Abstract
The replication of eukaryotic genomes follows a temporally staged program, in which late origin firing often occurs within domains of altered chromatin structure(s) and silenced genes. Histone deacetylation functions in gene silencing in some late-replicating regions, prompting an investigation of the role of histone deacetylation in replication timing control in Saccharomyces cerevisiae. Deletion of the histone deacetylase Rpd3 or its interacting partner Sin3 caused early activation of late origins at internal chromosomal loci but did not alter the initiation timing of early origins or a late-firing, telomere-proximal origin. By delaying initiation relative to the earliest origins, Rpd3 enables regulation of late origins by the intra-S replication checkpoint. RPD3 deletion suppresses the slow S phase of clb5Δ cells by enabling late origins to fire earlier, suggesting that Rpd3 modulates the initiation timing of many origins throughout the genome. Examination of factors such as Ume6 that function together with Rpd3 in transcriptional repression indicates that Rpd3 regulates origin initiation timing independently of its role in transcriptional repression. This supports growing evidence that for much of the S. cerevisiae genome transcription and replication timing are not linked.
Duplication of eukaryotic chromosomes is a temporally regulated process that involves accurate replication of DNA sequences coupled with assembly of chromatin. In general, transcriptionally active euchromatin replicates before the structurally condensed and transcriptionally silent heterochromatin. The significance of this epigenetic phenomenon remains to be determined; however, the temporal dynamics of chromosomal replication are thought to be mechanistically linked to the establishment and inheritance of gene expression programs encoded in the chromatin structure. Altered replication timing is associated not only with changes in gene expression, but also with chromosome instabilities and human cancers, suggesting that temporal staging of replication may contribute to the fidelity of genome duplication (reviewed in reference 21).
DNA replication initiates at specific sites distributed along each chromosome that are termed origins of replication (reviewed in references 4 and 20). Individual origins exhibit characteristic activities defined by their timing of activation in S phase and their likelihood of activation in each S phase (origin efficiency). The activation characteristics of individual origins, their relative chromosomal locations, and normal impediments to replication fork progression determine the overall dynamics of genome replication. Recent genome-wide studies of origin locations and replication timing have revealed much about the replication dynamics and organization of the Saccharomyces cerevisiae genome (36, 54). Nevertheless, differential origin efficiency and activation timing depends upon a largely undefined relationship between chromatin structure and replication initiation mechanisms.
In S. cerevisiae, origin relocation experiments have demonstrated that certain chromosomal regions impose delayed activation and/or reduced efficiency on origins (17). For example, replication origins near telomeres and the silent mating type loci are generally late replicating and/or inefficient (14, 28, 36). These chromosomal regions contain cis-acting sequences that recruit the SIR chromatin proteins, including the Sir2 histone deacetylase (reviewed in reference 39). Histone deacetylation by Sir2 enables the assembly of a higher-order chromatin structure that represses the transcription of genes within the region. Similarly, this heterochromatic structure can delay or block origin activation. Disruption of SIR chromatin in a sir3 mutant results in earlier initiation of a replication origin within a Y′ subtelomeric element but does not affect the initiation of more-internally located late origins (44). Also, mutation of SIR2 increases origin efficiency within the ribosomal DNA repeats, further suggesting a link between histone modification and origin function (31). Non-SIR-dependent chromatin also influences initiation timing through the action of undefined cis-acting sequences that flank internal chromosomal origins; however, the factors involved in this regulation remain unknown (18).
We are interested in the role of histone acetylation in the regulation of origin initiation timing, particularly of late-firing origins that are not located within SIR chromatin. The Rpd3 histone deacetylase is conserved among eukaryotes and has been characterized largely with respect to its function as a gene-specific transcriptional repressor (reviewed in reference 45). In S. cerevisiae, Rpd3 exists in a large protein complex that contains the Sin3 corepressor. This complex is recruited to certain promoters where it deacetylates the adjacent histone(s) to repress transcription of the target gene. However, genome-wide analysis of histone acetylation levels in RPD3 mutant cells indicates that deletion of RPD3 results in hyperacetylation of histones in many regions of the yeast genome, but not in SIR chromatin regions (37). Thus, Rpd3 has been proposed to act in an untargeted manner to deacetylate histones throughout much of the genome while using specific targeting factors to mediate gene-specific transcriptional repression. The widespread function of Rpd3 in histone deacetylation suggests that it could regulate initiation timing of replication origins. In this study we examined the role of Rpd3-Sin3 in regulating replication initiation of individual origins. Rpd3 delays initiation of many late-replicating origins and, thus, plays an important role in determining the overall replication dynamics of the yeast genome.
MATERIALS AND METHODS
Strains and plasmids.
Most gene deletions were constructed using the HIS5 or KANMX selectable markers as described previously (51). Correct chromosomal constructs were confirmed by PCR. All strains used in this study were in a W303 background. The following strains were derived from OAy470 (3). JAy22 (rpd3Δ::HIS5) and JAy30 (clb5Δ::KANMX) were derived directly from OAy470. JAy24 (clb5Δ::KANMX) was derived from JAy22. Rad53-HA3 was introduced into OAy470 with plasmid p404-RAD53-HA, yielding strain FHy20. DGy140 (rpd3Δ::KANMX) was derived from FHy20. OAy618 contains Polɛ-HA3 and has been described elsewhere (3). OAy781 (rpd3Δ::HIS5), JAy43 (ume6Δ::HIS5), and JAy44 (sin3Δ::HIS5) were derived from OAy618. DGy189 (clb5Δ::URA3) was derived from JAy22 by deleting CLB5 using plasmid ESD262 (42). DGy197 (cdc45Δ::CDC45-HA) and DGy198 (cdc45Δ::CDC45-HA) were derived from JAy30 and DGy189, respectively, using p404-CDC45-HA to introduce Cdc45-HA3. JAy71 (smlΔ::HIS3 rad53Δ::KANMX rpd3Δ::HIS5 cdc45Δ::CDC45-HA) was derived from a cross of DGy312 (smlΔ::HIS3 rad53Δ::KANMX pMEC2::URA3) and DGy227 (rpd3Δ::HIS5 cdc45Δ::CDC45-HA). The following strains were not derived from OAy470 but rather the listed strain. DGy159 is a bar1Δ::LEU2 derivative of 122172 (mec1Δ::TRP1 smlΔ::HIS3; from R. Rothstein); DGy170 (rpd3Δ::KANMX) is derived from DGy159. DGy164 (bar1Δ::LEU2 rad53Δ::KANMX cdc45Δ::CDC45-HA), DGy166 (bar1Δ::LEU2 cdc45Δ::CDC45-HA), and JAy72 (bar1Δ::LEU2 rad53Δ::KANMX cdc45Δ::CDC45-HA) are derivatives of 232178 (smlΔ::HIS3 in a W303 background; from R. Rothstein).
Yeast methods.
Cells were cultured and synchronized as described previously, except that phthalic acid was omitted and α-factor (Sigma) was used at 5 nM (3). DNA content analysis was performed as described previously, except that cells were stained with 1 μM SYTOX Green (Molecular Probes) with a Becton-Dickinson fluorescence-activated cell scanner (FACScan) (2).
ChIP.
