Abstract
The CCAAT-displacement protein (CDP) has been implicated in developmental and cell-type-specific regulation of many cellular and viral genes. We previously have shown that CDP represses mouse mammary tumor virus (MMTV) transcription in tissue culture cells. Since CDP-binding activity for the MMTV long terminal repeat declines during mammary development, we tested whether binding mutations could alter viral expression. Infection of mice with MMTV proviruses containing CDP binding site mutations elevated viral RNA levels in virgin mammary glands and shortened mammary tumor latency. To determine if CDP has direct effects on MMTV transcription rather than viral spread, virgin mammary glands of homozygous CDP-mutant mice lacking one of three Cut repeat DNA-binding domains (ΔCR1) were examined by reverse transcription-PCR. RNA levels of endogenous MMTV as well as α-lactalbumin and whey acidic protein (WAP) were elevated. Heterozygous mice with a different CDP mutation that eliminated the entire C terminus and the homeodomain (ΔC mice) showed increased levels of MMTV, β-casein, WAP, and α-lactalbumin RNA in virgin mammary glands compared to those from wild-type animals. No differences in amounts of WDNM1, ɛ-casein, or glyceraldehyde-3-phosphate dehydrogenase RNA were observed between the undifferentiated mammary tissues from wild-type and mutant mice, indicating the specificity of this effect. These data show independent contributions of different CDP domains to negative regulation of differentiation-specific genes in the mammary gland.
The CCAAT displacement protein (CDP) family constitutes a highly conserved group of homeoproteins that are involved in cell growth, differentiation, and development. CDP (also known as Cut in Drosophila melanogaster, Clox in dogs, and Cux in mice) is a transcriptional repressor that contains four DNA-binding domains, three Cut repeats (CR1, -2, and -3), and a homeodomain (32). CDP is involved in suppressing transcription from many cellular genes, including c-myc, transforming growth factor β type II receptor, phox, CD8, immunoglobulin heavy chain, and T-cell receptor β-chain (3, 7, 12, 19, 22, 46), as well as viral genes, including those from mouse mammary tumor virus (MMTV) and human papillomaviruses (HPVs) (1, 55). In addition, CDP has been implicated as a tumor suppressor gene in human breast carcinomas and uterine leiomyomas (28, 47, 53).
Many genes regulated by CDP are expressed in highly differentiated cells. CDP DNA-binding activity is downregulated during B-cell and myeloid cell development (22, 46), and CDP expression in the kidney is inversely related to the degree of cellular differentiation (44). We have shown that CDP is a repressor of MMTV long terminal repeat (LTR) reporter gene expression in cultured cell lines and that CDP-binding activity to the LTR declines during mammary gland development (55). These results suggest that CDP functions as a transcriptional repressor that inhibits expression of differentiation-specific genes in several tissues.
The retrovirus MMTV is a paradigm for hormone-regulated gene expression in the mammary gland. Virus is transmitted exogenously from mothers to offspring through virally infected milk (11). Gut-associated B and T lymphocytes are infected by MMTV and amplified by virally encoded superantigen (Sag) prior to transmission of virus to mammary cells during puberty. Hormonal stimulation during pregnancy and lactation increases MMTV transcription and replication, allowing mutagenic integration events and the development of mammary tumors (11). MMTV also is transmitted through the germ line of most inbred mouse strains as endogenous proviruses (8). Because most viruses specify very few genes, they must rely on their host cells for replication. In particular, many retroviruses show cell-type-specific replication and tumorigenesis, and studies of such viruses have elucidated many aspects of diverse cellular processes.
Current evidence suggests that tissue-specific MMTV replication is controlled primarily at the transcriptional level using host factors (24). Lymphomagenic MMTV strains invariably have a deletion of 350 to 500 bp of negative regulatory elements (NREs) within the LTR U3 region (18). These deletions remove the binding sites for two homeodomain-containing transcriptional repressors, special AT-rich binding protein 1 (SATB1) and CDP (23, 24). Studies in transgenic mice showed that mutation of a SATB1-binding site within the MMTV LTR elevates viral transcription in lymphoid tissues (24). Two CDP binding sites have been mapped to the promoter-proximal portion of the MMTV NRE, and mutation of either of these binding sites elevated reporter gene expression from the MMTV LTR (55). Other functional CDP binding sites have been mapped to the promoter-distal NRE (54). These data indicate that negative regulation of transcription is a primary method for regulating tissue-specific MMTV expression.
Since CDP binding to MMTV NREs is high in virgin mammary glands, we predicted that loss of CDP binding would increase MMTV transcription in early stages of mammary development, thereby altering the kinetics of virally induced tumors. To test whether CDP has a direct role in MMTV-specific transcription and tumorigenesis, we constructed infectious proviruses with different CDP binding site mutations. Mice infected with these mutants showed increased MMTV expression in the virgin mammary gland and accelerated appearance of breast tumors. To determine specific effects by CDP on transcription in the mammary gland, we also analyzed gene expression in two different strains of CDP-mutant mice, lacking either CR1 or the entire C terminus including the homeodomain (42, 43). RNA levels of endogenous MMTV, whey acidic protein (WAP), and α-lactalbumin, but not glyceraldehyde-3-phosphate dehydrogenase (gapdh) or WDNM1, were elevated in virgin mammary glands of both mutant strains compared to wild-type controls. However, only animals with one-half the wild-type CDP levels had increased β-casein expression. These results indicate that different CDP domains contribute to negative regulation of differentiation-specific genes in the mammary gland.
MATERIALS AND METHODS
Plasmid constructions.
CDP binding site mutations were transferred into an infectious, MMTV hybrid provirus, HYB-MTV (41), that had been engineered to contain a hygromycin resistance cassette (30). To create HP691, HP692, HP837, and HP838, p691-LUC, p692-LUC, p837-LUC, and p838-LUC were digested with AseI, and the fragments containing the mutations were gel purified using spin columns (QIAGEN). Subsequently, these fragments were substituted into the AseI site of the HYB-MTV construct. The correct orientation was determined by KpnI restriction enzyme digestion.
Cell culture and transfections.
Conditions for growth of XC rat fibroblasts have been described previously (54). Cells (2 × 105) were plated in each well of six-well plates and incubated overnight until the cells were ca. 50% confluent. The test plasmid containing the hygromycin resistance gene in 0.5 ml of Dulbecco's modified Eagle's medium (DMEM) without serum was added to 12 μl of DMRIE-C reagent (Invitrogen) in an equal amount of medium, mixed, and incubated at room temperature for 45 min. The cells were washed with DMEM without serum, the DNA solution was incubated with the cells for 8 h at 37°C, and then 1 ml of complete DMEM with 20% fetal bovine serum was added. After further incubation for 48 h, cells were selected in 0.5 mg of hygromycin (Invitrogen)/ml for 3 weeks. Three wells of the six-well plates containing ca. 150 colonies were pooled and assayed. Expression levels were normalized for DNA uptake in each pool using a quantitative PCR assay with primers specific for the C3H MMTV LTR (155+, 5′ GGC ATA GCT CTG CTT TGC 3′; 548-, 5′ TAC TTC TAG GCC TGT GGT CA 3′) (49). DNAs from transfected cells were tested by PCR for transfer of C3H LTR sequences from the 3′ LTR to the 5′ LTR. PCR was performed using primers specific for Mtv1 (plus strand starting at +883; 5′ ACA GTA GAG AGG AGG CCA AAA G 3′; minus strand, Gag620-, starting at +2254 of the proviral DNA, 5′ CCT CCA AAT CAT CCC AAT CCT C 3′). PCR also was performed using primers specific for C3H MMTV (plus strand starting at +910, 5′ GGC TGG ACT AAT AGA ACA TTA TTC 3′; minus strand, Gag620-).
RNase protection assays.
RNase protection assays were performed as described by Wrona et al. (48). To prepare a C3H LTR-specific riboprobe, a DNA fragment containing C3H LTR sequence from +843 to +1099 was cloned into a pGEMT-Easy vector (Promega). The plasmid was linearized with NdeI prior to in vitro transcription. Riboprobe templates for gapdh were purchased from Ambion. Riboprobes were prepared using Riboprobe System-T7 (Fisher Scientific).
Animals and virus infection experiments.
