid (YTH) assay.
The screening of an activation domain (AD) cDNA library generated from human peripheral blood mRNA using the C-terminal 647 amino acids of BLM as a bait was performed as described previously (38). Standard yeast transformation and interaction tests were used to map the interaction domains for BLM and hp150. Yeast strain L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ) was used.
In vitro binding assay.
Different amounts (1.25, 2.5, and 5 pmol) of purified recombinant BLM, which was diluted in protein storage buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 25% [vol/vol] glycerol) were dotted on a nitrocellulose membrane (MSI NitroBind); bovine serum albumin (BSA) and UvrD were dotted as control proteins. After being air dried for 15 min, the membrane was blocked for 2 h at room temperature in Tris-buffered saline (TBS) supplemented with 2.5% (wt/vol) milk and 0.05% (vol/vol) Tween 20 and then incubated for 3 h at 4°C, with different proteins that were 35S labeled using the TNT quick transcription-translation system (Promega), in 1 ml of TBS with 0.1% (wt/vol) BSA and 0.05% (vol/vol) Tween 20. Aliquots (20 μl) of the in vitro transcription-translation reaction mixture were used for each incubation. After extensive washing with TBS containing 0.05% Tween 20, the membranes were dried and exposed to a PhosphorImager (Molecular Dynamics).
Co-IP.
Portions (200 μg) of nuclear extract from BJAB cells, as well as untreated or HU-treated HeLa cells, were incubated with either 8 μl of anti-hp150 antibody (MAb1) (42) or 2 μg of control immunoglobulin G (IgG) in immunoprecipitation (IP) buffer (20 mM HEPES [pH 7.5], 120 mM KCl, 5 mM MgCl2, 0.1% [vol/vol] NP-40, protease inhibitors) at 4°C for 3 h in a total reaction volume of 200 μl. Then 40 μl of IP buffer-equilibrated protein G-Sepharose beads (Amersham Pharmacia Biotech) was added, and the mixture was incubated for 3 h at 4°C. The beads were washed five times with wash buffer (IP buffer supplemented with an additional 20 mM KCl) before the protein complexes bound to the beads were eluted and split into two portions for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A 10-μg portion of nuclear extract was used as the input control. Western blot analysis with an anti-BLM antibody, IHIC33 (53), and an anti-hp150 polyclonal antibody (gifts from Geneviève Almouzni) were used to detect proteins immunoprecipitated by MAb1. Enhanced chemiluminescence was performed as specified by the manufacturer (Amersham Pharmacia Biotech). Two other anti-hp150 antibodies were also used in the IP assays: MAb anti-CAF-1 p150 (Novus Biologicals) and Ab-3 (Oncogene Research Products). C-18 (anti-BLM; Santa Cruz) was used for the reverse IP in the above-described IP buffer containing 100 to 150 mM KCl. Nuclear extracts from BS cells were used as a negative control for the C-18 reverse IP.
Cellular fractionation.
Exponentially growing HeLa cells, BS cells (GM08505), and BS cells containing the BLM cDNA (PSNF5 cells) (53) were harvested in phosphate-buffered saline (PBS) and washed twice with buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM dithiothreitol [DTT], protease inhibitors) before the pellet was resuspended in 3 volumes of buffer A+ (buffer A containing an additional 0.1% NP-40). After incubation for 5 min, the cells were centrifuged and the pellet was resuspended in 3 volumes of buffer C (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 25% [vol/vol] glycerol, 1 mM PMSF, 5 mM DTT, other proteinase inhibitors) and incubated on ice for 15 min. Finally, after the mixture was pelleted, the supernatant was taken as the soluble fraction of nuclear proteins (the concentration of NaCl was adjusted to 120 mM with buffer D, which contains 20 mM HEPES [pH 7.9], 1 mM PMSF, 5 mM DTT, and proteinase inhibitors) and the pellet was either subjected to ultrasonication to dissolve the insoluble fraction of nuclear proteins in SDS sample buffer or digested with DNase I in DIG buffer (50 mM NaCl, 10 mM Tris [pH 6.8], 3 mM MgCl2, 1 mM EGTA, 0.5% Triton, 100 μg of DNase I per ml, 50 μg of RNase A per ml, 1.2 mM PMSF) for 30 min at room temperature before the chromatin fraction was separated from the nuclear matrix fraction.
Cell synchronization and immunofluorescence microscopy.
HeLa cells, BS cells, and PSNF5 cells (BS cells containing the BLM cDNA [BLM−/− + pBLM]) were grown on chamber slides (Lab-Tek) to up to 50% confluence before being subjected to synchronization or treatment with variant DNA-damaging agents. For a G1/S block, 5 mM thymidine and 2 μg of aphidicolin per ml were added to the cells for 28 and 24 h, respectively; for S-phase synchronization, 2 mM HU was used for 24 h; for a G2/M block, 1 μg of nocodazole per ml was incubated with the cells for 24 h. Synchronized cells or cells taken 20 h after treatment with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or 4 to 18 h after UV treatment were permeabilized with 0.5% (vol/vol) Triton in PBSS (PBS supplemented with 5 mM MgCl2) plus 5 mM PMSF for 5 min at room temperature before being fixed in 2% paraformaldehyde for 20 min. After 20 min of further treatment with 0.5% (vol/vol) Triton, and following washing with PBSST (PBSS with 0.1% [vol/vol] Tween 20), the slides were blocked for 20 min in PBSST plus 5% (wt/vol) BSA before being incubated with the primary antibodies anti-BLM (1:200) (32), anti-RPA (1:50; Ab-1 [Oncogene Research Products]), anti-BRCA1 (1:100; Ab-1 [Oncogene Research Products]), anti-hp150 polyclonal antibody (1:2,000), and anti-hp150 MAb1 (1:100; Ab-3 [Oncogene research products]) for 2 h at room temperature. To visualize the primary antibodies, Cy3-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch) and 5-(4,6-dichlorotriazin-2-yl)aminofluorescein (DTAF)-coupled goat anti-mouse IgG (1:100; ANAWA) or fluorescein isothiocyanate-coupled goat anti-mouse IgG (1:100; Jackson ImmunoResearch) were used, and 1 μg of 4,6-diamidino-2-phenylindole (DAPI) per ml was added to stain the DNA. The stained cells were mounted in Vectashield (Vector Laboratories) and observed under a Leica TCS 4D confocal microscope. Imaris software was used to process the images.
Chromatin assembly assay.