Chromatin immunoprecipitation (ChIP) for hemagglutinin (HA)-tagged proteins was performed as described previously, except immunoprecipitated DNA was purified using a QiaQuick PCR purification kit (Qiagen) (1). 16B12 (Covance-Babco) and/or 12CA5 (Roche) HA antibodies were used. Analysis of histone acetylation utilized anti-acetyl H4 K5 and anti-acetyl H2A K7 antibodies (Upstate) and a modified lysis buffer (46). For all amplifications, 22 to 25 cycles of PCR were performed using 1/50 of the immunoprecipitated DNA and 1/50,000 of the total input DNA. Primer DNA sequences are available upon request. DNA was analyzed by 6% polyacrylamide gel electrophoresis (PAGE) or 2.3% Tris-borate-EDTA agarose electrophoresis stained with SYBR Green (Molecular Probes) and quantified with a fluorimager (Molecular Dynamics, Storm 860, or Bio-Rad FX Scanner) and ImageQuant or QuantityOne software. The percent bound is the ratio of immunoprecipitated to total DNA based on the quantification of the respective PCR products and the dilution factor.
Analysis of replication structures.
Two-dimensional (2-D) gel electrophoresis was performed as described at the website http://fangman-brewer.genetics.washington.edu. Nascent DNA strand analysis was as described elsewhere (41). AlkPhos Direct (Amersham Pharmacia) was used for probe labeling and detection for the experiments shown below in Fig. 2 and 7 (ARS305, ARS603, and ARS1413). All other probes were radioactively labeled using a MegaPrime DNA labeling kit (Amersham Pharmacia) and detected on a phosphorimager (Molecular Dynamics, Storm 860, or Bio-Rad FX scanner). For 2-D gels, the following DNA fragments were analyzed at each origin: ARS1, 4.7-kb NcoI(/BamHI); ARS305, 5.8-kb EcoRI; ARS319, 3.2-kb XbaI; ARS603, 3.6-kb NcoI/BamHI; ARS1413, 5.3-kb EcoRI; ARS716, 4.9-kb EcoRI/XbaI; ARS1007, 5.3-kb XbaI(/EcoRI); and ARS1212, 5.8-kb EcoRI(/XbaI). Analysis of different origins digested with the same enzyme(s) was accomplished by stripping and reprobing the blot(s). ImageQuant was used to quantify the “bubble” arc signal and the 1N spot to determine relative origin efficiency [(bubble arc)/(1N spot)].
FIG. 2.
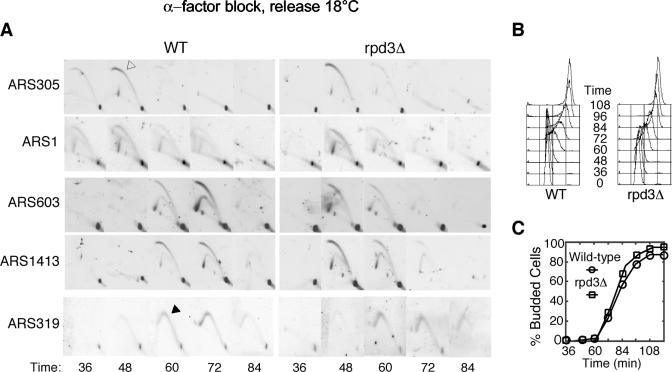
Rpd3 regulates internal late-firing origins. Wild-type (OAy618) and rpd3Δ (OAy781) cells were synchronized in late G1 at 23°C with α-factor and released into S phase at 18°C. At the indicated intervals, cells were fixed for analysis. (A) 2-D agarose gel electrophoresis analysis of ARS305, ARS1, ARS603, ARS1413, and ARS319. An open arrow indicates a bubble arc, and a Y arc is marked with a filled arrow. (B) DNA content was analyzed by FACScan. (C) The percentage of budded cells was determined by microscopic analysis of at least 100 cells at each time point.
FIG. 7.
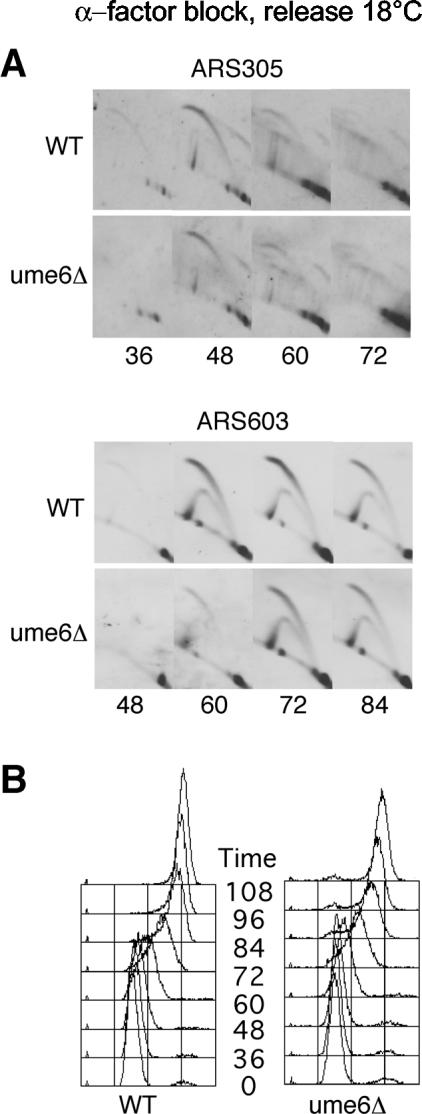
Ume6 is not required for regulation of origin initiation timing. Wild-type (OAy618) and ume6Δ (JAy43) cells were analyzed for replication structures at ARS305 and ARS603 (A) or by DNA content analysis (B) as described in the legend for Fig. 2.
RESULTS
Rpd3-Sin3 histone deacetylase complex delays activation of internal late origins.
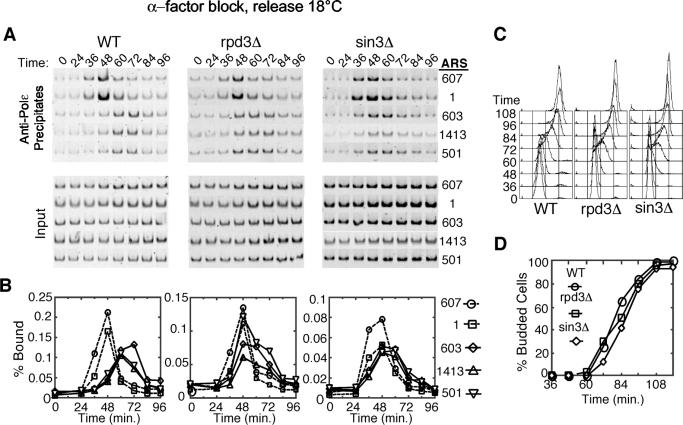
The critical role of histone acetylation and deacetylation as a mechanism for regulating chromatin structure and the function of Rpd3 in deacetylation of histones prompted us to analyze the effects of RPD3 and SIN3 deletion on origin initiation dynamics in S. cerevisiae. We used ChIP to monitor the temporal association of DNA polymerase ɛ (Polɛ) with representative early and late replication origins, which reflects the initiation time of these origins (2). G1 cells were released synchronously into S phase at 18°C, allowing clear distinction between initiation events at early versus late origins (Fig. 1). In wild-type cells, Polɛ associations with the early origins ARS607 and ARS1 were maximal at 48 min, whereas Polɛ associations with the late origins ARS603, ARS1413, and ARS501 peaked at 60 to 72 min (Fig. 1A and B). This pattern was consistently reproducible, including slightly earlier Polɛ association with ARS607 than with ARS1 and slightly later Polɛ association with ARS603 than with ARS501 and ARS1413. In rpd3Δ and sin3Δ cells, timing of Polɛ association with early origins was indistinguishable from that of the wild-type cells. However, in the rpd3Δ and sin3Δ cells, Polɛ association with ARS603, ARS1413, and ARS501 was well advanced, each reaching its maximum about 48 min following G1 release. The timing of peak Polɛ association with these late origins was similar to that with the early origins, indicating that these normally late-firing origins were being activated significantly earlier in the rpd3Δ and sin3Δ cells. The earlier association of Polɛ with ARS603 was lost upon mutational inactivation of ARS603, indicating that this association resulted from replication initiation and confirming that its timing was advanced in the rpd3Δ and sin3Δ cells (data not shown). Because wild-type, rpd3Δ, and sin3Δ cells exhibited very similar cell cycle progression (Fig. 1C and D) and early origin activation timing, our results indicate that the Rpd3-Sin3 deacetylase complex differentially regulates the initiation timing of these late-firing origins.