BALB/cJ mice (from Jackson Laboratories) were bred and maintained in the Animal Resources Center at the University of Texas at Austin. Sentinel mice were tested for pathogens at intervals to eliminate spurious effects on T-cell deletion during MMTV infection. XC cells expressing MMTV proviral constructs were treated with dexamethasone (DEX; 10−6 M) for 48 h, harvested in phosphate-buffered saline (PBS), and washed twice with PBS. For each injection, 2 × 107 XC cells were resuspended into 0.5 ml of PBS and injected into mice intraperitoneally at 5 to 6 weeks of age. Mice were observed weekly for the appearance of mammary tumors. The ΔCR1 and ΔC mice were kindly provided by Richard Scheuermann and Ellis Neufeld on an outbred background (43); these animals were backcrossed to BALB/c mice for at least nine generations before mammary tissues were obtained. RNA was extracted as described previously (50). Tumor DNAs were tested for the presence of the virus used for inoculation by PCR with primers specific for C3H MMTV (155+ and 548-). Primers specific for the 838 mutation also were used (155+ and minus strand starting at +838 in the LTR, 5′ TAA CCC ACC TAT CCC AGT TC 3′). Primers specific for the 692 mutation were as follows (155+ and minus strand starting at + 693, 5′TTC CTG TTC CTA GAT AGA TGT AG 3′).
Antibody staining and FACS analysis.
Lymphocytes were isolated and analyzed as described previously (31). Antibodies used for staining were obtained from Pharmingen. Samples were analyzed using a FACSCalibur flow cytometer and CELLQuest software (Becton Dickinson). Instrument settings were determined using fluorescence-activated cell sorter (FACS) Brite calibration beads (Becton Dickinson). Lymphocyte populations were selected for analysis based on their characteristic forward and side scatter, and at least 20,000 lymphocytes were analyzed. To determine the percentage of T cells positive for a particular Vβ chain, the number of CD4-positive, T-cell receptor Vβ-positive events was divided by the sum of the number of single-positive events for CD4 and the number of double-positive events.
Immunoprecipitations and Western blotting.
Tissue culture cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (15). Particulate material was removed by centrifugation at 16,000 × g in a microcentrifuge for 10 min at 4°C. The lysates (100 μg each) were precleared by addition of 150 μl of a 50% (vol/vol) suspension of formalin-fixed Staphylococcus aureus (Sigma) and 10 μl of preimmune serum and then incubated at 4°C for a minimum of 1 h on a roller wheel. After brief centrifugation, supernatants were incubated with 1 μl of immune or preimmune serum and 40 μl of protein A-agarose beads (50% in RIPA buffer; Sigma) at 4°C for 12 h on a roller wheel. Immune complexes were washed three times with RIPA buffer, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, and boiled for 5 min. After centrifugation, supernatants were immediately analyzed by Western blotting using antibodies specific for MMTV CA (1:500 dilution; National Cancer Institute Biological Carcinogenesis Branch Repository, National Institutes of Health) or actin (1:150 dilution; Sigma) as previously described (30). Proteins were visualized using the ECL Western blotting detection system (Amersham Pharmacia Biotech). Western blot assays also were performed with nuclear extracts from tissues of ΔCR1 and ΔC mice using CDP-specific antibodies as previously described (55).
Semiquantitative and real-time RT-PCR.
Total RNA (10 μg) was treated with DNase I and then incubated with SuperScript II reverse transcriptase (Invitrogen) for 1 h at 42°C, followed by heat inactivation for 15 min at 70°C. The cDNA was used to measure the expression of different mammary gland differentiation-specific genes by reverse transcription-PCR (RT-PCR) using Jumpstart REDAccuTaq DNA polymerase (Sigma) and 25 μM concentrations of primers specific for β-casein (+, 5′ CCT TGC CAG TCT TGC TAA TC 3′; −, 5′ GAA TGT GGA GTG GCA G 3′); WAP (+, 5′ TCA GTT CAG TCC ATG TTC CA 3′; −, 5′ GGA GCA TTC TAT CTT CAT TGG 3′); ɛ-casein (+, 5′ AAA TGG AAT CTG TTG AAG CTC 3′; −, 5′ CTG GTA TTG ATG GAG AAG C 3′); WDNM1 (+, 5′ ATG AAC AAT GCA CAG GAG AT 3′; −, 5′ GTC TAA GGA GGA GCC AAG CA 3′); α-lactalbumin (+, 5′ GCA GCA CAG AGT ACG GAC 3′; −, 5′ CTC AGG GCT TCT CAC AAC G 3′); and MMTV Env (+), 5′ CCT TGC GAA GAG CCT TGA C 3′; LTR(−), 5′ GAG TTC AAC CAT TTC TGC TG 3′). The gapdh primers were gapdh(+) (CAT GTT TGT GAT GGG TGT GAA CCA) and gapdh(−) (GTT GCT GTA GCC GTA TTC ATT GTC).
The resulting reaction mixtures were analyzed on 1% agarose gels and stained with ethidium bromide, and the band intensities were measured semiquantitatively using gel documentation Quantity One software version 4.4.0 (Bio-Rad). A series of cDNA dilutions were used to ensure that the amplification of the PCR product was in the linear range. The identity of bands for different PCR products was confirmed by sequencing. As an internal control, the RNA levels of different genes were normalized to those of gapdh and plotted relative to the levels in wild-type mice (assigned a value of 1). Some cDNAs (1 μl) also were used in a real-time PCR containing 20 μl of SYBR Green PCR master mix (Applied Biosystems) and 10 μM concentrations of primers. The results were analyzed using ABI Prism 7700 sequence detection system software (Applied Biosystems). The standard curve method was used for quantitation. As an internal control, the levels were normalized to those of gapdh.
EMSAs.
Nuclear extracts from mammary glands were used in electrophoretic mobility shift assays (EMSAs) with an end-labeled probe (pNRE4) containing four copies of a 22-bp sequence spanning the imperfect inverted repeat in the promoter-proximal MMTV NRE as previously described (55). Specificities of shifted bands were determined by antibody inhibition experiments using 0.1 μl of anti-CDP or preimmune serum (24).
Statistical analysis.
Results were analyzed by using SPSS version 10 (SPSS, Inc.).
RESULTS
Infection of XC cells with CDP binding site mutants.
Previous results have indicated that the homeodomain-containing transcription factor CDP is a repressor of MMTV transcription in mammary cells (55). We also have demonstrated that CDP is regulated during development of the mammary gland, since full-length CDP levels are highest in the virgin mammary gland and lowest in lactating mammary gland (55), a tissue with the highest levels of MMTV expression (13). Such results suggested that mutations that interfere with CDP binding to the MMTV LTR also would increase viral expression early during mammary gland development. Therefore, we engineered mutations into two different CDP binding sites within the 3′ LTR of an infectious MMTV provirus, HYB-MTV (41) (Fig. 1A).
FIG. 1.
Experimental design for MMTV proviruses with CDP binding site mutations. (A) Scheme for analysis of CDP binding site mutants. The 5′ half of the hybrid infectious provirus is composed of the 5′ LTR and gag-pol genes from the endogenous Mtv-1 provirus, whereas the 3′ end is composed of the env and 3′ LTR from a C3H MMTV provirus. An inverted triangle represents the CDP binding site mutations within the U3 region of the LTR. Transfection of proviral DNA leads to integration and transcription from the 5′ LTR by RNA polymerase II followed by RNA packaging into virions. Subsequent infection will allow RT of viral RNA so that the 3′ LTR mutations are duplicated in the 5′ LTR of the provirus. BALB/c mice were injected with XC cells containing the stably transfected proviruses and analyzed for T-cell deletion and the appearance of tumors. (B) Positions of mutations introduced in the distal NRE (dNRE) and proximal NRE (pNRE) of MMTV proviruses.
Prior experiments established that mutations in the promoter-distal and promoter-proximal NREs that disrupted CDP binding elevated reporter gene expression (54). Two such CDP binding site mutations in the distal NRE (691 and 692) and two mutations in the proximal NRE (837 and 838 [previously named 838S4]) were substituted into the infectious MMTV provirus, HYB-MTV, as described in Materials and Methods (for sequence, see Fig. 1B). These recombinant hybrid proviruses (HP691, HP692, HP837, and HP838) were transfected into rat XC fibroblasts. After selection for antibiotic resistance, a large pool of colonies was tested for expression of the hybrid viruses. Because RT of the transfected proviruses should duplicate in the 5′ LTR any mutations introduced in the 3′ LTR, we performed PCR with DNA extracted from provirus-transfected or untransfected XC cells. These results confirmed that RT transferred the predicted wild-type or mutant C3H sequences to the 5′ LTR of introduced proviruses (data not shown).
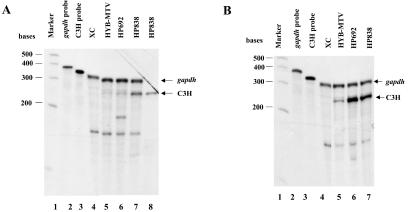
RNA was extracted from pooled colonies and subjected to RNase protection assays (RPAs) using a probe specific for the C3H MMTV LTR. The results showed that all mutants were expressed in XC cells at levels greater than the wild-type HYB-MTV (Fig. 2A and data not shown). Using phosphorimager analysis, mutant MMTVs showed two- to fivefold overexpression of RNA after normalization for the amount of gapdh RNA present in the same reaction.