An in vitro chromatin assembly assay with a human cell-free system was utilized to analyze the effect of recombinant BLM on DNA repair-coupled chromatin assembly (15). Briefly, UV (500 J/m2)-irradiated circular pBluescript plasmid DNA was incubated for 3 h at 37°C with cytosolic extract derived from HeLa cells (30) supplemented with purified recombinant CAF-1 complex or with nuclear extract from either HeLa or BS cells (47) in the presence or absence of recombinant native BLM or BLM preincubated with anti-BLM antibody (32). The extent of supercoiling was examined by agarose gel electrophoresis followed by drying of the gel and exposure to X-ray film. Incorporation of [α-32P]dCTP indicates newly synthesized DNA after nucleotide excision repair.
RESULTS
BLM and hp150 interact in vitro and in vivo.
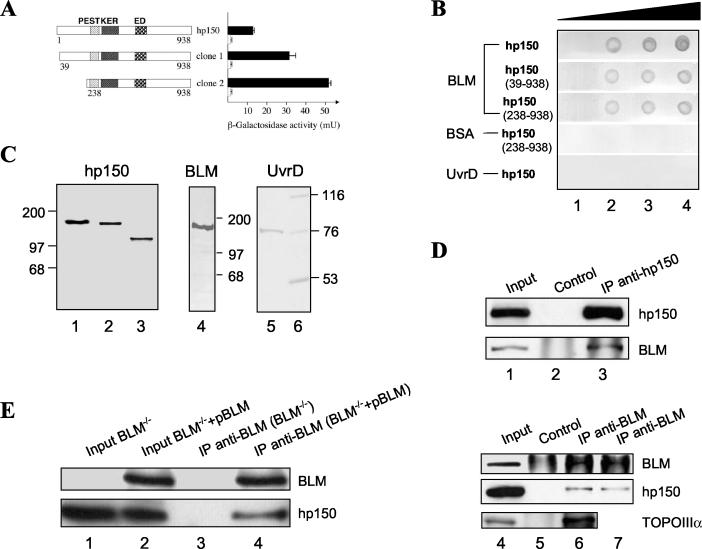
To identify proteins that interact with BLM, we screened an AD cDNA library generated from human peripheral blood mRNA, using the C-terminal 647 amino acids of BLM as a bait (38). Two interacting clones contained amino acids 39 to 938 and 238 to 938 of hp150 fused in frame to the AD (Fig. 1A). To ascertain the specificity of this interaction, we analyzed whether either of the AD-hp150 fusions could interact with a nonspecific control protein, yeast Dna2p. The resulting phenotype of the transformed yeast was His3− LacZ−, indicating that hp150 of human CAF-1 does not interact with Dna2p. We conclude that hp150 specifically binds to the C terminus of BLM in the YTH system (Fig. 1A).
FIG. 1.
BLM and hp150 interact both in vitro and in vivo. (A) Interaction of BLM and hp150 analyzed by the YTH assay. The left panel illustrates the AD-hp150 constructs. The PEST, KER, and ED regions of hp150 are indicated. The right panel shows the corresponding β-galactosidase activities with either LexA-BLM(770-1417) (black bars) or LexA-Dna2 (white bars). (B) In vitro binding assay. Different amounts (lane 1, 0 pmol; lane 2, 1.25 pmol; lane 3, 2.5 pmol; lane 4, 5 pmol) of purified recombinant BLM protein, BSA, or UvrD were dotted on a nitrocellulose membrane and incubated with in vitro-transcribed and -translated full-length hp150, hp15039-938, and hp150238-938, respectively, as indicated on the left. Signals were detected by autoradiography using a PhosphorImager. (C) An autoradiogram (left panel) of in vitro-transcribed and -translated full-length hp150 (lane 1), hp15039-938 (lane 2), and hp150238-938 (lane 3) used in the in vitro binding assay. Purified recombinant BLM protein (middle panel, lane 4) and UvrD protein (right panel, lane 5) were subjected to SDS-PAGE and stained with Coomassie blue. The positions of molecular mass standards are also indicated (lane 6). (D) BLM was coimmunoprecipitated from BJAB human nuclear extracts with an anti-hp150 MAb (MAb1, lane 3) but not with the control IgG (lane 2). One-tenth the amount of the same nuclear extract was used as input control (lane 1). Reciproical co-IP is shown in lanes 4 to 7: lane 4, input; lane 5, IP with the control IgG; lane 6, IP with an anti-BLM antibody (C-18) in 100 mM KCl salt condition; lane 7, IP with an anti-BLM antibody (C-18) in 150 mM salt condition. The known BLM-interacting protein, topoisomerase IIIα (TOPOIIIα), was also efficiently coimmunoprecipitated using the same anti-BLM antibody (lane 6). (E) Co-IP with anti-BLM antibody (C-18) from BS cell nuclear extracts (BLM−/−)and BS cells containing the BLM cDNA (BLM−/− + pBLM). hp150 could be coimmunoprecipitated in the presence (lane 4) but not in the absence (lane 3) of BLM. Lanes 1 and 2 are the inputs of the two different nuclear extracts.
To substantiate this interaction further, we tested whether BLM and hp150 could interact in vitro. To this end, full-length recombinant BLM protein was immobilized on a nitrocellulose filter, which was then incubated with in vitro-translated hp150 or its derivatives (Fig. 1C). As shown in Fig. 1B, the two truncated forms of hp150 found in the YTH screen, as well as the full-length hp150, interacted with BLM. Since in vitro-translated hp150 and the two N-terminally truncated forms did not interact with either of two control proteins, BSA and the bacterial helicase UvrD (Fig. 1B), we conclude that the interaction between BLM and hp150 is specific and direct, not requiring additional accessory proteins. This direct interaction was also confirmed by an in vitro co-IP assay by incubating the purified recombinant BLM with in vitro-translated hp150 and then with the MAb against hp150 (data not shown, available on request).
