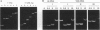Abstract
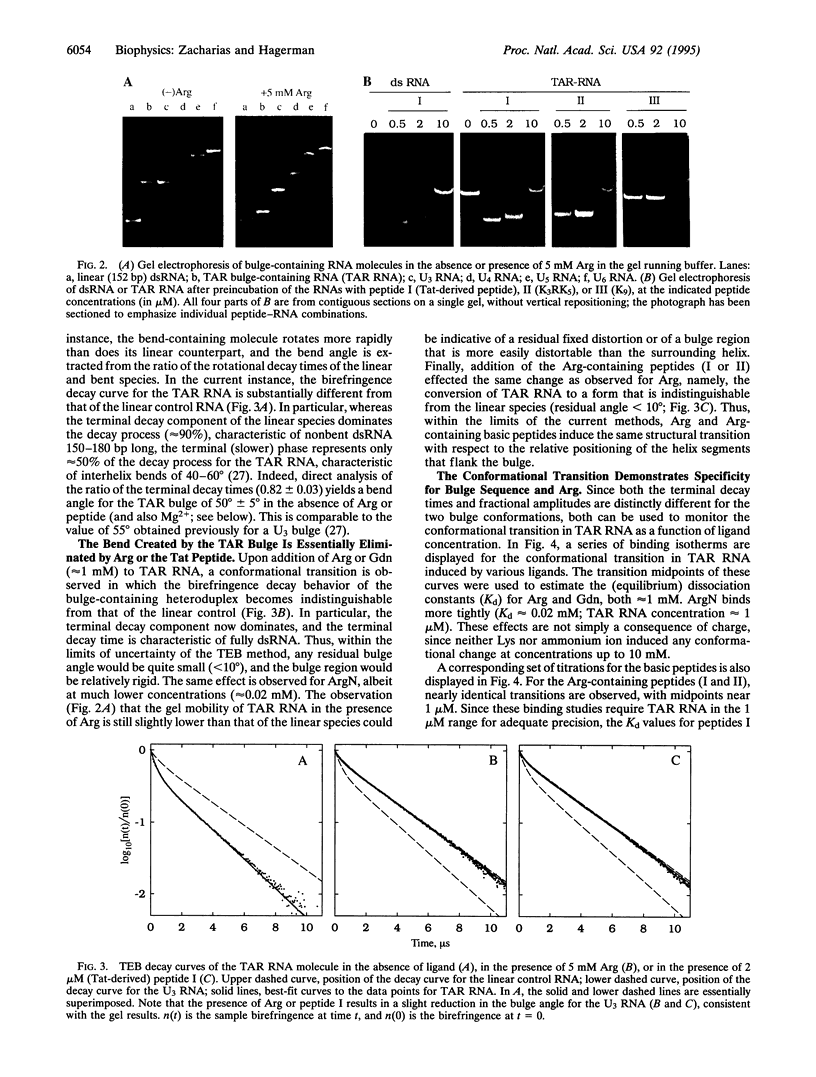
The trans-activation response element (TAR) found near the 5' end of the viral RNA of the human immunodeficiency virus contains a 3-nt bulge that is recognized by the virally encoded trans-activator protein (Tat), an important mediator of transcriptional activation. Insertion of the TAR bulge into double-stranded RNA is known to result in reduced electrophoretic mobility, suggestive of a bulge-induced bend. Furthermore, NMR studies indicate that Arg causes a change in the structure of the TAR bulge, possibly reducing the bulge angle. However, neither of these effects has been quantified, nor have they been compared with the effects of the TAR-Tat interaction. Recently, an approach for the quantification of bulge-induced bends has been described in which hydrodynamic measurements, employing the method of transient electric birefringence, have yielded precise estimates for the angles of a series of RNA bulges, with the angles ranging from 7 degrees to 93 degrees. In the current study, transient electric birefringence measurements indicate that the TAR bulge introduces a bend of 50 degrees +/- 5 degrees in the absence of Mg2+. Addition of Arg leads to essentially complete straightening of the helix (to < 10 degrees) with a transition midpoint in the 1 mM range. This transition demonstrates specificity for the TAR bulge: no comparable transition was observed for U3 or A3 (control) bulges with differing flanking sequences. An essentially identical structural transition is observed for the Tat-derived peptide, although the transition midpoint for the latter is near 1 microM. Finally, low concentrations of Mg2+ alone reduce the bend angle by approximately 50%, consistent with the effects of Mg2+ on other pyrimidine bulges. This last observation is important in view of the fact that most previous structural/binding studies were performed in the absence of Mg2+.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amiri K. M., Hagerman P. J. Global conformation of a self-cleaving hammerhead RNA. Biochemistry. 1994 Nov 15;33(45):13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Churcher M. J., Lamont C., Hamy F., Dingwall C., Green S. M., Lowe A. D., Butler J. G., Gait M. J., Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993 Mar 5;230(1):90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling U., Reid L. S., Barnett R. W., Ma M. Y., Climie S., Sumner-Smith M., Sonenberg N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: model for TAR RNA tertiary structure. J Virol. 1992 May;66(5):3018–3025. doi: 10.1128/jvi.66.5.3018-3025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Gast F. U., Amiri K. M., Hagerman P. J. Interhelix geometry of stems I and II of a self-cleaving hammerhead RNA. Biochemistry. 1994 Feb 22;33(7):1788–1796. doi: 10.1021/bi00173a023. [DOI] [PubMed] [Google Scholar]
- Gast F. U., Hagerman P. J. Electrophoretic and hydrodynamic properties of duplex ribonucleic acid molecules transcribed in vitro: evidence that A-tracts do not generate curvature in RNA. Biochemistry. 1991 Apr 30;30(17):4268–4277. doi: 10.1021/bi00231a024. [DOI] [PubMed] [Google Scholar]