FIG. 2.
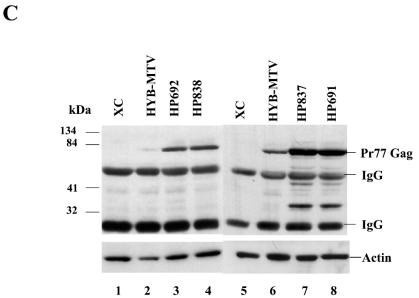
MMTV proviruses with CDP binding site mutations show elevated viral RNA and protein levels in transfected cells. (A and B) Total RNA was harvested from XC cells without (A) or with treatment of 10−6 M DEX for 24 h (B). RNA samples (50 μg in lanes 4 to 7) from normal XC cells (lane 4), XC cells stably transfected with HYB-MTV (lane 5), HP692 (lane 6), or HP838 (lane 7) were subjected to an RPA using riboprobe specific for the C3H LTR (5 × 105 cpm) and a gapdh riboprobe (5 × 103 cpm) as an RNA loading control. Lane 1 shows RNA molecular weight standards; lanes 2 and 3 show the position of the gapdh and LTR probes, respectively. Lane 8 (panel A) shows RNA from HP838-transfected XC cells hybridized to C3H LTR riboprobe in the absence of the gapdh riboprobe. (C) CDP-binding mutants show elevated viral protein levels. After treatment with 10−6 M DEX for 24 h, XC cell lysates containing wild-type virus (HYB-MTV) (lanes 2 and 6) or mutant provirus with a CDP binding site mutation in the proximal NRE (HP838 and HP837) (lanes 4 and 7) or in the distal NRE (HP692 and HP691) (lanes 3 and 8) were harvested and incubated with protein A. Precleared lysates (40 μg each) were analyzed by Western blotting using MMTV CA-specific antibody. XC rat cells (lanes 1 and 5) lack endogenous MMTVs.
Because the standard MMTV LTR promoter is upregulated in the presence of glucocorticoids, stable transfectants were treated with 10−6 M DEX for 48 h prior to extraction of RNA. RPAs then were repeated for all four mutants and quantitated by phosphorimaging (Fig. 2B and data not shown). All mutants showed two- to fivefold RNA overexpression relative to the wild-type virus transfectants. This level of overexpression is in good agreement with results obtained in stable transfections of reporter plasmids containing these mutations in the MMTV LTR promoter (54). Therefore, RPAs in the presence and absence of glucocorticoids indicated that four mutations in two different sites that disrupted CDP binding to the MMTV LTR resulted in an increase in RNA expression, presumably due to relief of CDP-mediated transcriptional repression.
To further characterize the effect of CDP binding site mutations on MMTV protein expression, transfected cells grown in the presence of DEX were lysed and immunoprecipitated with antibody specific for MMTV Gag proteins, and precipitates were analyzed by Western blotting using the same antisera (Fig. 2C). The mutant viruses showed three- to fivefold elevation of Gag protein expression in transfected cells after normalization for actin expression in the same lysate. Together with results from RPAs, these data indicated that mutations in at least two CDP binding sites in the MMTV LTR increased viral RNA and protein expression in the context of an infectious provirus.
In vivo infection with MMTVs containing CDP binding site mutations.
To determine if the CDP binding site mutations that upregulated virus expression in vitro would affect virus expression in vivo, we inoculated XC transfectants into susceptible BALB/c mice as initially described by Shackleford and Varmus (41) (Fig. 1). Only the HP692 and HP838 mutants were analyzed, since these mutations allowed preservation of Sag protein expression that is necessary to allow efficient milk-borne virus transmission from the gut to the mammary gland (16).
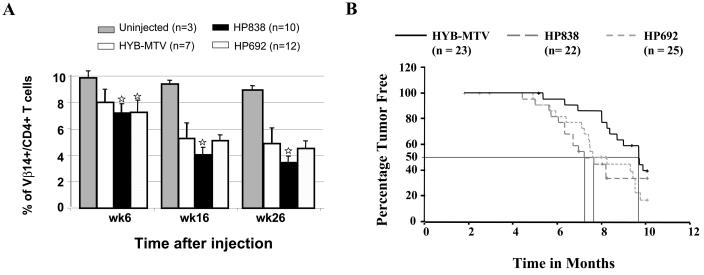
Expression of the MMTV-specific protein Sag leads to virus amplification in the lymphoid population and deletion of Sag-specific T cells (9). To follow the infection in wild-type and mutant-infected mice, we tested for Sag-specific deletion of Vβ14+CD4+ T cells at several different times after inoculation of infected cells. As expected, wild-type virus-infected animals showed specific deletion at 6 weeks postinoculation compared to uninfected BALB/c mice; a greater deletion was observed in both HP692 and HP838 mutant-infected mice (Fig. 3A). Deletion of T cells increased with time, but at 16 and 26 weeks postinfection HP838-infected mice showed significantly greater deletion (P < 0.05) relative to wild type-infected or HP692-infected mice. The Tukey honestly significant difference mean comparison post-hoc tests showed that the difference in the degree of deletion between the HP838-infected and HYB-MTV-infected mice was statistically significant (P < 0.05). These experiments confirmed that the CDP binding site mutants were infectious for BALB/c mice, that the introduced mutations did not affect the sag gene open reading frame, and that the CDP binding site mutation at +838 affected MMTV viral replication in lymphocytes.
FIG. 3.
Kinetics of infection and tumorigenesis by wild-type or CDP binding site mutants. (A) Deletion of C3H MMTV-specific Sag-reactive T cells in mice infected with wild-type or mutant viruses. BALB/c mice were injected intraperitoneally with XC cells that were stably transfected with wild-type MMTV proviruses (HYB-MTV) or CDP binding site mutants (HP692 and HP838), and lymphocytes were tested at 6, 16, and 26 weeks postinjection. Peripheral blood lymphocytes were stained with fluorescein isothiocyanate-conjugated antibodies to Vβ14 and phycoerythrin-conjugated antibodies to CD4. The lymphocyte population was determined by forward and side scatter using FACS analysis. Stars indicate values that were statistically different (P < 0.05) from those for wild-type virus-injected animals by Student's t tests and Tukey honestly significant difference mean comparison post-hoc tests at the same times after inoculation. (B) Mammary tumor development in mice infected by CDP binding site mutants. BALB/c weanling mice were injected intraperitoneally with XC cells that were stably transfected with wild-type hybrid MMTV proviruses (HYB-MTV) or CDP binding site mutants (HP692 and HP838) and monitored for tumor development. There was no detectable difference in the histology of mammary tumors induced by the wild-type virus compared to the mutants. Tumorigenesis results were analyzed by the Kaplan-Meier survival analysis technique using a log-rank test of significance. Symbols (+) represent animals that died for reasons other than mammary tumors and may represent more than one animal.
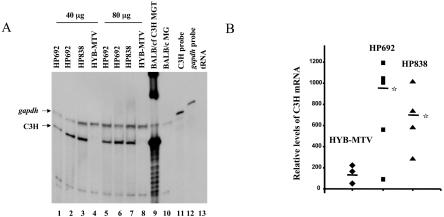
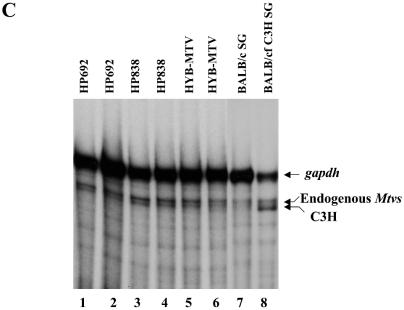
To determine if CDP-binding mutations in the LTR elevated MMTV expression, we quantified virus replication in vivo (Fig. 4). CDP effects on the LTR should be greatest in undifferentiated compared to differentiated mammary tissues because of high DNA-binding activities for the MMTV NREs and lack of hormonal stimulation (55). Therefore, total RNA from virgin mammary glands was extracted and used for RPAs 6 weeks after injection. A C3H-specific riboprobe protected a band of the correct size using RNA from an MMTV-infected mammary gland tumor (Fig. 4A, lane 9) but not when using RNA from an uninfected BALB/c mammary gland (Fig. 4A, lane 10). However, the intensity of the C3H-specific band from mice injected with HP692 (lanes 1, 2, 5, and 6) and HP838 (lanes 3 and 7) was 5.8- and 4.0-fold higher, respectively, than that obtained with the wild-type virus (lanes 4 and 8) after quantitation by phosphorimager and normalization to gapdh RNA (Fig. 4B). Both mutant viruses had statistically higher RNA levels than the wild-type virus in the virgin mammary glands, whereas RNA from salivary glands and hearts of the same mice showed no detectable differences in expression levels (Fig. 4C and data not shown). Therefore, CDP binding site mutations elevated MMTV RNA expression specifically in undifferentiated mammary glands.