To gain insight into the BLM-hp150 interaction in vivo, we performed co-IP experiments with nuclear extracts of human BJAB cells, using a MAb directed against hp150 (Fig. 1D). Endogenous BLM was effectively coprecipitated by the hp150 antibody (Fig. 1D, lower panel, lane 3) but was not detected when the hp150 antibody was omitted (lane 2). Similar results were obtained with two other anti-hp150 antibodies (data not shown). The reciprocal co-IP experiment was also carried out, in which an anti-BLM polyclonal antibody (C-18; Santa Cruz) was used to immunoprecipitate hp150 from HeLa nuclear extracts. As shown in Fig. 1D, hp150 could be specifically coimmunoprecipitated with anti-BLM (middle panel, lanes 6 and 7) but not with the control IgG antibody (lane 5). The results of this co-IP experiment have indicated that in soluble HeLa nuclear cell extracts, less than 10% of total hp150 is associated with BLM (data not shown, available upon request). Moreover, in addition to hp150, anti-BLM antibody efficiently coprecipitated topoisomerase IIIα (Fig. 1D, bottom panel, lane 6), a protein known to interact with BLM (53). Importantly, the anti-BLM antibody (C-18) could not immunoprecipitate hp150 from BS (BLM−/−) cells (Fig. 1E, lane 3) whereas it efficiently immunoprecipitated hp150 from BS cells containing the BLM cDNA (BLM−/− + pBLM) (lane 4). We conclude from these different protein interaction experiments that BLM associates with hp150 of CAF-1 in yeast, in vitro, and in human cells.
Mapping of interaction regions on BLM and hp150.
To identify the region of BLM that mediates the interaction with hp150, we generated a series of BLM deletion mutants. These deletion mutants were tested for interaction with the full-length hp150 in the YTH assay (Fig. 2A). Two independent regions in BLM were found to interact with hp150. The first interacting region resides at the N terminus of BLM and comprises amino acids 131 to 333, and the second region is located in the middle of the helicase domain and comprises amino acids 782 to 952 of BLM (Fig. 2A). The relatively weaker interaction of BLM amino acids 131 to 333 and BLM amino acids 782 to 952 with hp150 compared to that of full-length BLM could be due to ineffective folding of these two short forms. A similar YTH approach combined with an in vitro binding assay was used to map the BLM interaction region on hp150 (Fig. 2B). One region of hp150, comprising amino acids 551 to 714 containing the ED domain (amino acids 564 to 641) (24) and the dimerization domain (amino acids 642 to 678) (39), was found to be necessary and sufficient for interaction with BLM. Interestingly, this region is different from the region in hp150 that interacts with proliferating-cell nuclear antigen in vitro (33) and from the region critical for the interaction with p60, another subunit of CAF-1 (24).
BLM and hp150 colocalize on DNA replication block or DNA-damaging signals.
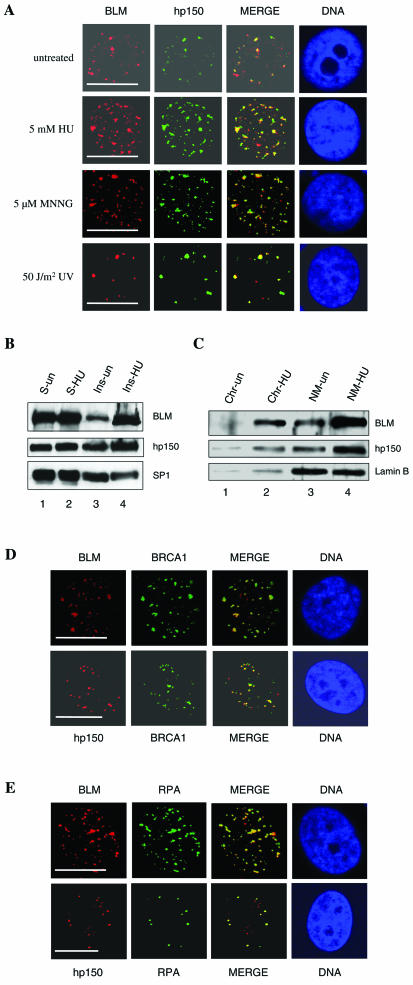
Next, we analyzed whether BLM and hp150 colocalize within the nucleus of intact human cells by indirect immunofluorescence of exponentially growing HeLa cells. Previous studies using HeLa cells have shown that CAF-1 is present at replication foci, since it localizes at sites of 5-bromodeoxyuridine incorporation (25, 45). In these cells, BLM forms nuclear foci corresponding to PML nuclear bodies (21, 52). The observation of nuclear foci containing BLM with PML, RPA, and 5-bromodeoxyuridine led to the suggestion that these sites represent sites of DNA synthesis and repair (6). In untreated HeLa cells, we detected some nuclear foci that were enriched in both BLM and hp150. Under these conditions, only 7% of the cells contained a mean of 1.8 colocalizing BLM and hp150 foci per nucleus (Fig. 3A and Table 1). This observation suggested that the two proteins might associate at sites of endogenous damage during the replication process. We therefore asked whether the number of colocalizing BLM-hp150 foci increased in response to treatment of the cells with HU. Indeed, following a 20-h HU treatment, the proportion of cells containing BLM-hp150 colocalizing nuclear foci increased significantly, such that 40% of the population contained a mean of 7.6 colocalizing foci per nucleus (Fig. 3A; Table 1). We then tested whether other DNA-damaging agents resulted in increased BLM-hp150 colocalization. At 4 h after treatment of HeLa cells with UV light (50 J/m2), 25% of the cells contained a mean of 4.2 BLM-hp150 colocalizing foci per nucleus. In addition, after exposure of HeLa cells to MNNG for 20 h, 36% of the HeLa cells contained a mean of 6.9 BLM-hp150 colocalizing foci (Fig. 3A; Table 1), demonstrating that this significant increase in BLM-hp150 colocalization is irrespective of the type of DNA damage present.
FIG. 3.
BLM and hp150 colocalize within distinct nuclear foci in response to DNA-damaging signals. (A) Indirect immunofluorescence of BLM (red) and hp150 (green) in untreated and HU-, MNNG-, and UV-treated HeLa cells. Representative pictures are shown for a portion of cells analyzed. For details, see Table 1. In the merged image, colocalization is seen as yellow. The panel labeled DNA indicates the position of the nucleus, as judged by DAPI staining. Bars, ∼10 μm. (B) The amount of BLM and hp150 increases after HU treatment in the insoluble fraction of nuclear proteins. Untreated (lanes 1 and 3)and HU-treated (lanes 2 and 4) HeLa nuclear proteins were resolved into soluble (lanes 1 and 2) and insoluble (lanes 3 and 4) fractions, and these were subjected to SDS-PAGE followed by Western blotting for BLM (top panel), hp150 (middle panel), and SP1 (bottom panel [loading control]). (C) The increased amount of BLM and hp150 is mainly associated with the nuclear matrix. After DNase 1 digestion, the insoluble nuclear fraction was separated into two fractions: chromatin (lanes 1 and 2) and nuclear matrix (lanes 3 and 4). Lanes 1 and 3 contain untreated HeLa cells; lanes 2 and 4 contain HU-treated HeLa cells. Lamin B is shown as a fractionation and loading control. (D) BLM and hp150 colocalize with BRCA1 after HU treatment of HeLa cells. Red foci represent either BLM or hp150, and green foci reveal BRCA1 staining. DNA is revealed by DAPI staining. Bars, ∼10 μm. (E) BLM and hp150 colocalize with RPA after HU treatment of HeLa cells. Red represents either BLM or hp150, as indicated, and green indicates RPA protein. DNA indicates the position of nucleus. Bars, ∼10 μm. See Results and Discussion for details.