- Hamy F., Asseline U., Grasby J., Iwai S., Pritchard C., Slim G., Butler P. J., Karn J., Gait M. J. Hydrogen-bonding contacts in the major groove are required for human immunodeficiency virus type-1 tat protein recognition of TAR RNA. J Mol Biol. 1993 Mar 5;230(1):111–123. doi: 10.1006/jmbi.1993.1129. [DOI] [PubMed] [Google Scholar]
- Hauber J., Cullen B. R. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol. 1988 Mar;62(3):673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kebbekus P., Draper D. E., Hagerman P. Persistence length of RNA. Biochemistry. 1995 Apr 4;34(13):4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989 Oct 20;59(2):283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Loret E. P., Georgel P., Johnson W. C., Jr, Ho P. S. Circular dichroism and molecular modeling yield a structure for the complex of human immunodeficiency virus type 1 trans-activation response RNA and the binding region of Tat, the trans-acting transcriptional activator. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9734–9738. doi: 10.1073/pnas.89.20.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Chen L., Frankel A. D., Williamson J. R. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Riordan F. A., Bhattacharyya A., McAteer S., Lilley D. M. Kinking of RNA helices by bulged bases, and the structure of the human immunodeficiency virus transactivator response element. J Mol Biol. 1992 Jul 20;226(2):305–310. doi: 10.1016/0022-2836(92)90947-i. [DOI] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Roy S., Parkin N. T., Rosen C., Itovitch J., Sonenberg N. Structural requirements for trans activation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by tat: importance of base pairing, loop sequence, and bulges in the tat-responsive sequence. J Virol. 1990 Mar;64(3):1402–1406. doi: 10.1128/jvi.64.3.1402-1406.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Shen Z., Hagerman P. J. Conformation of the central, three-helix junction of the 5 S ribosomal RNA of Sulfolobus acidocaldarius. J Mol Biol. 1994 Aug 19;241(3):415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- Tan R., Frankel A. D. Circular dichroism studies suggest that TAR RNA changes conformation upon specific binding of arginine or guanidine. Biochemistry. 1992 Oct 27;31(42):10288–10294. doi: 10.1021/bi00157a016. [DOI] [PubMed] [Google Scholar]
- Tao J., Frankel A. D. Electrostatic interactions modulate the RNA-binding and transactivation specificities of the human immunodeficiency virus and simian immunodeficiency virus Tat proteins. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1571–1575. doi: 10.1073/pnas.90.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Frankel A. D. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P. J. Bulge-induced bends in RNA: quantification by transient electric birefringence. J Mol Biol. 1995 Mar 31;247(3):486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]