FIG. 1.
Rpd3 and Sin3 delay activation of late-firing replication origins. Wild-type (OAy618), rpd3Δ (OAy781), and sin3Δ (JAy44) cells were synchronized in late G1 with α-factor at 23°C and released into S phase at 18°C. At the indicated intervals, cells were fixed for analysis. (A) DNA Polɛ association with early (ARS607 and ARS1) and late (ARS603, ARS1413, and ARS501) replication origins was analyzed by ChIP. Origin-specific PCR analysis of immunoprecipitated (Precipitates) and total (Input) DNA is shown. (B) Quantification of the data shown in panel A plotted as the percent bound. (C) DNA content of cells was analyzed by FACScan. (D) The percentage of budded cells was determined by microscopic analysis of at least 100 cells at each time point.
We also examined the timing of origin activation using 2-D agarose gel electrophoresis (2-D gel) to observe initiation (bubble) structures in time courses comparable to those for the Polɛ ChIP analyses. In wild-type cells, replication bubble arcs for the early origins ARS305 and ARS1 were most prominent at 48 min and were observed at 60 and 72 min for the late origin ARS1413 and predominantly at 72 min for ARS603 (Fig. 2A). In rpd3Δ cells, bubble arcs were observed at the early origins predominantly at 48 min, as in the wild-type cells. However, initiation structures at the late origins ARS603 and ARS1413 were observed significantly earlier in rpd3Δ than in the wild-type cells, almost simultaneously with the early origins; the majority of initiation events at ARS603 and ARS1413 occurred at about 48 min. Introduction into the rpd3Δ cells of a plasmid expressing wild-type RPD3 restored late-initiation timing to ARS603, ARS1413, and ARS501, whereas expression of RPD3 with a point mutation in its catalytic deacetylase domain did not (data not shown). Together, these results indicate that Rpd3-Sin3 deacetylase activity modulates the execution time of a critical step in the replication initiation process at a subset of chromosomal replication origins.
ARS603 and ARS1413 are internal chromosomal origins, and ARS501 is internal to a subtelomeric Y′ element (about 20 kb from the telomere). Initiation timing of ARS1413 and ARS501 (ARS603 was not tested) is not affected directly by SIR3, which regulates origins within subtelomeric X and Y elements (9, 44). To determine whether Rpd3 regulates initiation of a subtelomeric origin and an origin within SIR chromatin, we analyzed the replication timing of ARS319 (an efficient, late-firing origin within the X element adjacent to the right telomere of chromosome III) and HML (which contains four inactive, late-replicating ARS elements). The proximity of ARS319 to the chromosome end precludes analysis of bubble structures (because they very rapidly convert into Y structures as one replication fork completes replication of the telomere); therefore, the appearance of replication fork Y structures instead of bubbles was analyzed as an indicator of replication initiation timing (Y arcs) (Fig. 2A). Replication timing of ARS319 and HML was not altered in rpd3Δ cells (Fig. 2A; data not shown for HML). Thus, Rpd3-Sin3 appears to regulate initiation timing of internal, late origins that are not under SIR regulation. Moreover, the specificity of this regulation argues against global effects on cell cycle progression or replication initiation due to changes in gene transcription as a result of RPD3 deletion.
Deletion of RPD3 results in alterations of chromatin structure that likely are responsible for the advanced initiation timing of the late origins analyzed in this study. Deacetylation of origin-proximal histones could also potentially influence the efficiency of origin activation. We analyzed replication efficiency of the origins analyzed above using 2-D gel analysis of unsynchronized, logarithmically growing cells to compare the ratio of bubble to fork structures. We were unable to discern a difference in the initiation frequency of the efficient origins analyzed above in rpd3Δ cells (data not shown). Nor did we observe differences in firing of inefficient origins, including ARS304, ARS308, and the four HML origins (data not shown).
Late origins elude the intra-S replication checkpoint in rpd3Δ cells.
When cells enter S phase in the presence of the replication inhibitor hydroxyurea (HU) or the DNA-damaging agent methyl methanesulfonate (MMS), early origins fire, but firing of late origins is inhibited in a checkpoint-dependent manner (32, 41, 43, 49). These studies support a model in which replication forks emanating from early-activated origins stall due to nucleotide depletion (HU) or DNA lesions (MMS) and generate a checkpoint signal that inhibits subsequent origin initiation. It follows that earlier timing of normally late origins relative to the earliest origins should permit their initiation before an intra-S checkpoint is activated. We therefore tested whether the advanced initiation timing of late origins in rpd3Δ cells allowed these origins to bypass this checkpoint.
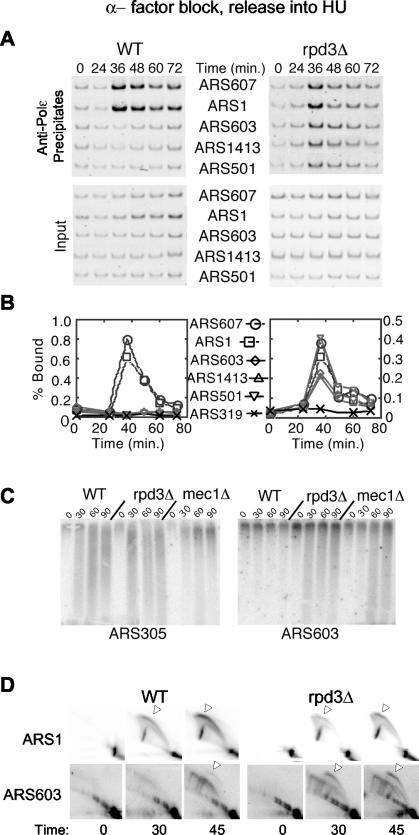
Wild-type and rpd3Δ cells synchronized in G1 were released into medium containing HU, and origin initiation was analyzed with ChIP of Polɛ, nascent DNA strand detection, and 2-D gels. In wild-type cells, Polɛ associated with the early origins at 36 min but was blocked from loading onto the late origins (Fig. 3A and B). In the rpd3Δ cells, Polɛ associated with the early origins and internal, late origins concurrently at 36 min, indicating that these normally late origins initiated early and escaped checkpoint inhibition. Association of Polɛ with the subtelomeric origin ARS319 was not detectable. These results suggest that the internal, late origins initiated replication, escaping checkpoint inhibition, while ARS319, which was not advanced in initiation, did not escape checkpoint control.
FIG. 3.