TABLE 1.
Proportion of cells containing foci and number of nuclear foci containing both BLM and hp150 in HeLa cellsa
| Treatment | % of cells containing:
|
Mean no. of foci/cell (range)
|
% of cells containing BLM and CAF-1 colocalizing foci
|
Mean no. of BLM and CAF-1 colocalizing foci/cell | ||||
|---|---|---|---|---|---|---|---|---|
| BLM foci | CAF-1 foci | BLM foci | CAF-1 foci | 0 foci | 1-5 foci | >5 foci | ||
| None | 36 | 9 | 4.2 (0-27) | 3.1 (0-29) | 93 | 6 | 1 | 1.8 |
| HU (5 mM) | 85 | 68 | 22.2 (0-51) | 29.6 (0-54) | 60 | 32 | 8 | 7.6 |
| MNNG (5 M) | 94 | 57 | 26.8 (0-66) | 23.3 (0-49) | 64 | 29 | 7 | 6.9 |
| UV | 45 | 37 | 6.8 (0-30) | 6.3 (0-32) | 75 | 23 | 2 | 4.2 |
In each case, 150 cells were scored.
To examine the BLM-hp150 interaction on inhibition of DNA replication, we treated HeLa cells with HU, an agent that inhibits ribonucleotide reductase and thereby halts DNA replication fork progression, and coimmunoprecipitated proteins from soluble nuclear extracts with an anti-hp150 MAb. This treatment did not appear to dramatically change the amount of the BLM-hp150 complex (data not shown). Similar results were observed following treatment of HeLa cells with the alkylating agent MNNG (data not shown). Next, we examined if the amount of BLM and/or hp150 increases in the fraction of insoluble nuclear proteins after HU treatment. To that end, HeLa cell nuclear proteins were resolved into soluble (Fig. 3B, lanes 1 and 2) and insoluble (lanes 3 and 4) fractions, which were subjected to SDS-PAGE followed by Western blotting for BLM (upper panel), hp150 (middle panel), or SP1 (lower panel), the last of which was used as a loading control (28). Interestingly, the amount of both BLM and hp150 increased, although to different extents, in the insoluble (lane 4), but not the soluble (lane 2) fraction of nuclear proteins after HU treatment of HeLa cells. To determine the fraction of insoluble nuclear proteins in which BLM and CAF1-hp150 were associated, we separated chromatin from the nuclear matrix fraction. As shown in Fig. 3C, most of the functional BLM and hp150 were found to be associated with the nuclear matrix fraction of the insoluble nuclear structure, as judged by the presence of the nuclear matrix marker protein lamin B. To define further the sites in the nucleus where BLM and hp150 colocalize, additional immunofluorescence experiments were performed with antibodies to proteins involved in replication, repair, and recombination. RPA is a nuclear protein that interacts with BLM (8) and localizes at sites of DNA replication in nuclei of different cell types (3, 9). BRCA1, another BLM-interacting protein (50), is required for efficient homologous recombination and serves as a signal processor for DNA damage responses (46). We found that following treatment with HU, UV, or MNNG, both BLM and hp150 colocalized with BRCA1 in discrete nuclear foci (Fig. 3D and data not shown). Similarly, both BLM and hp150 co-localized with RPA in HU-, UV-, or MNNG-treated HeLa cells (Fig. 3E and data not shown). Taken together, these immunofluorescence data suggest that after treatment of cells with DNA-damaging agents or in response to endogenous damage, a proportion of BLM and hp150 colocalize at sites of ongoing DNA synthesis and repair.
BLM-hp150 colocalization is cell cycle dependent.
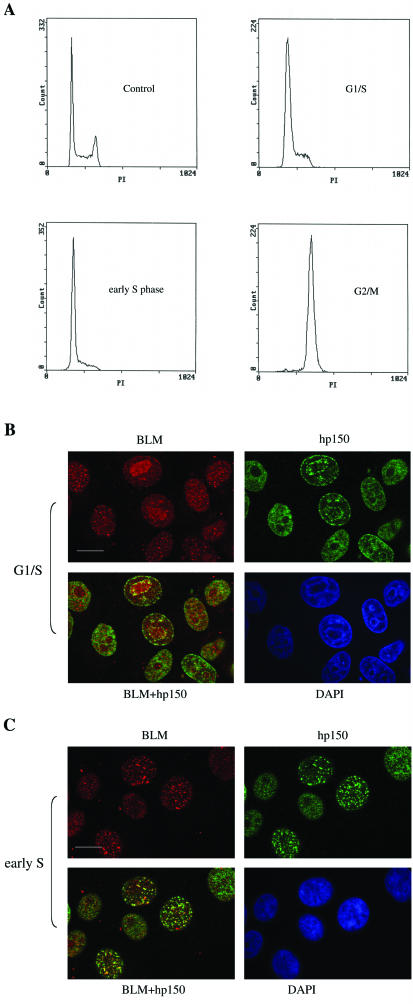
Since the subcellular localization patterns for BLM and CAF-1 are not identical during a normal, unperturbed cell cycle (25, 55), we set out to determine the stage of the cell cycle at which BLM and hp150 colocalize to the greatest extent. For this purpose, we synchronized HeLa cells in the G1/S, early S, and G2/M phases (Fig. 4A) and immunostained the cells with anti-BLM and anti-hp150 antibodies. Figure 4B and C show the pattern of BLM-hp150 colocalization in G1/S- and S-phase cells, respectively. As before, only a portion of cells exhibited BLM-hp150 colocalizing foci: only 3% of the G1/S phase cells contained BLM-hp150 colocalizing foci, while 38% of the S-phase cells displayed BLM-hp150 colocalization. During the G2/M stage, no cells showed any BLM-hp150 colocalization (data not shown). This reveals that colocalization between BLM and hp150 occurs most frequently during the S phase, where the majority of DNA replication and repair events are occurring.
