Abstract
The Cdx1 gene product is essential for normal anterior-posterior vertebral patterning. Expression of Cdx1 is regulated by several pathways implicated in anterior-posterior patterning events, including retinoid and Wnt signaling. We have previously shown that retinoic acid plays a key role in early stages of Cdx1 expression at embryonic day 7.5 (E7.5), while both Wnt3a signaling and an autoregulatory loop, dependent on Cdx1 itself, are involved in later stages of expression (E8.5 to E9.5). This autoregulation is reflected by the ability of Cdx1 to affect expression from proximal Cdx1 promoter sequences in tissue culture. However, this region is devoid of a demonstrable Cdx response element(s). We have now found that Cdx1 and LEF1, a nuclear effector of Wnt signaling, synergize to induce expression from the Cdx1 promoter through previously documented LEF/T-cell factor response elements. We also found a direct physical interaction between the homeodomain of Cdx1 and the B box of LEF1, suggesting a basis for this synergy. Consistent with these observations, analysis of Cdx1 Wnt3avt compound mutants demonstrated that Wnt and Cdx1 converged on Cdx1 expression and vertebral patterning in vivo. Further data suggest that Cdx-high-mobility group box interactions might be involved in a number of additional pathways.
Somites are derived from segmentation of the paraxial mesoderm in the caudal embryo and subsequently differentiate into the dermamyotome and sclerotome, the latter being the anlage of the vertebrae. Many vertebrae exhibit morphological differences along the anterior-posterior axis, such as the ribs characteristic of thoracic vertebrae. These distinct morphological characteristics are indicative of patterning events which dictate vertebral identity along the anterior-posterior axis. A number of signaling molecules, such as retinoic acid, are well documented to affect vertebral anterior-posterior patterning. All such effectors typically impact on the expression of Hox genes, and a wealth of gain- and loss-of-function experiments clearly demonstrate a critical role for Hox gene products in vertebral patterning (see, e.g., references 9, 11, 13, 19, and 27).
The 39 murine Hox genes are distributed in four clusters, Hoxa to Hoxd, which have likely evolved by duplication of an ancestral complex related to the HOM-C genes of Drosophila melanogaster (17, 18, 20). In the mouse, Hox expression is initiated at embryonic day 7.5 (E7.5) in the primitive streak, with transcripts subsequently expanding anteriorly in the neural tube and mesoderm to eventually reach a predetermined rostral limit (14, 45, 50). The onset and rostral limit of expression are generally related to the location of a given Hox gene within its complex, with more 3′ members initiated earlier and reaching more rostral limits of expression than 5′ paralogs. This results in staggered domains of Hox expression along the anterior-posterior axis, which have been suggested to comprise a Hox code (5, 23, 33).
While Hox genes are expressed initially in the primitive streak and subsequently in the somites and prevertebrae, grafting experiments in the chicken embryo demonstrate that vertebral anterior-posterior patterning is imparted before overt segmentation of the paraxial mesoderm, likely during or shortly after gastrulation (34, 44). Paraxial mesoderm from such transplants also retains the Hox expression patterns characteristic of its original axial position, suggesting that the molecular program dictating anterior-posterior patterning is imparted during this early stage of vertebral ontogenesis.
Considerable effort has been directed to a better understanding of the mechanisms involved in establishing Hox gene expression. Recently, members of the vertebrate Cdx family, Cdx1, Cdx2, and Cdx4, have emerged as important players in this process. Cdx genes encode homeodomain transcription factors related to the Drosophila gene caudal. A number of studies suggest a key role for Cdx members in vertebral patterning by direct regulation of Hox expression. In particular, loss-of-function studies in the mouse have shown that Cdx1 null mutants, Cdx2 heterozygotes, and Cdx1−/− Cdx2+/− compound mutants all exhibit skeletal defects which are associated with a posterior shift in the anterior boundaries of expression of a number of Hox genes (8, 55, 60). Furthermore, consensus Cdx binding motifs have been identified in the promoters of many Hox genes (2, 55), some of which direct spatial expression in vivo (7). Gain- and loss-of-function studies in chicken and frog embryos also support a role for Cdx members in anterior-posterior patterning of both mesoderm and neurectoderm through regulation of Hox expression (3, 30).
A number of signaling pathways, including retinoic acid and some members of the fibroblast growth factor and Wnt/wingless families, affect posterior embryonic patterning, at least in part through regulation of Hox expression. However, the means by which these signaling molecules impact Hox expression is incompletely understood. A number of studies have demonstrated that Cdx family members respond to fibroblast growth factor, Wnt, and retinoic acid (3, 28, 29, 30, 47, 49), suggesting that Cdx proteins serve to convey these signals to the Hox genes. Of particular relevance to the present study, we (28, 49) and others (29) have shown that Cdx1 is directly regulated by both retinoic acid and Wnt3a in the caudal embryo.
The Wnt signaling pathway is involved in many developmental processes (reviewed in references 42 and 62). Activation of the canonical Wnt signaling pathway results in stabilization of cytoplasmic β-catenin, which subsequently translocates to the nucleus and associates with LEF/T-cell factor (TCF) transcription factors (LEF1, TCF1, TCF3, and TCF4) to induce expression of target genes. Wnt3a is expressed in an overlapping manner with Cdx1 in the primitive streak and tailbud of murine embryos (41, 56). Wnt3a homozygous null embryos (56) and the Wnt3a hypomorph vestigial tail (vt) (24) exhibit vertebral abnormalities which occur concomitant with reduced expression of Cdx1 and certain Hox genes (29, 49). This effect is likely mediated through two LEF/TCF response elements present on the proximal Cdx1 promoter, which function in tissue culture (38, 49) and are also essential for normal expression of a Cdx1 transgenic reporter in vivo (39). Among the Wnt nuclear effectors, LEF1 and TCF1 are most likely responsible for conveying the Wnt3a signal to Cdx1, based on their overlapping patterns of expression (46, 56) and the finding that LEF1 TCF1 double null mutants have phenotypes similar to those of Wnt3a null mice (21, 56).