FIG. 4.
Expression of wild-type and mutant MMTVs in virgin mammary glands of inoculated mice. (A) Total RNA was extracted from virgin mammary glands 6 weeks after injection with virus-producing XC cells. RNA samples were subjected to an RPA using riboprobes specific for the C3H MMTV LTR (5 × 105 cpm) and gapdh (1 × 104 cpm) as an RNA loading control. Lane 9 contains 20 μg of mammary gland tumor (MGT) RNA from a BALB/c mouse that was infected by foster nursing on a C3H mother (BALB/cfC3H). Lane 10 contains 40 μg of BALB/c mammary gland (MG) RNA. Lanes 11 and 12 show the position of the LTR and gapdh probes, respectively. Lane 13 contains 40 μg of yeast tRNA. (B) Quantitation of relative levels of C3H riboprobe-protected bands from RPAs. RNA samples from individual HYB-MTV- (n = 3), HP692- (n = 5), and HP838- (n = 4) injected mice were analyzed by RPAs, and the intensities of C3H and gapdh riboprobe protected bands were quantitated using a phosphorimager and normalized to gapdh levels. The average for each group is indicated by a horizontal bar. Stars indicate values that were statistically different (P < 0.05) from those of wild-type virus-injected animals by Student's t test. (C) Total RNA was extracted from salivary glands 6 weeks after injection with virus-producing XC cells. RNA samples (70 μg in lanes 1 to 6) from HP692 (lanes 1 and 2), HP838 (lanes 3 and 4), and HYB-MTV (lanes 5 and 6) virus-injected mice were subjected to an RPA as described for panel A. Lane 7 contains 70 μg of salivary gland (SG) RNA from a BALB/c mouse. Lane 8 contains 35 μg of salivary gland RNA from a BALB/c mouse infected by foster nursing on a C3H mother (BALB/cfC3H).
Infected mice also were monitored for the appearance of mammary tumors (Fig. 3B). With wild-type-infected animals, mammary tumors were first detected 5.5 months after the infection, and 50% of mice developed tumors by 9.6 months. However, in mutant-injected mice, breast tumors were first detected at 5 months after infection and 50% of HP692-injected animals developed tumors after 7.5 months. The HP838-injected mice first developed mammary tumors after 4.5 months, and 50% of the mice developed tumors after 7.2 months. Statistical tests (Kaplan-Meier survival analysis) were performed to analyze the difference in tumor-free time between each group of infected animals. Such analysis showed that HYB-MTV-infected mice had significantly higher tumor-free time (P < 0.05) than HP692-infected mice. The HP838-infected mice also showed a trend toward accelerated mammary tumorigenesis; however, this was not statistically significant, presumably due to the relatively high mortality that was unrelated to mammary tumors (e.g., birthing problems). These results suggested that mutations of CDP binding sites in the promoter-distal NRE accelerated MMTV-induced mammary cancers.
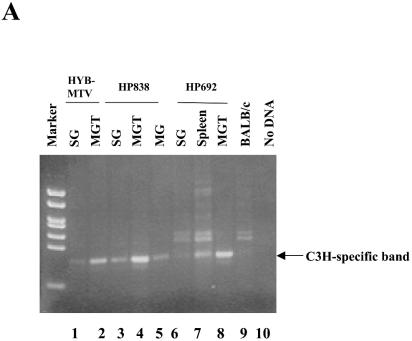
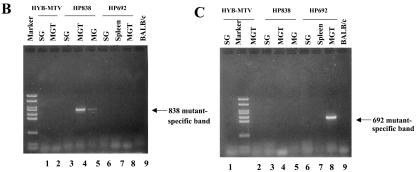
To determine viral levels in different tissues of wild-type- and mutant-infected mice, we performed PCR using DNA extracted from infected mice. Using primers that distinguish between the infecting C3H wild-type or mutant viruses and the endogenous MMTVs, a PCR product was detected both in salivary glands and a mammary tumor obtained from HYB-MTV-infected mice (Fig. 5A, lanes 1 and 2), but also the same tissues from HP838-infected (lanes 3, 4, and 5) and HP692-infected (lanes 6, 7, and 8) mice. As expected, DNA from uninfected BALB/c mice was negative for C3H MMTV-specific sequences in PCR assays (lane 9). To determine if the viruses in mammary tumors retained the original mutations, we also tested salivary gland and mammary tumor DNA by PCR with mutation-specific primers. Primers specific for the 838 mutation detected DNA in mammary gland and mammary tumors of HP838-infected mice (Fig. 5B, lanes 4 and 5), whereas no products were detectable in DNA extracted from tissues of mice infected with HYB-MTV (wild type) or HP692, or uninfected mice. Similarly, PCRs with primers specific for the 692 mutation only detected products using DNA from a mammary gland tumor from HP692-infected mice (Fig. 5C, lane 8), but not other tissues or tumors from mice infected with the wild-type virus or other mutants. Together with previous results, these data indicated that the mammary tumors induced by the HP692 and HP838 proviruses retained the original mutations.
FIG. 5.
In vivo infection by CDP binding site mutants detected using PCR. Genomic DNA was extracted from different tissues in mice that were injected with XC transfectants of wild-type (HYB-MTV; lanes 1 and 2) or CDP binding site mutant proviruses (HP692, lanes 6 to 8; HP838, lanes 3 to 5). (A) PCR products detected with primers specific for C3H MMTV. The C3H-specific band was 393 bp. (B) PCR products detected with primers specific for the HP838 mutation. The HP838-specific band was 703 bp. (C) PCR products detected with primers specific for the HP692 mutation. The HP692-specific band was 561 bp. Reaction mixtures contained 100 ng of DNA, except for those shown in panel A, lanes 1 and 6 (200 ng) and lane 7 (400 ng). DNA from an uninjected BALB/c mouse was used as a negative control (lane 9). MG, mammary gland; MGT, mammary gland tumor; SG, salivary gland.
Increased endogenous MMTV expression in CDP-mutant mice.
Our results with CDP-binding mutations in the MMTV LTR suggested that reduced CDP levels would elevate viral transcription in undifferentiated mammary glands, where CDP-binding activity is highest (55). Thus, we measured MMTV expression in CDP-mutant mice. Previous experiments have shown that mice harboring a knockout of the CR1 DNA-binding domain of the Cutl1 gene (ΔCR1) have a defect in milk composition and production of ɛ-casein (43), suggesting that CDP has a critical role in regulation of mammary gland differentiation-specific genes. Experiments using either splenic or thymic nuclear extracts also indicated that ΔCR1 homozygous and heterozygous mice produce a truncated protein by exon skipping that retains DNA-binding activity for the gp91-phox promoter (43). Since the ΔCR1 mutation originally was characterized on a mixed background, we bred the mutation onto the BALB/c background. Characterization of progeny from heterozygous parents revealed that only 4.6% of pups were homozygous for the mutation, compared to the expected frequency of 25% (Table 1). These results indicate that the ΔCR1 mutation was more lethal in the BALB/c background, since a normal segregation ratio was observed prior to inbreeding (43). Although equal numbers of female and male ΔCR1 homozygotes were observed, these animals often had reduced life span and fertility. Results from EMSAs revealed that the ΔCR1 protein binds to the promoter-proximal NRE of the MMTV LTR in the presence or absence of the wild-type protein when using nuclear extracts from virgin mammary glands (data not shown).
TABLE 1.
Genotyping of ΔCR1 weanling mice by PCRa
| Animals tested | No. of wild type/ no. tested (%) | No. of heterozygotes/no. tested (%) | No. of homozygotes/no. tested (%) |
|---|---|---|---|
| Females | 108/308 (35.1) | 184/308 (59.7) | 16/308 (5.2) |
| Males | 125/340 (36.8) | 201/340 (59.1) | 14/340 (4.1) |
| Total | 233/648 (36.0) | 385/648 (59.4) | 30/648 (4.6) |
Typing of progeny from crosses of heterozygous females and males inbred on the BALB/c background.