FIG. 4.
BLM-hp150 colocalization is cell cycle dependent. (A) Fluorescence-activated cell sorter analysis of HeLa cells, which were either unsynchronized (control) or synchronized at different cell cycle stages: G1/S, early S, and G2/M (see Materials and Methods). (B) HeLa cells blocked at the G1/S phase boundary (see Materials and Methods) were immunostained for BLM and hp150. DAPI staining was for nuclear DNA. Bar, ∼10 μm. (C) Colocalization of BLM and hp150 in S-phase HeLa cells (see Materials and Methods and the Results section for more information). Bar, ∼10 μm.
BLM inhibits CAF-1-mediated chromatin assembly coupled to DNA repair.
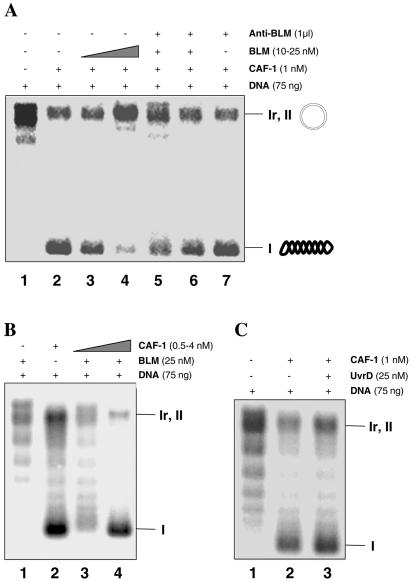
Next, we investigated the functional consequences of the interaction between BLM and hp150 by analyzing whether BLM affects the activity of hp150 in terms of its critical role in chromatin assembly. To address this, we used the in vitro chromatin assembly assay coupled to repair of UV-damaged DNA. This assay takes advantage of simultaneous analysis of DNA repair and chromatin assembly processes on the same damaged circular DNA template (15). HeLa cytosolic extracts complemented by recombinant CAF-1 efficiently mediated DNA repair-linked chromatin assembly, as indicated by the appearance of radioactively labeled form I DNA molecules corresponding to supercoiled, repaired DNA (Fig. 5A, lane 2). In this reaction, chromatin assembly that occurs independently of DNA synthesis or repair is very inefficient and appears to be CAF-1 independent (data not shown). Addition of purified BLM alone, in the absence of recombinant CAF-1, did not result in the formation of supercoiled molecules (Fig. 5B, lane 1). However, preincubation of CAF-1 complex with recombinant BLM led to a significant decrease in the amount of supercoiled product (Fig. 5A, lanes 3 and 4), indicating that BLM acted as an inhibitor of CAF-1-mediated chromatin assembly coupled to DNA repair. When recombinant BLM was preincubated with anti-BLM antibody before addition of the mixture to the chromatin assembly reaction mix, the BLM no longer inhibited CAF-1-dependent DNA repair-coupled chromatin assembly (Fig. 5A, lanes 5 and 6). This is most probably due to an inability of the antibody-complexed BLM to interact productively with hp150. No effect on CAF-1-mediated chromatin assembly was observed when anti-BLM antibody alone was added to the reaction mix (lane 7). Importantly, chloroquine gel analysis demonstrated that there was no difference in the topology of the DNA repaired by the cytosolic extracts in the presence or absence of BLM (data not shown), thus excluding the possibility that BLM-mediated inhibition of chromatin assembly is due to the effect of BLM on topoisomerases in our system.
FIG. 5.
BLM inhibits CAF-1-mediated chromatin assembly coupled to DNA repair. (A) Addition of BLM inhibits CAF-1 activity. Each lane contains 75 ng of UV-irradiated circular pBluescript plasmid DNA, as indicated above the figure. Purified CAF-1 complex is present in lanes 2 to 7 but not in lane 1 which serves as a negative control. The concentration of recombinant BLM was 10 nM in lane 3 and 25 nM in lanes 4 to 6. In lanes 5 and 6, BLM protein was incubated with 1 μl of anti-BLM antibody before being added to the chromatin assembly reaction mix. Lane 7 is a control of anti-BLM antibody. The positions of the relaxed/nicked (Ir, II) and supercoiled (I) DNA are indicated on the right. (B) An excess of CAF-1 holoenzyme complex restores BLM-mediated inhibition of chromatin assembly in vitro. A 75-ng portion of UV-irradiated plasmid DNA was used in each lane; 25 nM BLM was added in lanes 1, 3 and 4, but not in lane 2. CAF-1 was present only in lanes 3 and 4, at 0.5 and 4 nM, respectively. The positions of the relaxed/nicked (Ir, II) and supercoiled (I) DNA are indicated on the right. (C) Inhibition of CAF-1 activity is BLM specific. Bacterial helicase UvrD does not inhibit CAF-1-mediated chromatin assembly coupled to DNA repair (lane 3). Lanes 1 and 2 are negative and positive controls, respectively.
Next, we asked whether the inhibitory effect of BLM on CAF-1-mediated chromatin assembly could be relieved by the addition of an excess of purified CAF-1 holoenzyme complex. Figure 5B (lane 4) shows that a significant and CAF-1 concentration-dependent relief of BLM-mediated inhibition of chromatin assembly was detected on the addition of the CAF-1 holoenzyme complex. Most importantly, this inhibition of CAF-1-mediated chromatin assembly is specific for BLM, since the bacterial helicase UvrD, which does not interact with hp150 (Fig. 1B), showed no inhibitory effect on CAF-1-mediated chromatin assembly (Fig. 5C, lane 3). Collectively, these results argue for a role of BLM in inhibiting CAF-1-dependent chromatin assembly coupled to DNA repair through a direct protein-protein interaction.
We also tested how p150 affects BLM helicase activity. Although hp150 inhibited BLM-catalyzed DNA unwinding, this appeared to be a nonspecific effect, probably mediated via the binding of hp150 to the DNA substrate, because hp150 also inhibited UvrD (data not shown, available on request).
BLM is required for an optimal CAF-1 response to HU.