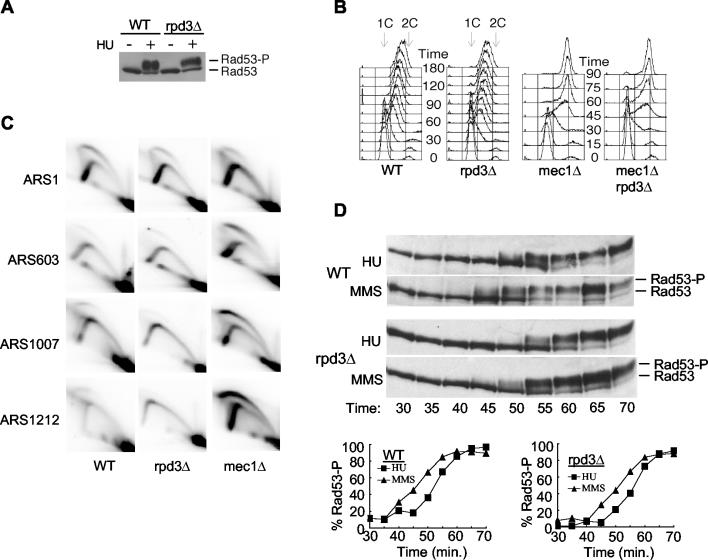
Late origins escape checkpoint inhibition in rpd3Δ cells treated with HU. Wild-type (OAy618) and rpd3Δ (OAy781) cells (A and B) or wild-type (OAy470), rpd3Δ (JAy22), and mec1Δ (DGy159) cells (C and D) were synchronized in late G1 with α-factor at 23°C and released into S phase in the presence of 200 mM HU at 23°C. Samples were collected at the indicated intervals. DNA Polɛ association with origins was analyzed by ChIP (A) and quantified (B) as described in the Fig. 1 legend. (C) Nascent DNA strand analysis of ARS305 and ARS603. (D) Replication structures detected by 2-D gel analysis.
Analysis of nascent DNA strands using denaturing agarose gel electrophoresis supported the results of the ChIP analysis. As expected, newly synthesized DNA was detected at the early origin ARS305 in wild-type, rpd3Δ, and mec1Δ cells (Fig. 3C). When the blots were reprobed to analyze ARS603 and ARS1413, nascent strands were not detected at these origins in wild-type cells but were detected in the mec1Δ mutant cells due to loss of the checkpoint (data not shown for ARS1413) (41). In rpd3Δ cells, nascent strands were detected at the late origins, indicating escape from the checkpoint (Fig. 3C). Analysis of replication structures with 2-D gels also confirmed that late origins escaped checkpoint inhibition in rpd3Δ cells. Bubble arcs at the late origin ARS603 were significantly more abundant in rpd3Δ cells than in wild-type cells (Fig. 3D). Together, these data demonstrate that late origins escape checkpoint inhibition in response to HU and support the notion that the RPD3-mediated delay of origin initiation is necessary for effective origin regulation by the intra-S checkpoint pathway.
The intra-S checkpoint pathway is intact in rpd3Δ cells.
Escape from checkpoint inhibition might reflect a loss of checkpoint function rather than an alteration of initiation timing in rpd3Δ cells. To address this we determined whether the intra-S checkpoint signaling pathway was intact by examining two consequences of its activation: Rad53 phosphorylation and a reduced rate of genome replication (32, 40, 47). In the presence of HU, rpd3Δ cells displayed normal Rad53 phosphorylation, based on the mobility shift of Rad53 as analyzed by sodium dodecyl sulfate-PAGE (Fig. 4A). Next, we analyzed the replication rate of cells in the presence of MMS. Replication was similarly slowed in the wild-type and rpd3Δ cells, whereas mec1Δ cells, which are defective in the checkpoint, exhibited much less inhibition of the overall rate and extent of replication (Fig. 4B). Thus, the intra-S checkpoint pathway is intact in rpd3Δ cells.
FIG. 4.
The checkpoint pathway is intact in rpd3Δ cells. (A) Wild-type (FHy20) and rpd3Δ (DGy140) cells were grown for 1 h at 30°C in the presence or absence of 200 mM HU. Protein extracts prepared by trichloroacetic acid precipitation were analyzed by Western blotting using anti-HA antibody (16B12) to detect Rad53-HA (33). (B) Wild-type (OAy470), rpd3Δ (JAy22), mec1Δ (DGy159), and mec1Δ rpd3Δ (DGy170) cells were synchronized in late G1 with α-factor at 23°C and released into S phase at 30°C in yeast extract-peptone-dextrose containing 0.033% MMS. At the indicated intervals, cells were fixed for DNA content analysis. (C) Cells were treated as for panel B and fixed for analysis at 25, 32, 39, 46, and 53 min after release from α-factor. Cells from all time points were pooled to prepare S-phase DNA for 2-D gel analysis. (D) Wild-type (FHy20) and rpd3Δ (DGy140) cells synchronized at 23°C with α-factor were released into S phase at 18°C, and samples were collected at the indicated times. Rad53 phosphorylation was analyzed by Western blotting as for panel A and was quantified (percent Rad53-P) by direct analysis of the chemiluminescent signal using a Bio-Rad ChemiDoc system and QuantityOne software. Percent phosphorylation was calculated as (Rad53-P)/(Rad53-P + Rad53), where Rad53-P includes all slower-migrating forms of Rad53.
The more-rapid genome replication in mec1 cells treated with MMS has been ascribed to precocious activation of normally inefficient origins and/or loss of late origin inhibition in the presence of MMS (32, 41, 43, 48). Because late origins were not efficiently inhibited by HU in rpd3Δ cells, we anticipated that late origins would escape inhibition by MMS treatment also. However, the low rate of genome duplication in MMS-treated rpd3Δ cells suggested either that late origins were inhibited by MMS or that the more-rapid replication in the mec1Δ cells was due primarily to activation of normally inefficient origins rather than loss of late origin regulation. To distinguish between these possibilities, we analyzed late origin firing in cells released into S phase in the presence of MMS. Samples from five time points spanning S phase were collected and pooled, allowing a composite analysis of all initiation events during this period. In addition to the early origin ARS1 and the late origin ARS603, we analyzed two additional, nontelomeric late origins, ARS1007 and ARS1212, which are recently characterized late origins that conveniently could be analyzed using the same DNA digest for 2-D gel analysis (36, 54). As expected, early origin ARS1 fired in each of the analyzed strains (Fig. 4C). Also, mec1Δ cells exhibited robust firing of late origins ARS603, ARS1007, and ARS1212, whereas firing of these origins was inhibited in wild-type cells. Consistent with the low replication rate in MMS, late origin firing of rpd3Δ cells was inhibited as in wild-type cells. Analysis of individual time points (rather than the pooled time course) yielded similar results (data not shown). Thus, deletion of RPD3 permits late origins to escape checkpoint inhibition triggered by nucleotide depletion with HU, but not that triggered by DNA damage with MMS.
This unexpected difference in late origin firing prompted us to test the kinetics of checkpoint activation in response to replication inhibition by HU versus DNA damage by MMS. We analyzed the kinetics of Rad53 phosphorylation by monitoring the mobility shift of Rad53 with sodium dodecyl sulfate-PAGE in wild-type and rpd3Δ cells released synchronously into S phase at 18°C in the presence of HU or MMS. In wild-type and rpd3Δ cells, phosphorylation of Rad53 increased about 5 min sooner in the presence of MMS compared to that in HU (Fig. 4D). Although the difference in onset of Rad53 phosphorylation was relatively minor, the increase in Rad53 phosphorylation coincided closely with the timing of late origin activation in rpd3Δ cells. For example, ARS603 initiates at about 48 min in rpd3Δ cells released from G1 into S phase at 18°C (Fig. 1 and 2), at which time phosphorylation of Rad53 is just beginning in HU-treated cells (Fig. 4D, 50 min). In contrast, Rad53 phosphorylation has already begun to increase by 45 min and reaches about 50% of the maximal level by 50 min in MMS-treated rpd3Δ cells (Fig. 4D). In similar experiments conducted at 23 and 30°C, the onset of Rad53 phosphorylation was also detected earlier in MMS than in HU cultures (data not shown). Thus, checkpoint activation was slightly more rapid in the presence of MMS than with HU in both wild-type and rpd3Δ cells, and this difference likely accounts for the ability of MMS but not HU to inhibit late origin firing in rpd3Δ cells.