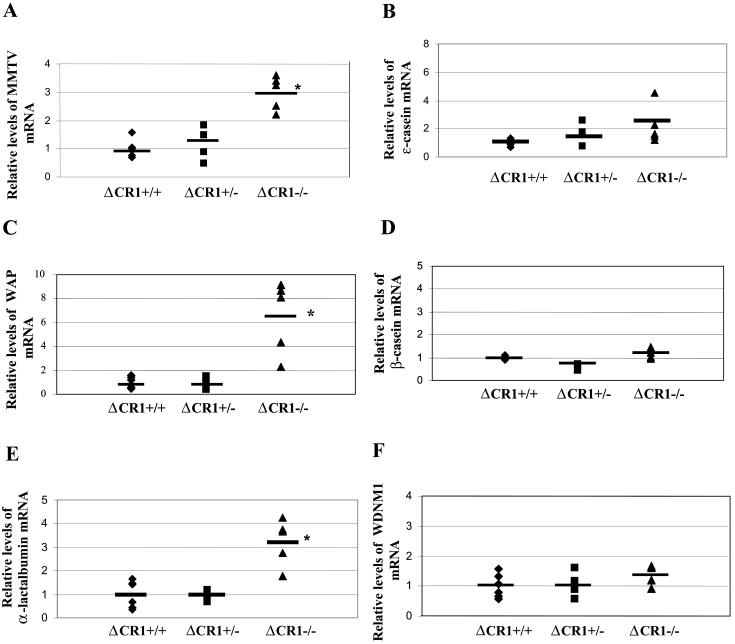
If CDP is a transcriptional repressor of genes known to be upregulated in differentiated mammary gland, e.g., MMTV genes, then expression of endogenous MMTVs should be elevated in the undifferentiated mammary tissue of CDP-mutant mice. Therefore, semiquantitative RT-PCR was used to assess expression of endogenous MMTV using RNA extracted from virgin mammary glands of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) ΔCR1 mice on the BALB/c background. Using tissue from wild-type animals to establish relative MMTV RNA levels of 1.0, virgin glands from heterozygous and homozygous mutant animals had relative viral RNA levels of 1.3 and 3.1, respectively, after normalization for levels of gapdh RNA in each sample (Fig. 6A). Similar results were obtained using the more sensitive real-time RT-PCR method, except that virgin mammary glands of heterozygous and homozygous mutant animals showed relative MMTV RNA levels of 1.4 and 5.5, respectively, compared to wild-type animals (relative value of 1) (data not shown).
FIG. 6.
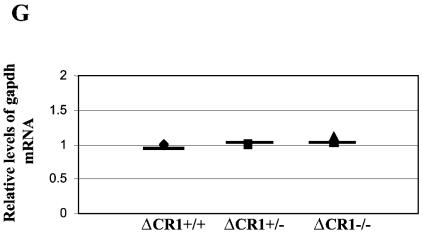
Increased RNA levels of endogenous MMTVs and mammary-specific genes in virgin mammary glands of ΔCR1 mice. RNA levels in virgin mammary glands of individual animals (each represented by a symbol) were detected by semiquantitative RT-PCR. Six wild-type (+/+), four ΔCR1 heterozygous (+/−), and five homozygous ΔCR1 (−/−) mice were tested. The average for each group of age-matched mice is indicated by a horizontal line. The average for the wild-type animals was assigned a value of 1, and the RNA levels of the heterozygous and homozygous ΔCR1 mice were assigned values relative to the wild-type levels after normalization for gapdh levels in individual samples. No significant differences in gapdh levels were observed between mammary glands from wild-type or mutant animals (G). RNA levels of the following genes are shown: endogenous MMTV (A), ɛ-casein (B), WAP (C), β-casein (D), α-lactalbumin (E), WDNM1 (F), and gapdh (G) genes. An asterisk indicates that the values for the wild-type and mutant mice were statistically different (P < 0.05) as determined by the two-tailed Student t test.
The ΔC mutation, which removes the entire CDP C terminus, including the homeodomain (ΔC), and gives high postnatal mortality in the homozygous state (42), also was bred onto the BALB/c background. By comparison to previous results where mating of heterozygous animals led to 5.4% homozygous ΔC progeny in a mixed background, we obtained 0.8% homozygous pups from the same cross (Table 2). Therefore, two different Cutl1 mutations appeared to give a more severe phenotype on the inbred BALB/c background.
TABLE 2.
Genotyping of ΔC weanling mice by PCRa
| Animals tested | No. of wild type/ tested (%) | No. of heterozygotes/no. tested (%) | No. of homozygotes/no. tested (%) |
|---|---|---|---|
| Females | 96/211 (45.5) | 114/211 (54.0) | 1/211 (0.5) |
| Males | 85/175 (48.6) | 88/175 (50.3) | 2/175 (1.1) |
| Total | 181/386 (46.9) | 202/386 (52.3) | 3/386 (0.8) |
Typing of progeny from crosses of heterozygous females and males inbred on the BALB/c background.
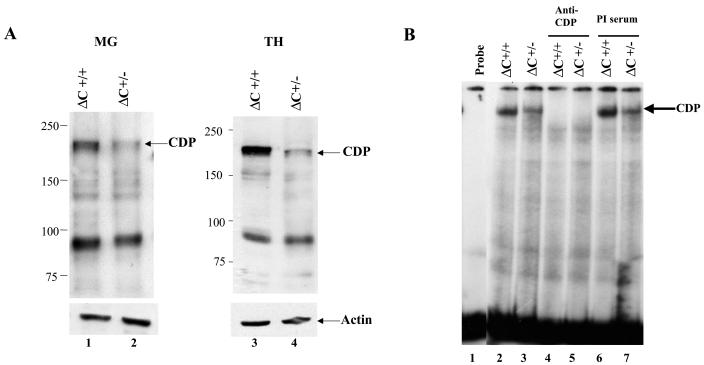
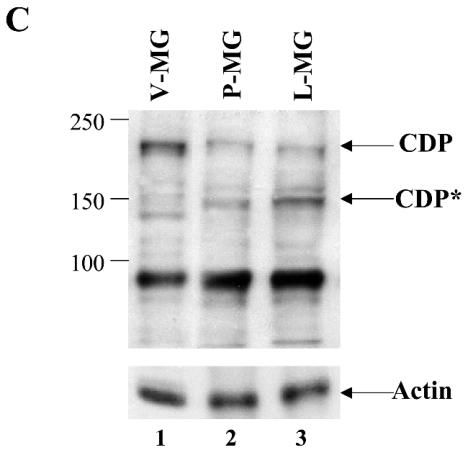
Previous results differ with respect to the localization of the ΔC protein (26, 42). To examine the location of the ΔC protein in tissues from animals on the inbred BALB/c background, nuclear extracts from thymi and virgin mammary glands of wild-type and ΔC heterozygous mice were analyzed by Western blotting (Fig. 7A). In agreement with previous results from lung extracts of mixed background mice (26), approximately one-half the level of wild-type protein was detected in both tissues tested. However, the mutant protein of ca. 123 kDa was not observed. Using the more sensitive EMSA, we also tested nuclear extracts from virgin mammary glands for the presence of binding activity to the promoter-proximal NRE in the MMTV LTR (Fig. 7B). These assays also revealed only full-length CDP in heterozygous extracts as determined by relative mobility and reactivity with anti-CDP, but not preimmune, sera. Furthermore, our previous results using BALB/c mice indicated that full-length CDP levels decline during mammary differentiation, followed by appearance of a 150-kDa truncated protein during pregnancy and lactation, as determined by Western blotting (55). Analysis of ΔC heterozygous females also revealed a decrease in full-length CDP protein in nuclear extracts prepared at different stages of mammary development (Fig. 7C). The ΔC protein was not detected and did not appear to affect the decline in wild-type CDP levels or the differentiation-specific appearance of the 150-kDa protein.
FIG. 7.
CDP protein levels and DNA-binding activity in ΔC mutant mice. (A) Western blotting of nuclear extracts from tissues of ΔC wild-type and heterozygous animals. Extracts from virgin mammary glands (MG; 50 μg; lanes 1 and 2) or thymi (TH; 25 μg; lanes 3 and 4) were analyzed on a 6% polyacrylamide gel containing sodium dodecyl sulfate followed by Western blotting and detection with CDP-specific antibody. The same blots were stripped and incubated with antiactin as a loading control (lower panel). (B) DNA-binding activity of nuclear extracts from virgin mammary glands of ΔC mice. Extracts (5 μg) from wild-type (lanes 2, 4, and 6) or heterozygous (lanes 3, 5, and 7) animals were incubated in the presence or absence of anti-CDP or preimmune (PI) serum as indicated prior to incubation with a labeled MMTV pNRE4 probe (ca. 3 fmol) (24). Complexes were separated on a 4% nondenaturing polyacrylamide gel prior to autoradiography. (C) CDP levels in ΔC-heterozygous animals during mammary gland development. Nuclear extracts (75 μg) of mammary glands from virgin (V-MG), pregnant (P-MG), or lactating (L-MG) mice were analyzed by Western blotting with CDP-specific antibody as described for panel A. The blot was stripped and incubated with actin-specific antibody to serve as a protein loading control (lower panel). The 150-kDa differentiation-specific form of CDP is indicated as CDP*.