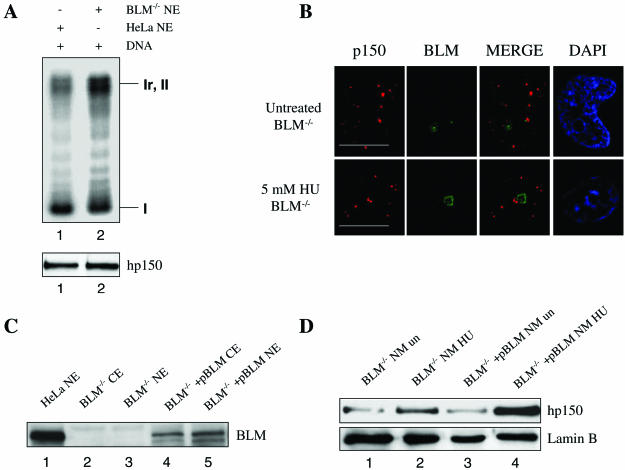
To further study the in vivo functional relationship between BLM and CAF-1, we examined CAF-1 function in BLM-deficient cells. To determine whether CAF-1-mediated chromatin assembly is affected in BS cells, we prepared nuclear extracts from BS cells and compared them with the nuclear extracts from wild-type (HeLa) cells for their ability to mediate chromatin assembly. The presence and absence of BLM in these cell lines was controlled by Western blotting (Fig. 6C). As shown in Fig. 6A, the endogenous CAF-1 activity was not obviously altered in BS cells (lane 2) compared with the CAF-1 activity from wild-type cells (lane 1).
FIG. 6.
BLM is required for a proper CAF-1 response to HU treatment. (A) Chromatin assembly assay using either the HeLa cell nuclear extracts (lane 1) or nuclear extracts prepared from BLM−/− cells (lane 2). The amounts of nuclear extract added to the reaction mixes are approximately equal, as shown at the bottom by Western blotting with an anti-hp150 antibody. (B) Immunostaining of hp150 revealed normal CAF-1 foci in morphology in both untreated and HU-treated BS cells. Bars, ∼10 μm. (C) Western blot showing that no BLM is detected in BS cells (lanes 2 and 3). Lane 1 is the positive control. Lanes 4 and 5 show that BLM is present in PSNF5 cells corrected with the full-length BLM cDNA. (D) Western blot showing that in BS cells, HU treatment did not result in as great an increase of nuclear matrix-associated hp150 (lanes 1 and 2) compared with PSNF5 cells corrected with the BLM cDNA (lanes 3 and 4). Lanes 1 and 3 contain untreated cells; lanes 2 and 4 contain HU-treated cells. Lamin B serves as the loading control.
Next, we investigated whether CAF-1 forms normal nuclear foci in untreated or HU-treated BS cells. Immunofluorescence analysis of the subcellular localization of hp150 indicated that CAF-1 formed normal nuclear foci in both untreated and HU-treated BS cells (Fig. 6B). In untreated BS cells, the percentage of cells containing CAF-1 foci (6.7%) did not significantly differ compared with that in untreated PSNF5 cells [BS cells containing the BLM cDNA (BLM−/− + pBLM)] (7.1%), suggesting that BLM is not required for recruitment of CAF-1 to nuclear foci. In HU-treated BS cells, the percentage of cells containing CAF-1 foci (18%) increased compared with that in untreated BS cells (6.7%). In PSNF5 cells which contain the BLM cDNA, the percentage of cells containing CAF-1 foci (43%) also increased in the presence of HU compared to that in untreated cells (7.1%). However, this increase in the percentage of CAF-1-foci containing cells observed in HU treated PSNF5 cells was greater than that seen in BS cells alone. This result suggests that BLM is required for an optimal CAF-1 response to HU and is consistent with the cellular fractionation experiment showing an increase in the amount of hp150 associated with nuclear matrix in PSNF5 cells containing the BLM cDNA compared to that in BS cells alone (Fig. 6D).
DISCUSSION
To maintain genome stability, both genetic and epigenetic information has to be correctly inherited from mother cells after cell division. BLM and other members of the RecQ helicase family play a crucial role in maintaining genome stability through their ability to suppress inappropriate recombination (4, 19). Consistent with this notion, BS cells show increased rates of homologous recombination and the aberrant resolution of this event (14, 41). However, the action of BLM per se does not suffice to preserve genome integrity. The newly synthesized DNA, either newly-replicated or repaired, has to be assembled into chromatin to make sure that the genome is also epigenetically stable, and CAF-1 plays a critical role in this process (17). Recent results clearly showed not only that CAF-1 is directly involved in DNA replication (20) but also that it ensures the maintenance of epigenetic information by acting at the sites of DNA repair (17). The results presented in this paper suggest a cross talk between the genetic and epigenetic pathways in the maintenance of genome stability. In particular, we have shown that BLM physically and functionally associates with hp150, the largest subunit of CAF-1. The results of our interaction region mapping experiments indicate that BLM interacts with hp150 via two separate sites on BLM, the first located near the N terminus of BLM (amino acids 131 to 333) and the second located in the middle of the helicase domain (amino acids 782 to 952) of BLM. Furthermore, we have shown that one region of hp150 (amino acids 551 to 714) containing the ED domain and the dimerization domain is necessary and sufficient for interaction with BLM.
Our results show that the proportion of detergent-resistant BLM and CAF-1 foci that colocalize in the nucleus increases after replication fork inhibition and treatment of cells with DNA-damaging agents. Together with the nuclear fractionation experiments, this suggests that the function of the BLM-hp150 interaction is associated with the matrix fraction of nuclear proteins. The increase in the number of foci containing both BLM and hp150 after HU treatment suggests that the BLM-hp150 complex concentrates in the vicinity of stalled replication forks. These replication forks probably contain nonduplex DNA structures, which might provoke a DNA damage response. In addition, BLM and hp150 are recruited together to discrete foci after DNA damage induction with MNNG or UV light. One interpretation of these phenomena is that in each case, the foci represent DNA repair processes that are initiated at a replication fork. In the case of HU treatment, as well as of UV- and MNNG-induced damage, stalled DNA replication may also be accompanied by a recombinational DNA damage response. The colocalization of BLM and hp150 at sites containing RPA and BRCA1 further supports the contention that the BLM-hp150 complex plays a repair role during DNA synthesis. Although the results of our indirect immunofluorescence experiments show that BLM is not required for recruitment of CAF-1 to nuclear foci, BLM seems to be required for an optimal CAF-1 response to HU. That the physical interaction between BLM and hp150 is of functional significance is indicated by the inhibitory effect of BLM on CAF-1-mediated chromatin assembly coupled to repair. This inhibitory effect appeared specific because a control helicase, UvrD, was unable to inhibit chromatin assembly.