We also note that rpd3Δ cells exhibit a slight but reproducible delay in onset of Rad53 phosphorylation in both HU and MMS compared to that in wild-type cells (Fig. 4D). This could be due to slightly delayed cell cycle onset in rpd3Δ cells; however, we detected no delay in the onset of replication (by DNA content analysis) or budding (data not shown). This delay in Rad53 phosphorylation could contribute to the escape of late origins from HU-induced checkpoint inhibition in rpd3Δ cells; however, advanced initiation timing is critical. By 72 min, when ARS603 fires in wild-type cells (Fig. 1), Rad53 is predominantly in its activated, phosphorylated form in rpd3Δ cells (Fig. 4D).
Histone deacetylation by Rpd3 appears to be independent of Rad53 and Mec1 function.
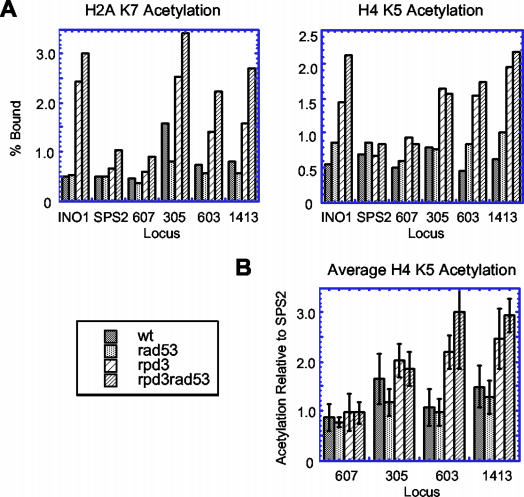
In addition to the RPD3 mutant, a RAD53 mutant strain exhibits deregulated origin firing time (41, 43), raising the possibility that checkpoint proteins mediate the function of Rpd3 in histone deacetylation at origins. To address this we analyzed Rpd3-dependent histone deacetylation by examining acetylation levels of histones H2A and H4 using ChIP (46). We compared H2A K7 and H4 K5 acetylation in wild-type, rpd3Δ, rad53Δ, and rpd3Δ rad53Δ cells at early and late origins and at sequences known to be (INO1) or not be (SPS2) deacetylated by Rpd3 (38). This analysis detected acetylation of one to two histones flanking each side of these sequences. In rpd3Δ cells, acetylation levels of H2A K7 and H4 K5 at INO1, ARS305, ARS603, and ARS1413 increased relative to that in wild-type cells, while little or no increase was observed at SPS2 and ARS607 (Fig. 5A). Similar results were seen when an average H4 K5 acetylation from three experiments was calculated for each locus relative to that at SPS2 (Fig. 5B). Thus, it appears that Rpd3 deacetylates histones flanking origins ARS305, ARS603, and ARS1413. Deletion of RAD53 had little to no effect on acetylation of H2A K7 and H4 K5 (Fig. 5). Similar results were obtained in mec1Δ cells (data not shown). In cells lacking RAD53, deacetylation surrounding these origins was still dependent on Rpd3, as acetylation levels of these sequences increased upon deletion of RPD3 (Fig. 5). Thus, Rpd3-mediated deacetylation of histones H2A K7 and H4 K5 surrounding replication origins appears to be independent of Rad53 and Mec1 function.
FIG. 5.
Histone deacetylation by Rpd3 functions independently of Rad53. (A) ChIP analysis of histone acetylation in asynchronous cultures of wild-type (DGy166), rad53Δ (DGy164), rpd3Δ (JAy72), and rad53Δ rpd3Δ (JAy71) cells. (B) Average H4 K5 acetylation from three experiments as in panel A was calculated for each locus relative to that of SPS2. SPS2 serves as an internal control, as its acetylation is not affected by Rpd3 and enables comparison between different experiments in which the absolute levels of chromatin immunoprecipitated can vary significantly.
Rpd3 regulates many origins.
Late replicating origins appear to contribute significantly to the timely duplication of the genome, as clb5Δ mutant cells, which are deficient in activation of late origins, exhibit a prolonged period of DNA replication (13, 16, 25, 42). In cells lacking the S phase cyclin Clb5, activation of early origins occurs efficiently and is dependent on Clb6, which effectively activates early but not late origins, suggesting its activity is most abundant early in S phase (13). If Rpd3 determines the activation timing of a significant proportion of the late origins in the cell, deletion of RPD3 might increase the replication rate in clb5Δ cells if earlier initiation of late origins facilitated their activation when Clb6 is thought to be more abundant. Indeed, clb5Δ rpd3Δ cells replicated their DNA with approximately wild-type kinetics, significantly faster than clb5Δ cells (Fig. 6A). Wild-type and clb5Δ rpd3Δ cells were about half finished replicating the genome at 72 min and fully duplicated at about 120 min, whereas the clb5Δ cells were about half replicated at about 96 min and did not complete replication in this time course. These data are consistent with the idea that earlier firing of late origins enables their activation by Clb6 and that initiation timing of many such origins is determined by Rpd3.
FIG. 6.
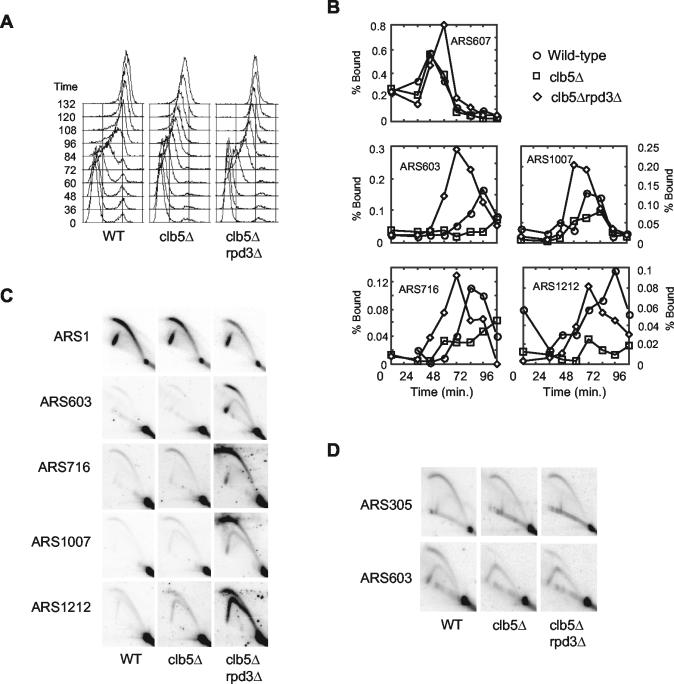
Advanced timing of late origin activation upon deletion of RPD3 restores normal S-phase kinetics in clb5Δ cells. Wild-type (OAy617), clb5Δ (DGy197), and rpd3Δ clb5Δ (DGy198) cells were synchronized in late G1 at 23°C with α-factor and released into S phase at 18°C. (A) At the indicated intervals, cells were fixed for DNA content analysis. (B) Association of Cdc45-HA with origin DNA was monitored by ChIP at the indicated times. (C) Wild-type (OAy470), clb5Δ (JAy30), and rpd3Δ clb5Δ (JAy24) cells were synchronized in late G1 at 23°C with α-factor and released at 23°C into medium containing 200 mM HU for 60 min. Replication structures were analyzed by 2-D agarose gel electrophoresis. (D) Replication structures from asynchronous cultures (30°C) of wild-type (OAy470), clb5Δ (JAy30), and rpd3Δ clb5Δ (DGy189) cells were analyzed by 2-D agarose gel electrophoresis.