To determine if deregulation of MMTV expression in the mammary gland was specific to the ΔCR1 mutation, we also tested endogenous MMTV RNA levels in the undifferentiated mammary tissue of ΔC mice. Because of the severity of the phenotype observed in ΔC homozygotes (Table 2), virgin mammary glands only were available from wild-type and ΔC heterozygous mice. Semiquantitative RT-PCR revealed that endogenous MMTV expression was increased 2.5-fold in ΔC +/− animals compared to wild-type mice (P < 0.05) (Fig. 8A). These experiments indicated that endogenous MMTV expression in the mammary gland, like that of milk-borne MMTV, was regulated by full-length CDP levels in two independent strains of Cutl1-mutant mice.
FIG. 8.
Increased RNA levels of endogenous MMTVs and mammary-specific genes in virgin mammary glands of ΔC mice. RNA levels in virgin mammary glands of individual animals (each represented by a symbol) were detected by semiquantitative RT-PCR. Four wild-type and six heterozygous ΔC mice were tested. The average for each group of age-matched mice is indicated by a horizontal line. The average for the wild-type animals was assigned a value of 1, and the RNA levels of the heterozygous ΔC mice were assigned values relative to the wild-type levels after normalization for gapdh levels in individual samples. No significant differences in gapdh levels were observed between mammary glands from wild-type or mutant animals (G). RNA levels of the following genes are shown: endogenous MMTV (A), ɛ-casein (B), WAP (C), β-casein (D), α-lactalbumin (E), WDNM1 (F), and gapdh (G) genes. An asterisk indicates that the values for the wild-type and mutant mice were statistically different (P < 0.05) as determined by the two-tailed Student t test.
Increased expression of specific cellular genes in the mammary gland of CDP-mutant mice.
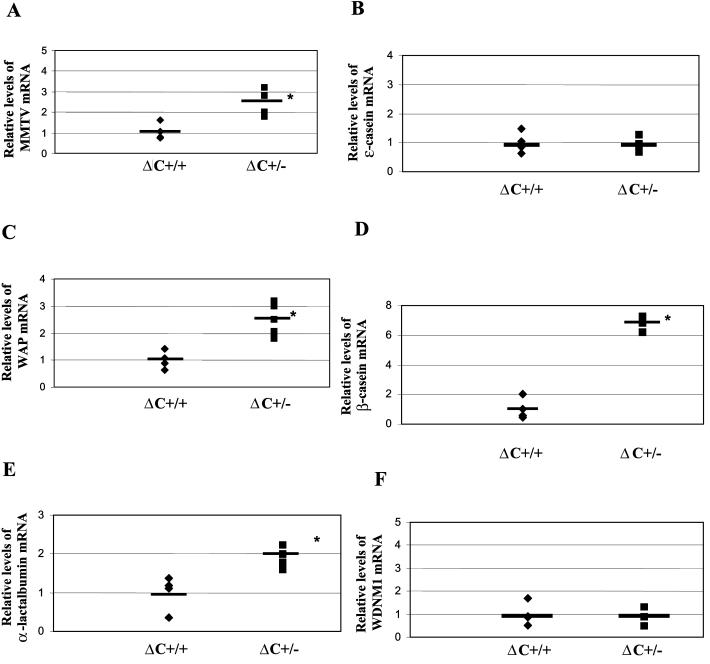
Retroviruses rely on cellular RNA polymerase II for transcription of integrated proviral DNA (35). These viruses encode relatively few genes and, therefore, transcriptional regulation of retroviral expression depends on factors encoded by the host genome. Since transcription of endogenous and milk-borne exogenous MMTVs appears to be developmentally controlled by CDP during mammary gland differentiation, it is likely that CDP also controls the expression of mammary-specific cellular genes. Thus, we determined the RNA levels of five such genes, the ɛ-casein, WAP, β-casein, α-lactalbumin, and WDNM1 genes, in the virgin mammary glands of ΔCR1 mice (Fig. 6B to F) (6, 17, 29, 45, 51).
RNA derived from individual wild-type, ΔCR1 heterozygous, and homozygous mice was subjected to semiquantitative RT-PCR and normalized for the level of gapdh RNA. Levels of α-lactalbumin and WAP were both increased three- to sixfold in virgin mammary glands of homozygous mutant mice relative to those from wild-type mice (P < 0.05) (Fig. 6C and E). This effect was tissue specific, since we did not observe elevation of these genes in the spleens or thymi of the same animals (data not shown). In contrast, levels of ɛ-casein, β-casein, and WDNM1 RNA were not significantly different in the virgin mammary glands of wild-type and ΔCR1 (−/−) animals (P > 0.05). The steady-state RNA levels of all genes tested in undifferentiated mammary tissues of ΔCR1 heterozygous mice was not detectably different from that of wild-type mice (Fig. 6).
The levels of these same genes also were tested in ΔC heterozygous mice (Fig. 8). Levels of β-casein RNA were most profoundly affected in virgin mammary glands (ca. sevenfold increases in ΔC heterozygotes compared to wild-type animals) (P < 0.05) (Fig. 8D). Western blotting confirmed that β-casein protein was elevated in lactating mammary glands of ΔC heterozygotes compared to that in wild-type mice (P < 0.05) (data not shown). Levels of WAP and α-lactalbumin RNA also were significantly higher in ΔC (+/−) undeveloped mammary tissues relative to those in wild-type mice (Fig. 8C and E, respectively). This effect was tissue specific, since upregulation of the same genes was not detected in spleens and thymi (data not shown). Like ΔCR1 virgin glands, the levels of ɛ-casein and WDNM1 RNA were unaffected compared to wild-type ani-mals. The quantity of gapdh RNA did not vary significantly in undeveloped mammary glands of wild-type and CDP-mutant mice (Fig. 6G and 8G). These results indicate that CDP is a negative regulator of specific cellular and viral genes that are developmentally expressed during mammary gland differentiation.
DISCUSSION
CDP binding site mutations enhance MMTV expression and tumorigenesis.
Our previous results indicated that CDP is a transcriptional repressor of MMTV expression in mouse mammary cells (55) and rat fibroblasts (54). CDP overexpression in these cells suppressed MMTV LTR-reporter activity, and mutations that inhibited CDP binding to the MMTV LTR increased viral transcription (54, 55). We also have shown that CDP is developmentally regulated in the mammary gland and that CDP-binding activity for the MMTV LTR is high in virgin mammary gland, where MMTV expression normally is repressed (55). Conversely, CDP-binding activity for the LTR is undetectable in lactating mammary gland, a tissue that shows the highest levels of MMTV transcription (24, 55). These results suggested that introduction of CDP binding site mutations into the LTR of an infectious MMTV clone would accelerate the development of MMTV-induced mammary cancers by increasing viral transcription and insertional mutagenesis at early stages of mammary gland development.
To test this hypothesis, we selected several CDP binding site mutations in the MMTV U3 region that had been shown to increase reporter gene expression from the MMTV LTR (54). We confirmed that these mutations greatly reduced CDP binding to the MMTV LTR prior to introduction into the 3′ LTR of HYB-MTV, an infectious MMTV proviral clone (40). Subsequently, the mutant proviruses, HP692 and HP838, were stably transfected into rat fibroblast cells and shown to overexpress both viral RNA and proteins (Fig. 2). Inoculation of these transfectants into susceptible BALB/c mice revealed that all inoculated viruses expressed the MMTV-encoded protein Sag, as demonstrated by the deletion of a specific T-cell subset (Fig. 3A), a clear indicator of MMTV infection. One mutant (HP838) accelerated MMTV-induced T-cell deletion (Fig. 3A), and the other mutant (HP692) significantly decreased the mammary tumor latency of the infected mice compared to those inoculated with the wild-type virus (Fig. 3B). As anticipated, both mutants also specifically elevated MMTV expression in virgin mammary glands (Fig. 4). Therefore, the mutation in the distal NRE at +692 may function to promote viral replication in the mammary gland itself, whereas the +838 mutation may increase viral expression in B cells, a cell type facilitating MMTV transmission to breast tissue (11). These results suggest that the functions of CDP binding in the proximal and distal NREs are not equivalent, although mutations in these binding sites showed similar effects on MMTV expression in transfected fibroblast cells. Because there appear to be at least eight CDP binding sites in the MMTV LTR (54), it is possible that CDP prevents the binding of different enhancer factors that are responsible for regulation of virus expression in various cell types.