Given all these observations, we propose that the BLM-hp150 interaction is important during times when DNA synthesis is perturbed in human cells. On the basis of our results, we propose a model suggesting a functional link between BLM and hp150 in restoring functional replication forks following fork stalling. Progression of a replication fork might be impeded by DNA secondary structures such as G-quadruplexes and DNA hairpins, thus causing the regression of the fork and formation of a four-way junction, a structural analog of a Holliday junction. It has been suggested previously that BLM and other RecQ helicases process such lesions that arise at replication forks (or are generated by the translocating fork itself) and, in doing so, reset the fork by “reverse” branch-migrating the four-way junction. This has the effect of preventing cleavage of the four-way junction and hence suppressing subsequent “promiscuous” recombination (10, 19, 23, 51). A second potential role for BLM at replication forks has been suggested by our recent observation that BLM and topoisomerase IIIα together catalyze the dissolution of a recombination intermediate containing a double Holliday junction (54). Such double Holliday junctions are predicted to arise during postreplication gap filling when lesions on the lagging-strand template are bypassed. It is known that CAF-1-dependent chromatin assembly occurs on newly replicated DNA (43) and that CAF-1 is required in vitro for chromatin assembly following DNA repair (15). CAF-1 binds directly to PCNA, a DNA polymerase processivity factor that forms a sliding clamp around DNA, and the CAF-1-PCNA interaction promotes chromatin assembly (33). Although it remains unclear exactly how BLM and CAF-1 act after damage at a replication fork, our data suggest that the recruitment of BLM, probably not by CAF-1, may inhibit CAF-1, and hence chromatin assembly, in the vicinity of a stalled replication fork, perhaps to allow more efficient lesion processing. We envisage that such inhibition would continue until the point where normal replication structures are restored. Once the DNA lesion is bypassed and the replication fork is restored, the CAF-1 complex can be reactivated (it again assembles the newly synthesized DNA into chromatin) and DNA replication fork progression can resume.
This sequential scheme requires a level of regulation that may be achieved by posttranslational modification. Interestingly, BLM is phosphorylated following DNA damage (1, 2, 13), and UV damage induces changes in the amount of chromatin-bound phosphorylated CAF-1 complex (30). It is possible that either of these phosphorylation events or some other posttranslational modification(s) could be responsible for modulating both physical and functional interactions between BLM and CAF-1. The inhibitory effect of BLM on CAF-1 might also be modulated by associations with other interacting proteins. A candidate would be PCNA, which can bind to both CAF-1 (33) and BLM (S. L. Davies, J.-L. Li, and I. D. Hickson, unpublished data).
While the precise biochemical relationship between BLM and CAF-1 remains to be fully elucidated, the physical and functional interactions documented here provide a basis to further understand how BLM and hp150 function together to facilitate DNA repair and/or replication.
Acknowledgments
We are grateful to Geneviève Almouzni and her coworkers Catherine Green, Jill Mello, Jean-Pierre Quivy, and Danièle Roche for help with chromatin assembly reagents and techniques and for helpful discussions. We also thank Lovorka Stojic and Josef Jiricny for tips about MNNG treatment; Bo Zhang and Walter Schaffner for the BJAB nuclear extracts; Steve Matson for the UvrD protein; Raimundo Freire for the anti-BLM antibody; Heiko Blaser, Ingrid Stoffel, and Bruno Schmid for technical assistance; Mike Fetchko, Primo Schär, and Fritz Thoma for helpful discussions; and Giancarlo Marra for BLM−/− cells and extracts. We are also grateful to Elektronenmikroskopisches Zentrallaboratorium der Universität Zürich for providing confocal facilities for this study.
The IS group is financed by grants from the Stiftung für medizinische Forschung, Novartis Foundation, Olga Mayenfish Foundation, Sassella Foundation, Gebert Rüf Foundation, EU grant HPRN-CT-2002-00240, the Swiss Cancer League (OCS-01310-02-2003), the Swiss National Science Foundation (grants 31-58798.99 and 3100A0-100256/1), and the Zürcher Krebsliga. C.Z.B., J.-L.L. and I.D.H. are supported by Cancer Research UK and EU grant HPRN-CT-2002-00240. C.Z.B. was a Marie Curie Fellow of the European Union (HPMF-CT-2000-00952).
REFERENCES
- 1.Ababou, M., V. Dumaire, Y. Lecluse, and M. Amor-Gueret. 2002. Bloom's syndrome protein response to ultraviolet-C and hydroxyurea-mediated DNA synthesis inhibition. Oncogene 21:2079-2088. [DOI] [PubMed] [Google Scholar]
- 2.Ababou, M., S. Dutertre, Y. Lecluse, R. Onclercq, B. Chatton, and M. Amor-Gueret. 2000. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19:5595-5563. [DOI] [PubMed] [Google Scholar]
- 3.Adachi, Y., and U. K. Laemmli. 1992. Identification of nuclear pre-replication centers poised for DNA synthesis in Xenopus egg extracts: immunolocalization study of replication protein A. J. Cell Biol. 119:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachrati, C. Z., and I. D. Hickson. 2003. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 374:577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beamish, H., P. Kedar, H. Kaneko, P. Chen, T. Fukao, C. Peng, S. Beresten, N. Gueven, D. Purdie, S. Lees-Miller, N. A. Ellis, N. Kondo and M. F. Lavin. 2002. Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J. Biol. Chem. 277:30515-30523. [DOI] [PubMed] [Google Scholar]
- 6.Bischof, O., S.-H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom's syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braybrooke, J. P., J.-L. Li, L. Wu, F. Caple, F. E. Benson and I. D. Hickson. 2003. Functional interaction between the Bloom's-syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D). J. Biol. Chem. 278:48357-48366. [DOI] [PubMed] [Google Scholar]
- 8.Brosh, R. M., Jr. J. L. Li, M. K. Kenny, J. K. Karow, M. P. Cooper, R. P. Kureekattil, I. D. Hickson, and V. A. Bohr. 2000. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275:23500-23508. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso, M. C., H. Leonhardt, and B. Nadal-Ginard. 1993. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74:979-992. [DOI] [PubMed] [Google Scholar]
- 10.Chakraverty, R. K., and I. D. Hickson. 1999. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays 21:286-294. [DOI] [PubMed] [Google Scholar]
- 11.Davalos, A. R., and J. Campisi. 2003. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J. Cell Biol. 162:1197-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 13.Franchitto, A., and P. Pichierri. 2002. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg, E. C., U. K. Ehmann and J. I. Williams. 1979. Human diseases associated with DNA repair. Adv. Radiat. Biol. 8:85-174. [Google Scholar]
- 15.Gaillard, P.-H., E. M. Martini, P. D. Kaufman, B. Stillman, E. Moustacchi and G. Almouzni. 1996. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86:887-896. [DOI] [PubMed] [Google Scholar]
- 16.German, J., and N. A. Ellis. 1998. Bloom syndrome, p. 301-315. In B. Vogelstein and K. W. Kinzler (ed.), The genetic basis of human cancer. McGraw-Hill, New York, N.Y.