In clb5Δ cells, firing of most late origins is decreased but not eliminated (J. Aparicio, D. Gibson, and O. Aparicio, unpublished data). Therefore, the increased rate of genome replication in the clb5Δ rpd3Δ cells could be due to earlier and/or more efficient activation of a significant proportion of late origins. To determine the effect of RPD3 deletion on late origin firing in clb5Δ cells, we analyzed origin firing with ChIP of Cdc45, which associates with origins at the time of initiation. Wild-type, clb5Δ, and clb5Δ rpd3Δ cells were synchronized in G1 and released into S phase at 18°C. In wild-type cells, Cdc45 bound to early origin ARS607 at 36 min and to late origins ARS603, ARS716, ARS1007, and ARS1212 at 60 to 84 min (Fig. 6B; ARS716 is another recently characterized late origin that conveniently could be analyzed on the same blot [Fig. 6C]). In clb5Δ and clb5Δ rpd3Δ cells, Cdc45 bound to early origin ARS607 with timing similar to that in wild-type cells, although peak association occurred at 60 min in the clb5Δ rpd3Δ cells. Significantly decreased association of Cdc45 with all of the late origins reflected their deficient activation in clb5Δ cells. However, in clb5Δ rpd3Δ cells, association of Cdc45 with each late origin was restored and occurred earlier than in wild-type cells, despite the lack of CLB5 function. These data suggest that deletion of RPD3 advances the timing and increases the efficiency of many late origins in clb5Δ cells.
We confirmed that late origins were activated earlier in clb5Δ cells lacking RPD3 by using 2-D gel analysis of cells synchronized in G1 with α-factor and released into HU for 60 min (Fig. 6C; similar results were obtained 45 min after release [data not shown]). Firing of early origin ARS1 was observed in wild-type, clb5Δ, and clb5Δ rpd3Δ cells, while firing of the late origins was inefficient in wild-type and clb5Δ cells due to checkpoint inhibition as well as lack of Clb5 function in the clb5Δ cells. Initiation at ARS603, ARS716, ARS1007, and ARS1212 was readily observed in clb5Δ rpd3Δ cells (Fig. 6C). Initiation of these normally late origins is consistent with the assumption of an early firing program, which facilitates their activation by Clb6 while also eluding checkpoint control.
The earlier activation of late origins in clb5Δ rpd3Δ cells is likely sufficient to restore the wild-type rate of overall genome replication. In addition, earlier firing of these origins might be expected to increase their efficiency by reducing the likelihood of their being replicated (without initiation) by replication forks from proximal early origins. To determine if origin efficiency was increased in clb5Δ rpd3Δ cells, origin firing was examined in unsynchronized cells. Because the efficiency of ARS603 was more severely affected by deletion of CLB5 than other origins we have analyzed, we chose ARS603 for this analysis. Initiation of ARS603 was severely compromised in the absence of CLB5, and deletion of RPD3 increased ARS603 efficiency about 2.2- ± 0.1-fold (n = 2), based on measurement of the bubble arc intensity relative to the 1N spot (Fig. 6D). The incomplete suppression of the clb5Δ initiation defect by deletion of RPD3 may reflect a competition among early and normally late origins for a limiting amount of Clb6 or another replication factor(s) (Fig. 6C; note the reduced ARS1 firing in clb5Δ rpd3Δ cells in the presence of HU). The combined effects of earlier activation and increased efficiency of internal, late origins should lead to a substantial increase in the number of replication forks synthesizing DNA early in S phase. Thus, these data strongly suggest that initiation timing of late origins can have a significant impact on genome replication kinetics and that a considerable proportion of all late origins throughout the genome are under Rpd3 control.
Regulation of initiation timing by Rpd3-Sin3 does not require proteins important for transcriptional repression.
The regulation of origin timing by Rpd3 could share mechanisms with or otherwise arise from targeted, local deacetylation integral to transcriptional repression by Rpd3-Sin3. This is plausible because, in S. cerevisiae, replication origins generally are located within intergenic regions that frequently also contain gene promoters (30, 54). To address this we tested whether proteins involved in transcriptional regulation by Rpd3 influence origin timing. Ume6 and Ume1 are required for repression of meiotic genes by the Rpd3-Sin3 complex (24, 27, 34). Ume6, a DNA binding protein, interacts with Sin3 to target Rpd3 to promoters containing a URS1 sequence. Sequence scanning identified a potential URS1 element(s) (based on the consensus defined in reference 5) near each of four late-firing origins (ARS501, ARS603, ARS1413, and ARS1412), whereas no URS1 was identified near three of four early origins (ARS305, ARS1, and ARS307). This was consistent with the possibility that URS1-bound Ume6 recruits Rpd3-Sin3 to regulate origin initiation timing. To test this we analyzed origin initiation timing in ume6Δ cells. Deletion of UME6 did not alter initiation timing of the origins analyzed in this study (Fig. 7). Furthermore, deletion of a URS1 site in the DMC1 gene promoter did not alter timing of the proximal ARS501 (data not shown). These results indicate that Ume6 is not required for the recruitment of Rpd3 to chromatin for its role in regulation of initiation timing.
The precise mechanism by which Ume1 contributes to Rpd3-dependent transcriptional repression is not clearly understood; however, Ume1 interacts with Sin3 and Rpd3, likely serving to stabilize the DNA-associated complex. Also, Ume1 colocalizes with Rpd3 more extensively than does Ume6 in genome-wide chromatin binding analyses (26, 27). As with UME6, deletion of UME1 had no effect on initiation timing (data not shown). Likewise, we examined Tup1. The Tup1-Ssn6 repressor complex is targeted to promoters by a number of different DNA binding proteins, where it serves to mediate gene repression in a mechanism involving interactions with histone deacetylases, including Rpd3 (10, 52). However, we observed no alterations in origin timing in tup1Δ cells (data not shown). Thus, proteins that collaborate with Rpd3 to regulate gene expression are not individually responsible for the role of Rpd3 in origin timing regulation.
DISCUSSION
Rpd3-Sin3 regulates chromosomal replication timing.
We have identified the Rpd3-Sin3 histone deacetylase complex as a crucial factor that establishes the replication dynamics of the S. cerevisiae genome. This finding opens new avenues for investigating how chromatin structure influences chromosomal replication dynamics, which remains poorly understood. Rpd3-Sin3 delays replication initiation of most nontelomeric, late-firing origins that we have analyzed. In the absence of RPD3 or SIN3, normally late-firing origins initiated replication early in S phase, while the activation time of early origins, a telomeric origin, and origins located within a SIR chromatin domain (HML) remained unchanged (Fig. 1 and 2; data not shown for HML). Because the effect of RPD3 deletion is specific to a subset of origins, it is unlikely that these results are due to indirect effects on transcription of cell cycle or replication genes, which might universally alter origin function. Other factors may also delay initiation of the late origins we have analyzed, because in the absence of RPD3 the late origins still are slightly delayed relative to the earliest firing origins.