What is the role of CDP in MMTV replication? We suggest that CDP restricts MMTV replication during and between pregnancies so that MMTV production peaks only during lactation, a stage critical to viral transmission in the milk. CDP also antagonizes the normally positive effects of glucocorticoid receptor binding to the MMTV LTR during pregnancy (54), but this suppression is alleviated when CDP binding to the LTR disappears at lactation. Restriction of MMTV production at specific stages of mammary differentiation prevents excess integration events that result in mammary tumors, leading to increases in the life span of the mother and the number of offspring to which virus will be transmitted.
A similar type of developmental restriction of virus production by CDP has been observed with HPV (33, 34). HPV is not produced by undifferentiated skin cells that are located well below the skin surface, and this is believed to be due, in part, to CDP-mediated repression of viral transcription in undifferentiated keratinocytes (1). However, during differentiation of skin cells, CDP binding to HPV promoters declines, allowing higher levels of virus production (34). Since differentiated cells also are found at the surface of the skin, production of virions is maximal where virus infection can be spread by contact with other organisms. Thus, developmental restriction by CDP may be a common mechanism used by viruses whose expression is limited in undifferentiated cell types.
CDP regulates transcription of mammary-specific genes.
Viruses depend on cellular factors and processes to achieve replication. Studies of viruses have repeatedly revealed new aspects of these cellular processes. Regulation of MMTV by CDP in mammary tissue is not a process specified by the virus to control its replication. Rather, MMTV has adopted a regulatory process that evolved to regulate cellular differentiation events in the mammary gland.
Homeodomain proteins, including CDP, are known to control cell type specification in a number of tissues (14, 32, 43). In studies of the Drosophila CDP homologue, embryonic lethal mutations of the cut gene transform external sensory organs into internal sensory (chordotonal) organs (5), and similar mutations prevent development of other organs, e.g., Malpighian tubules (25). In mice, loss of Cutl1 results in lung and hair follicle developmental defects, myeloid hyperplasia, and reduced male fertility, and CDP overexpression causes multiorgan hyperplasia (14, 26, 42). Moreover, CDP has been reported to be a potential tumor suppressor gene in breast carcinomas and uterine leiomyomas (52, 53).
Several mechanistic insights into CDP function may be derived from our studies. First, our experiments show that the effects of CDP, a ubiquitously expressed transcription factor, are tissue specific. WAP and α-lactalbumin RNA levels were elevated in virgin mammary glands, but not spleens or thymi, of two different CDP-mutant mouse strains (Fig. 6 and 8 and data not shown), indicating the tissue specificity of CDP-mediated gene regulation. Second, these data are the first to show that individual CDP domains have differential effects on gene expression in the whole animal. For example, β-casein expression was elevated in the virgin mammary tissues of ΔC, but not ΔCR1, mice. Increased expression of β-casein in ΔC mice cannot be explained by a dominant-negative effect, since we and others have not observed interference with DNA-binding activity of the wild-type protein in virgin mammary gland, lung, and brain extracts (Fig. 7B) (26) and there is no detectable ΔC protein by Western blotting in nuclear extracts of thymus or mammary gland, where full-length CDP is clearly observed (Fig. 7A). Elevated β-casein expression in virgin mammary glands appears to result from reduced CDP expression in ΔC heterozygotes compared to wild-type animals. Thus, the four DNA-binding domains do not contribute equally to all CDP-regulated genes (2, 27). Third, previous studies have reported that ɛ-casein expression is reduced in the milk of homozygous ΔCR1 mice (43). However, our results showed that ɛ-casein RNA levels were not altered in the virgin mammary tissue of two different CDP-mutant strains. This suggests that altered amounts of ɛ-casein in milk of ΔCR1 females may result from CDP-induced deregulation at the posttranscriptional level. We have observed increased β-casein RNA in virgin mammary gland (Fig. 8D) and elevated β-casein protein levels in lactating mammary tissue of ΔC heterozygotes, but this has no apparent effect on the viability of nursing pups (data not shown). Fourth, mutations in CDP binding sites within an infectious MMTV provirus increased viral expression in the virgin mammary gland and decreased latency for mammary tumors compared to that of wild-type virus. These data reveal that CDP binding is required for control of MMTV expression in vivo.
Previous evidence indicates that CDP affects expression of several cellular genes, e.g., ɛ-casein, transforming growth factor-β type II receptor, and c-myc known to affect milk composition, mammary differentiation, or tumorigenesis (12, 19, 43). Homozygous mice lacking one of the Cut-repeat domains (ΔCR1) and heterozygous mice missing the C-terminal region (ΔC) both had increased levels of endogenous MMTV in the undifferentiated mammary gland (Fig. 6 and 8). Since endogenous viruses are resident in every mammary cell, these results clearly indicate that the effects of CDP binding site mutations on infectious MMTVs are not solely due to effects on transmission or spread of the virus, a result supported by the HP692 mutation, which had no effect on Sag-mediated deletion. In addition, these experiments argue that the CR1 domain is necessary for developmental control of MMTV transcription.
Development of the mammary gland proceeds in distinct stages marked by the appearance of specific genes. For example, β-casein and WDNM1 are upregulated earlier during mammary differentiation than ɛ-casein, α-lactalbumin, or WAP (37). The upregulation of specific milk genes in the virgin mammary tissues of CDP-mutant mice suggests that some of the “late” genes, including WAP and α-lactalbumin, as well as genes that appear earlier, e.g., β-casein, are regulated by CDP (Fig. 6 and 8). The β-casein promoter has several binding sites for the CCAAT-enhancer binding protein (C/EBP) (38), and the isoforms C/EBPβ and -δ reportedly bind to these sites to increase expression during pregnancy (10). C/EBPβ has been shown to control the proliferation and differentiation of mammary tissue and is required for activation of milk protein expression (36, 39). A different isoform, C/EBPɛ, appears to upregulate lactoferrin expression during myeloid cell differentiation and CDP, a protein initially described as having CCAAT-displacement activity on tissue-specific histone promoters (4), negatively regulates C/EBPɛ expression (21). More recently, CDP has been reported to interfere with C/EBP binding to the lactoferrin promoter and subsequent gene expression (20). Based on these observations, it is tempting to speculate that CDP differentially regulates the synthesis and/or function of different C/EBP isoforms during mammary gland development. Our preliminary data suggest that C/EBP overexpression elevates MMTV RNA levels and that CDP overexpression will antagonize this effect. Irrespective of the mechanism, our results strongly indicate that CDP negatively regulates transcription of many genes known to be expressed in differentiated mammary glands.
Acknowledgments
We acknowledge helpful discussions and comments on the manuscript by Jon Huibregtse and members of the Dudley laboratory. We also thank Dan Medina for analysis of mammary tumor histology.
This work was supported by grants CA34780 from the National Institutes of Health and DAMD17-01-1-0424 from the U.S. Army Breast Cancer Research Program.
REFERENCES
- 1.Ai, W., E. Toussaint, and A. Roman. 1999. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. J. Virol. 73:4220-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aufiero, B., E. J. Neufeld, and S. H. Orkin. 1994. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc. Natl. Acad. Sci. USA 91:7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banan, M., I. C. Rojas, W. H. Lee, H. L. King, J. V. Harriss, R. Kobayashi, C. F. Webb, and P. D. Gottlieb. 1997. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8α gene. J. Biol. Chem. 272:18440-18452. [DOI] [PubMed] [Google Scholar]
- 4.Barberis, A., G. Superti-Furga, and M. Busslinger. 1987. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 50:347-359. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer, R., S. Barbel, S. Sheperd, J. W. Jack, L. Y. Jan, and Y. N. Jan. 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51:293-307. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, S. M., J. M. Rosen, L. G. Hennighausen, U. Strech-Jurk, and A. E. Sippel. 1984. Comparison of the whey acidic protein genes of the rat and mouse. Nucleic Acids Res. 12:8685-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay, S., C. E. Whitehurst, and J. Chen. 1998. A nuclear matrix attachment region upstream of the T cell receptor β gene enhancer binds Cux/CDP and SATB1 and modulates enhancer-dependent reporter gene expression but not endogenous gene expression. J. Biol. Chem. 273:29838-29846. [DOI] [PubMed] [Google Scholar]
- 8.Cho, K., D. A. Ferrick, and D. W. Morris. 1995. Structure and biological activity of the subgenomic Mtv-6 endogenous provirus. Virology 206:395-402. [DOI] [PubMed] [Google Scholar]
- 9.Choi, Y., J. W. Kappler, and P. Marrack. 1991. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of mouse mammary tumour virus. Nature 350:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Doppler, W., T. Welte, and S. Philipp. 1995. CCAAT/enhancer-binding protein isoforms beta and delta are expressed in mammary epithelial cells and bind to multiple sites in the β-casein gene promoter. J. Biol. Chem. 270:17962-17969. [DOI] [PubMed] [Google Scholar]
- 11.Dudley, J. P. 1999. Mouse mammary tumor virus, p. 965-972. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology. Academic Press Ltd., London, United Kingdom.