- 17.Green, C. M., and G. Almouzni. 2003. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 22:5163-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hand, R., and J. German. 1975. A retarded rate of DNA chain growth in Bloom's syndrome. Proc. Natl. Acad. Sci. USA 72:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 20.Hoek, M., and B. Stillman. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100:12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow, J. K., R. K. Chakraverty, and I. D. Hickson. 1997. The Bloom's syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 272:30611-30614. [DOI] [PubMed] [Google Scholar]
- 23.Karow, J. K., A. Constantinou, J.-L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufman, P. D., R. Kobayashi, N. Kessler, and B. Stillman. 1995. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81:1105-1114. [DOI] [PubMed] [Google Scholar]
- 25.Krude, T. 1995. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res. 220:304-311. [DOI] [PubMed] [Google Scholar]
- 26.Langland, G., J. Kordich, J. Creaney, K. H. Goss, K. Lillard-Wetherell, K. Bebenek, T. A. Kunkel, and J. Groden. 2001. The Bloom's syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 276:30031-30035. [DOI] [PubMed] [Google Scholar]
- 27.Liao, S., J. Graham, and H. Yan. 2000. The function of Xenopus Bloom's syndrome protein homolog (xBLM) in DNA replication. Genes Dev. 14:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenmuth, D. M., A. J. van Wijnen, S. Hiebert, J. L. Stein, J. B. Lian, and G. S. Stein. 1997. Subcellular partitioning of transcription factors during osteoblast differentiation: developmental association of the AML/CBFa/PEBP2a-related transcription factor-NMP-2 with the nuclear matrix. J. Cell. Biochem. 66:123-132. [PubMed] [Google Scholar]
- 29.Lonn, U., S. Lonn, U. Nylen, G. Winblad, and J. German. 1990. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 50:3141-3145. [PubMed] [Google Scholar]
- 30.Martini, E., D. M. J. Roche, K. Marheineke, A. Verreault, and G. Almouzni. 1998. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin following UV irradiation of human cells. J. Cell Biol. 143:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moens, P. B., R. Freire, M. Tarsounas, B. Spyropoulos, and S. P. Jackson. 2000. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113:663-672. [DOI] [PubMed] [Google Scholar]
- 33.Moggs, J. G., P. Grandi, J.-P. Quivy, Z. O. Jonsson, U. Hübscher, P. B. Becker, and G. Almouzni. 2000. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20:1206-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohaghegh, P., and I. D. Hickson. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10:741-746. [DOI] [PubMed] [Google Scholar]
- 35.Neff, N. F., N. A. Ellis, T. Z. Ye, J. Noonan, K. Huang, M. Sanz, and M. Proytcheva. 1999. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of Bloom syndrome cells. Mol. Biol. Cell 10:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opresko, P. L., C. von Kobbe, J.-P. Laine, J. Harrigan, I. D. Hickson, and V. A. Bohr. 2002. Telemere-binding protein TRF2 binds to and stimulates the werner and bloom syndrome helicases. J. Biol. Chem. 277:4378-4386. [DOI] [PubMed] [Google Scholar]
- 37.Pedrazzi, G., C. Z. Bachrati, N. Selak, I. Studer, M. Petkovic, I. D. Hickson, J. Jiricny, and I. Stagljar. 2003. The Bloom's syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol. Chem. 384:1155-1164. [DOI] [PubMed] [Google Scholar]
- 38.Pedrazzi, G., C. Perrera, H. Blaser, P. Kuster, G. Marra, S. L. Davies, G.-H. Ryu, R. Freire, I. D. Hickson, J. Jiricny, and I. Stagljar. 2001. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 29:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quivy, J.-P., P. Grandi, and G. Almouzni. 2001. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 20:2015-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray, J. H., and J. German. 1983. The cytogenetics of the chromosome-breakage syndromes, p. 135-167. In J. German (ed.), Chromosome mutation and neoplasia. Alan R. Liss, Inc., New York, N.Y.
- 41.Sengupta, S., S. P. Linke, R. Pedeux, Q. Yang, J. Farnsworth, S. H. Garfield, K. Valerie, J. W. Shay, N. A. Ellis, B. Wasylyk, and C. C. Harris. 2003. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 22:1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, S., and B. Stillman. 1991. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J. Biol. Chem. 266:12041-12047. [PubMed] [Google Scholar]
- 43.Smith, S., and B. Stillman. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15-25. [DOI] [PubMed] [Google Scholar]
- 44.Stavropoulos, D. J., P. S. Bradshaw, X. Li, I. Pasic, K. Truong, M. Ikura, M. Ungrin, and M. S. Meyn. 2002. The bloom syndrome helicase BLM interacts with TRFs in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 11:3135-3144. [DOI] [PubMed] [Google Scholar]
- 45.Taddei, A., R. D., J. B. Sibarita, B. M. Turner, and G. Almouzni. 1999. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147:1153-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 47.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 48.von Kobbe, C., P. Karmakar, L. Dawut, P. Opresko, X. Zeng, R. M. Brosh Jr., I. D. Hickson, and V. A. Bohr. 2002. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 277:22035-22044. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X. W., A. Tseng, N. A. Ellis, E. A. Spillare, S. P. Linke, A. I. Robles, H. Seker, Q. Yang, P. Hu, S. Beresten, N. A. Bemmels, S. Garfield, and C. C. Harris. 2001. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 276:32948-32955. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, L., S. L. Davies, and I. D. Hickson. 2000. Roles of RecQ family helicases in the maintenance of genome stability. Cold Spring Harbor Symp. Quant. Biol. 65:573-581. [DOI] [PubMed] [Google Scholar]
- 52.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 53.Wu, L., S. L. Davies, P. S. North, H. Goulaouic, J. F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 54.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 55.Yankiwski, V., R. A. Marciniak, L. Guarente, and N. F. Neff. 2000. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA 97:5214-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]