The specificity of Rpd3's timing regulation to internal, late origins reported here contrasts with a previous report that found advanced replication timing of early and late origins in rpd3Δ cells (50). There appear to be two components to this difference. First, the rpd3Δ cells in the Vogelauer et al. study exhibited somewhat advanced cell cycle entry, as both S-phase entry and budding occurred earlier than in wild-type cells following release from α-factor arrest (50). Compared to the rpd3Δ cells, which entered the cell cycle synchronously, the wild-type cells were delayed in cell cycle entry and were much less synchronous, complicating interpretation of the data. We consistently observed no difference in the onset or length of S phase between rpd3Δ and wild-type cells released from α-factor at 18, 23, or 30°C, nor did we observe differences in budding kinetics (Fig. 1 and 2 and data not shown). Vogelauer et al. attributed much of the advancement in early origin firing to this advanced cell cycle entry and, after accounting for this difference, they concluded that Rpd3 exerted more influence on late than on early origins. Nevertheless, their conclusion that both early and late origin initiations were advanced relative to cell cycle entry differs from our findings. Our conclusion that Rpd3 functions differentially at early and late origins is supported by characterization of Rpd3's impact on checkpoint regulation. The firing of late origins in rpd3Δ cells treated with HU, which normally inhibits late origin firing, argues that late origins are being significantly advanced in their timing relative to the earliest firing origins, which generate the inhibitory signal (Fig. 3).
Mechanism of Rpd3 interaction with replication origins.
Our studies do not exclude that RPD3 can affect timing of early origins, origins near telomeres, or those within SIR chromatin, but they argue that its action predominates at certain chromosomal sequences or regions. Potential effects of Rpd3 on origins within SIR chromatin, for instance, may be masked by a dominant effect of SIR chromatin within these regions. The number of chromosomal origins affected by Rpd3 appears to be extensive, because RPD3 deletion advanced initiation timing of most late-firing, nontelomeric origins we have analyzed (eight of nine) (Fig. 1, 2, and 5 and data not shown) and substantially increased the rate of genome replication in clb5Δ cells, which are deficient in late origin activation. These findings are consistent with results of genome-wide binding analysis of Rpd3 that found it associates with many internal loci but is largely excluded from telomeric regions (26).
To gain insights into the mechanism of Rpd3 function, we examined proteins involved in Rpd3 transcriptional control of individual genes and found no evidence that deletion of these genes altered origin firing time. For example, deletion of UME6 derepresses DMC1 transcription about 20-fold but does not alter replication timing of the proximal ARS501 (data not shown) (53). Moreover, analysis of changes in gene expression resulting from RPD3 deletion of genes proximal to replication origins showed no greater effect (on average) for early versus late origins (data not shown and reference 5). Thus, advanced replication initiation in rpd3Δ cells is not due to local derepression of transcription, consistent with the conclusion of a recent study that no correlation exists between transcription levels and replication timing in S. cerevisiae (36). Our findings suggest that a distinct mechanism determines the interaction of Rpd3 with chromatin at promoters versus replication origins. There may be an origin-specific factor or, instead, origin regulation may derive, directly or indirectly, from the combined action of multiple factors that individually recruit Rpd3 to distinct, but perhaps origin-proximal, sites.
Whereas the recruiting mechanism appears to be distinct from that utilized in transcriptional control, the critical biochemical effect of Rpd3 that regulates origin timing is likely the same, deacetylation of histones. Direct examination of histone acetylation near origins showed increases in histone acetylation (H2A K7, H4 K5, and H4 K12) upon deletion of Rpd3 (Fig. 5) (50). Although no strict correlation between the observed effect of Rpd3 on origin timing and absolute levels of local histone acetylation has emerged, greater increases in acetylation are generally observed near late versus early origins upon Rpd3 deletion (Fig. 5) (50). At this stage, it is not clear whether any of the differences in acetylation levels that have been observed near origins are functionally related to initiation timing. Nor is it clear whether histone modifications must be proximal to an origin or can act over some distance. That local acetylation can modulate origin timing has been demonstrated through artificial targeting of the histone acetyltransferase, Gcn5, to a normally late-firing origin, resulting in higher local levels of histone acetylation and advanced initiation timing (50). These data suggest that, in general, histone deacetylation delays origin initiation. However, the overall patterns of histone acetylation may obscure a highly specific acetylation-deacetylation event, or series of events, which could be cell cycle specific and much less obvious. Indeed, establishment of late replication timing appears to occur within the M-to-G1 period of the cell cycle (12, 35). Alternatively, it remains possible that Rpd3 directly regulates the activity of a replication protein through deacetylation. Cdc45 is an intriguing candidate, because its association with replication origins is temporally regulated (2) and Rpd3 and Sin3 copurify with Cdc45, suggesting these coexist in a complex in vivo (19).
How might histone deacetylation regulate origin timing?
Histone deacetylation by Sir2 promotes the assembly of a higher-order chromatin structure that silences transcription of the underlying genes and blocks or delays origin firing (39). Targeting of SIR chromatin to an early origin delays its firing but also silences local transcription (55). Rpd3 appears to function quite differently, as no correlation is observed between its effect on gene expression and origin timing, and Rpd3 interacts with chromatin much more extensively throughout the genome. Hence, histone deacetylation by Rpd3 may alter the interaction of origins with initiation factors without forming a heterochromatic structure. The cell cycle establishment of origin timing coincides closely with assembly of the prereplicative complex (pre-RC) at origins in early G1 (11, 12, 35). Although pre-RC assembly (defined as MCM association) occurs simultaneously at early- and late-programmed origins (2), critical targets of activation in the pre-RC could be occluded at late origins, prohibiting their activation upon S-phase entry. However, a limited accessibility must be temporary, because later in S phase origin activation occurs. Thus, temporal regulation of origins might involve, first, the establishment of an inhibitory state likely dependent on histone deacetylation, followed by a reversal or modification to overcome the inhibition in mid-S phase. Histone acetylation or acetylation of replication factors is an intriguing possibility, as MYST-family histone acetyltransferases have been implicated in origin function (7, 15, 23).
A different, but not exclusive, hypothesis is that histone acetylation-deacetylation influences the subnuclear localization of chromatin. Late replicating origins associate with the nuclear periphery in early G1, roughly coincident with the stable establishment of the late-replicating state (12, 22, 35). Perhaps more highly acetylated chromatin is better poised for immediate assembly into a limiting or regulated number of replication “factories” (8). Hence, some origins fire late because they have to wait for a replication factory to become available as early replicons complete DNA synthesis. Checkpoint proteins may participate in a mechanism that regulates the number of concurrently active replication forks, as deletion of RAD53 allows normally inactive origins to fire and late origins to fire earlier. The same signaling pathway that inhibits late origin firing in response to replication inhibition or DNA damage may function in unperturbed cells to delay late origin firing while replication forks from early origins are active. As some active forks terminate, the inhibitory signal (possibly single-stranded DNA at an active replication fork) also diminishes, permitting the next staging of origins to commence at a recently recycled replication factory. Moreover, the replication process itself, or perhaps histone acetylation, may relocalize unfired origins, enabling their activation. Thus, chromatin structure may establish a spatial hierarchy of origins relative to replication sites within the nucleus, determining their relative firing order, and the checkpoint pathway modulates the execution of this sequence during S phase.
The idea that the number of simultaneously active replication forks is limited may explain why deletion of RPD3 increases the replication rate of clb5Δ cells but not of wild-type cells. In wild-type cells, efficient activation of early origins would rapidly meet the replication fork limit and, thus, the earlier firing of a late origin in rpd3Δ cells would serve only to replace the activation of another potential origin. However, in clb5Δ cells where fewer total origins are activated because of diminished Clb activity, earlier firing of late origins by RPD3 deletion would enable more total origin activation (until the fork limit is reached) and faster genome duplication.