- 12.Dufort, D., and A. Nepveu. 1994. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol. Cell. Biol. 14:4251-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durban, E. M., J. E. Knepper, D. Medina, and J. S. Butel. 1990. Influence of mammary cell differentiation on the expression of proteins encoded by endogenous BALB/c mouse mammary tumor virus genes. Virus Res. 16:307-323. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, T., L. Gambardella, M. Horcher, S. Tschanz, J. Capol, P. Bertram, W. Jochum, Y. Barrandon, and M. Busslinger. 2001. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 15:2307-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilead, Z., Y. H. Jeng, W. S. Wold, K. Sugawara, H. M. Rho, Harter, M. L., and M. Green. 1976. Immunological identification of two adenovirus 2-induced early proteins possibly involved in cell transformation. Nature 264:263-266. [DOI] [PubMed] [Google Scholar]
- 16.Golovkina, T. V., A. Chervonsky, J. P. Dudley, and S. R. Ross. 1992. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell 69:637-645. [DOI] [PubMed] [Google Scholar]
- 17.Hennighausen, L. G., A. Steudle, and A. E. Sippel. 1982. Nucleotide sequence of cloned cDNA coding for mouse epsilon casein. Eur. J. Biochem. 126:569-572. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, C. L., C. Fabritius, and J. Dudley. 1988. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J. Virol. 62:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, R. J., S. J. Antonia, K. L. Wright, N. S. Moon, A. Nepveu, and T. Munoz-Antonia. 1999. Human cut-like repressor protein binds TGFβ type II receptor gene promoter. Arch. Biochem. Biophys. 371:290-300. [DOI] [PubMed] [Google Scholar]
- 20.Khanna-Gupta, A., T. Zibello, H. Sun, P. Gaines, and N. Berliner. 2003. Chromatin immunoprecipitation (ChIP) studies indicate a role for CCAAT enhancer binding proteins alpha and epsilon (C/EBPα and C/EBPɛ) and CDP/cut in myeloid maturation-induced lactoferrin gene expression. Blood 101:3460-3468. [DOI] [PubMed] [Google Scholar]
- 21.Khanna-Gupta, A., T. Zibello, H. Sun, J. Lekstrom-Himes, and N. Berliner. 2001. C/EBP ɛ mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut). Proc. Natl. Acad. Sci. USA 98:8000-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lievens, P. M., J. J. Donady, C. Tufarelli, and E. J. Neufeld. 1995. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J. Biol. Chem. 270:12745-12750. [DOI] [PubMed] [Google Scholar]
- 23.Liu, J., A. Barnett, E. J. Neufeld, and J. P. Dudley. 1999. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Mol. Cell. Biol. 19:4918-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S., E. McLeod, and J. Jack. 1991. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics 127:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luong, M. X., C. M. van der Meijden, D. Xing, R. Hesselton, E. S. Monuki, S. N. Jones, J. B. Lian, J. L. Stein, G. S. Stein, E. J. Neufeld, and A. J. van Wijnen. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 22:1424-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon, N. S., G. Berube, and A. Nepveu. 2000. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J. Biol. Chem. 275:31325-31334. [DOI] [PubMed] [Google Scholar]
- 28.Moon, N. S., Z. W. Rong, P. Premdas, M. Santaguida, G. Berube, and A. Nepveu. 2002. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int. J. Cancer 100:429-432. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, B. W., and P. Leder. 1994. neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2-initiated tumors. Oncogene 9:3417-3426. [PubMed] [Google Scholar]
- 30.Mustafa, F., S. Bhadra, D. Johnston, M. Lozano, and J. P. Dudley. 2003. The type B leukemogenic virus truncated superantigen is dispensable for T-cell lymphomagenesis. J. Virol. 77:3866-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mustafa, F., M. Lozano, and J. P. Dudley. 2000. C3H mouse mammary tumor virus superantigen function requires a splice donor site in the envelope gene. J. Virol. 74:9431-9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 33.O'Connor, M. J., W. Stunkel, C. H. Koh, H. Zimmermann, and H. U. Bernard. 2000. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses through a conserved silencing element. J. Virol. 74:401-410. [PMC free article] [PubMed] [Google Scholar]
- 34.Pattison, S., D. G. Skalnik, and A. Roman. 1997. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 71:2013-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-261. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 36.Robinson, G. W., P. F. Johnson, L. Hennighausen, and E. Sterneck. 1998. The C/EBPβ transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 12:1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, G. W., R. A. McKnight, G. H. Smith, and L. Hennighausen. 1995. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development 121:2079-2090. [DOI] [PubMed] [Google Scholar]
- 38.Rosen, J. M., S. L. Wyszomierski, and D. Hadsell. 1999. Regulation of milk protein gene expression. Annu. Rev. Nutr. 19:407-436. [DOI] [PubMed] [Google Scholar]
- 39.Seagroves, T. N., S. Krnacik, B. Raught, J. Gay, B. Burgess, G. J. Darlington, and J. M. Rosen. 1998. C/EBPβ, but not C/EBPα, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 12:1917-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shackleford, G. M., C. A. MacArthur, H. C. Kwan, and H. E. Varmus. 1993. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc. Natl. Acad. Sci. USA 90:740-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shackleford, G. M., and H. E. Varmus. 1988. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc. Natl. Acad. Sci. USA 85:9655-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98:3658-3667. [DOI] [PubMed] [Google Scholar]
- 43.Tufarelli, C., Y. Fujiwara, D. C. Zappulla, and E. J. Neufeld. 1998. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 200:69-81. [DOI] [PubMed] [Google Scholar]
- 44.Vanden Heuvel, G. B., R. Bodmer, K. R. McConnell, G. T. Nagami, and P. Igarashi. 1996. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int. 50:453-461. [DOI] [PubMed] [Google Scholar]
- 45.Vilotte, J. L., and S. Soulier. 1992. Isolation and characterization of the mouse α-lactalbumin-encoding gene: interspecies comparison, tissue- and stage-specific expression. Gene 119:287-292. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Z., A. Goldstein, R. T. Zong, D. Lin, E. J. Neufeld, R. H. Scheuermann, and P. W. Tucker. 1999. Cux/CDP homeoprotein is a component of NF-μNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol. Cell. Biol. 19:284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster, M. A., N. Martin-Soudant, A. Nepveu, R. D. Cardiff, and W. J. Muller. 1998. The induction of uterine leiomyomas and mammary tumors in transgenic mice expressing polyomavirus (PyV) large T (LT) antigen is associated with the ability of PyV LT antigen to form specific complexes with retinoblastoma and CUTL1 family members. Oncogene 16:1963-1972. [DOI] [PubMed] [Google Scholar]
- 48.Wrona, T. J., M. Lozano, A. A. Binhazim, and J. P. Dudley. 1998. Mutational and functional analysis of the C-terminal region of the C3H mouse mammary tumor virus superantigen. J. Virol. 72:4746-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, L., T. J. Wrona, and J. P. Dudley. 1996. Exogenous mouse mammary tumor virus (MMTV) infection induces endogenous MMTV sag expression. Virology 215:113-123. [DOI] [PubMed] [Google Scholar]
- 50.Xu, L., T. J. Wrona, and J. P. Dudley. 1997. Strain-specific expression of spliced MMTV RNAs containing the superantigen gene. Virology 236:54-65. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimura, M., and T. Oka. 1989. Isolation and structural analysis of the mouse beta-casein gene. Gene 78:267-275. [DOI] [PubMed] [Google Scholar]
- 52.Zeng, W. R., S. W. Scherer, M. Koutsilieris, J. J. Huizenga, F. Filteau, L. C. Tsui, and A. Nepveu. 1997. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene 14:2355-2365. [DOI] [PubMed] [Google Scholar]
- 53.Zeng, W. R., P. Watson, J. Lin, S. Jothy, R. Lidereau, M. Park, and A. Nepveu. 1999. Refined mapping of the region of loss of heterozygosity on the long arm of chromosome 7 in human breast cancer defines the location of a second tumor suppressor gene at 7q22 in the region of the CUTL1 gene. Oncogene 18:2015-2021. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, Q., and J. P. Dudley. 2002. CDP binding to multiple sites in the mouse mammary tumor virus long terminal repeat suppresses basal and glucocorticoid-induced transcription. J. Virol. 76:2168-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]