Relationship between Rpd3 and checkpoint regulation of late origins.
The finding that late origins escaped checkpoint inhibition by HU in rpd3Δ cells was consistent with a mechanistic dependence of checkpoint inhibition on RPD3 function. However, we have shown that the intra-S checkpoint pathway is intact in the absence of RPD3, because Rad53 activation by HU occurs normally and late origins are effectively blocked when MMS is used to elicit the checkpoint. Therefore, we conclude that the loss of late origin inhibition in HU-treated rpd3Δ cells occurs because late origins initiate before the HU-generated inhibitory signal can act. However, the late origins did not initiate before inhibition by MMS took place. The susceptibility of late origins to inhibition in MMS-treated rpd3Δ cells indicates that these origins are still somewhat delayed relative to the earliest origins and that histone deacetylation by Rpd3 is not inherently required for checkpoint inhibition.
The more effective inhibition by MMS was unexpected and implies that MMS generates a more urgent inhibitory signal. This is plausible, as some degree of replication is likely required before the effect of HU (nucleotide depletion) is elicited, whereas replication forks should immediately encounter the alkylated DNA template generated by MMS treatment. In addition, different factors mediate Rad53 activation in response to MMS and HU; Mrc1 primarily mediates Rad53 activation in HU, whereas Rad9 primarily mediates Rad53 activation in MMS (reviewed in reference 29). Once activated by either pathway, Rad53 is thought to block late origin firing by inhibiting Cdc7-Dbf4, which is required for origin activation. We directly tested the kinetics of Rad53 activation by monitoring its phosphorylation and found that Rad53 activation occurred about 5 min earlier in response to MMS treatment than with HU treatment in wild-type and rpd3Δ cells (Fig. 4D). Although onset of Rad53 activation in rpd3Δ cells treated with HU was slightly delayed relative to that in wild-type cells, the similar kinetics of response suggested there is no defect in this signaling pathway that would explain the lack of inhibition in response to HU. Because the timing of Rad53 activation in MMS precedes late origin firing, whereas Rad53 activation in HU is approximately concurrent with late origin firing in rpd3Δ cells, the differential effects of MMS and HU likely derive from these differences in Rad53 activation kinetics.
What is the significance of temporal control of initiation by Rpd3?
Ultimately, by temporally distributing initiation events throughout S phase, Rpd3 may serve to ensure consistent and efficient activation of selected origins by delaying the activation of potential competing origins. Thus, the normal temporal pattern of replication timing may help to maintain a highly accurate and reproducible spatial pattern of genome and chromatin replication, which may be crucial to proper programming and inheritance of gene expression patterns. Delayed activation of some origins also enhances the cell's ability to regulate origin usage under conditions of replicative stress. Histone deacetylase inhibitors alter replication timing in mammalian cells, suggesting that origin regulation by histone deacetylation is conserved among eukaryotes (6). In further defining the role of Rpd3, it will be important to determine its cell cycle execution point, its critical biochemical target(s) in chromatin, how deacetylation alters initiation kinetics, and whether it functions similarly in higher eukaryotes.
Acknowledgments
We thank S. Haase and R. Lipford for critical reading of the manuscript and J. Diffley, M. Foiani, F. Hu, C. Newlon, R. Rothstein, E. Schwob, and K. Struhl for strains, plasmids, and protocols.
This work was supported by a Burroughs-Wellcome Career Award (992834) and National Institutes of Health grant (1RO1GM-CA65494-01A1) to O.M.A.
REFERENCES
- 1.Aparicio, O. M. 1999. Characterization of proteins bound to chromatin by immunoprecipitation from whole-cell extracts, p. 21.3.1-21.3.12. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 4. John Wiley and Sons, Inc., New York, N.Y.
- 2.Aparicio, O. M., A. M. Stout, and S. P. Bell. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. USA 96:9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bickmore, W. A., and A. D. Carothers. 1995. Factors affecting the timing and imprinting of replication on a mammalian chromosome. J. Cell Sci. 108:2801-2809. [DOI] [PubMed] [Google Scholar]
- 7.Burke, T. W., J. G. Cook, M. Asano, and J. R. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 8.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove, A. J., C. A. Nieduszynski, and A. D. Donaldson. 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davie, J. K., R. J. Trumbly, and S. Y. Dent. 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diffley, J. F., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., and D. M. Gilbert. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4:983-993. [DOI] [PubMed] [Google Scholar]
- 13.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 14.Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. G. Zhu, J. R. Broach, C. S. Newlon, and J. A. Huberman. 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 11:5346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrenhofer-Murray, A. E., D. H. Rivier, and J. Rine. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695-1706. [DOI] [PubMed] [Google Scholar]
- 17.Fangman, W. L., and B. J. Brewer. 1992. A question of time: replication origins of eukaryotic chromosomes. Cell 71:363-366. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, K. L., J. D. Diller, B. M. Ferguson, S. V. Nyland, B. J. Brewer, and W. L. Fangman. 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10:1595-1607. [DOI] [PubMed] [Google Scholar]
- 19.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert, D. M. 2002. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14:377-383. [DOI] [PubMed] [Google Scholar]
- 22.Heun, P., T. Laroche, M. K. Raghuraman, and S. M. Gasser. 2001. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 152:385-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iizuka, M., and B. Stillman. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274:23027-23034. [DOI] [PubMed] [Google Scholar]
- 24.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 25.Kuhne, C., and P. Linder. 1993. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 12:3437-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 27.Mallory, M. J., and R. Strich. 2003. Ume1p represses meiotic gene transcription in Saccharomyces cerevisiae through interaction with the histone deacetylase Rpd3p. J. Biol. Chem. 278:44727-44734. [DOI] [PubMed] [Google Scholar]
- 28.McCarroll, R. M., and W. L. Fangman. 1988. Time of replication of yeast centromeres and telomeres. Cell 54:505-513. [DOI] [PubMed] [Google Scholar]
- 29.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 30.Newlon, C. S., I. Collins, A. Dershowitz, A. M. S. Deshpande, S. A. Greenfeder, L. Y. Ong, and J. F. Theis. 1993. Analysis of replication origin function in chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 58:415-423. [DOI] [PubMed] [Google Scholar]
- 31.Pasero, P., A. Bensimon, and E. Schwob. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 33.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pemberton, L. F., and G. Blobel. 1997. Characterization of the Wtm proteins, a novel family of Saccharomyces cerevisiae transcriptional modulators with roles in meiotic regulation and silencing. Mol. Cell. Biol. 17:4830-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raghuraman, M. K., B. J. Brewer, and W. L. Fangman. 1997. Cell cycle-dependent establishment of a late replication program. Science 276:806-809. [DOI] [PubMed] [Google Scholar]
- 36.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 37.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 38.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 39.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 41.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 42.Schwob, E., and K. Nasmyth. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7:1160-1175. [DOI] [PubMed] [Google Scholar]
- 43.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, J. B., and D. E. Gottschling. 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 13:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 46.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 47.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 48.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 49.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 50.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 51.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 52.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis, C. Sarrauste de Menthiere, R. W. Davis, and R. E. Esposito. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99:13431-13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high- resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 55.Zappulla, D. C., R. Sternglanz, and J. Leatherwood. 2002. Control of replication timing by a transcriptional silencer. Curr. Biol. 12:869-875. [DOI] [PubMed] [Google Scholar]