In addition to regulation by Wnt and retinoic acid, Cdx1 is required to maintain its own expression in vivo. This autoregulation can be recapitulated in P19 embryocarcinoma cells in tissue culture, where Cdx1 can induce expression from a reporter vector comprised of proximal Cdx1 genomic sequences (49). However, consensus Cdx1 binding motifs have not been identified in these sequences, and the means by which this effect is mediated is unclear. We now present evidence that this autoregulation is mediated by direct physical interaction between Cdx1 and LEF1, with the latter binding to the Cdx1 promoter. This interaction occurs between the homeodomain of Cdx1 and the B box of LEF1. Analysis of an allelic series of Cdx1 Wnt3avt/vt mutants further supports an interaction between Wnt signaling and Cdx1 autoregulation in vivo. Finally, we present evidence that Cdx-high-mobility group (HMG) interactions may be involved in other regulatory pathways.
MATERIALS AND METHODS
Plasmid constructs.
The Cdx1 genomic sequences and derivation of reporter vectors have been described previously (28, 49). The Flag epitope-tagged Cdx1 vector was generated by subcloning the relevant coding sequences into a modified pCEP4 plasmid (Invitrogen), while the Flag-tagged retinoic acid receptor gamma (RARγ) fusion construct was generated by PCR and subcloned into pSG5 (Stratagene). The reporter construct bearing four Cdx binding elements from the Hoxb8 promoter was generated by subcloning EcMs79 sequences (7) into pTK109-Luc (43). Expression vectors for glutathione S-transferase (GST)-Cdx1 fusion proteins were generated by PCR and subcloned in pGEX4T-1 (Amersham-Pharmacia). The LEF1ΔBbox vector was generated by PCR and subcloned into pSG5. The Cdx1-LEF1 fusion constructs were made by PCR and subcloned into pCMV5β (Liliana Attisano) or pCEP4Flag. Further details of the derivation of these constructs are available upon request.
The pXPA17-5XUAS reporter vector, harboring five GAL4 binding elements, and the expression vector pCMXGAL4DBD, used to generate GAL4-Cdx1 fusion constructs, were gifts from Mark Featherstone. Hemagglutinin (HA) epitope-tagged LEF1 expression vectors were provided by Liliana Attisano, while the LEF1 DNA binding domain/β-catenin activation domain (LEF1ΔN-βCTA) and the LEF1 N-terminal deletion mutant (LEF1ΔN/pCS2) were provided by Andreas Hecht. The Flag-tagged β-catenin expression vector was provided by David Rimm, and the LEF/TCF reporter vector pGL3-OT and control pGL3-OF vector were provided by Bert Vogelstein.
Cell culture and transfections.
P19 embryocarcinoma cells were propagated under standard conditions. For transfection analysis, cells were passed into six-well plates and transfected the same day by the calcium phosphate precipitation method. Unless otherwise stated, transfections consisted of 1 μg of reporter construct and 0.5 μg each of expression vector(s) and empty vector (where required) to a final amount of 2 μg of DNA. The following day, the medium was replenished, and culture was continued for another 24 h. Cells were then disrupted in 250 μl of lysis buffer (0.1 M Tris [pH 8.0], 1% Igepal, 1 mM dithiothreitol) and assessed for luciferase activity with an AutoLumat LB953 luminometer (Berthold Technologies). Cos7 and F9 embryocarcinoma cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum. Transfections were performed under the same conditions as for P19 cells except that 100-mm tissue culture plates were used with a total of 25 μg of expression vector(s) per transfection.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays were performed as previously described (37) with anti-FlagM2 antibody (Santa Cruz Biotechnology). PCRs were performed for 30 cycles (96°C for 29 s, 60° C for 30 s, and 72° C for 1 min), with primers flanking the proximal Cdx1 LEF/TCF response elements, the Cdx1 retinoic acid response element, or exon 3. Amplification products were resolved on a 2% agarose gel, transferred to a Hybond N (Amersham-Pharmacia) membrane, and hybridized with an internal oligonucleotide probe specific for the predicted amplification product (oligonucleotide sequences available on request).
Immunoprecipitation assays.
Cells were harvested in immunoprecipitation buffer (20 mM Tris [pH 8], 25 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, and protease inhibitors) and immunoprecipitated with mouse monoclonal anti-HA antibody (Covance, Berkeley, Calif.) with 20 μl of protein A-G Plus-agarose (Santa Cruz). The beads were washed twice with immunoprecipitation buffer, and proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blots were performed by the ECL method (Amersham-Pharmacia) with anti-FlagM2 antibody (Santa-Cruz Biotechnology) at a 1:5,000 dilution.
GST fusion proteins.
Fifty milliliters of bacterial culture, transformed with the appropriate GST-Cdx1 plasmid, was grown to an optical density of 0.6 to 0.8 and treated with 0.5 mM isopropylthiogalactopyranoside (IPTG) for 2 to 3 h. Cells were recovered by centrifugation, resuspended in 1.5 ml of phosphate-buffered saline containing 1 mM dithiothreitol, 0.02% Triton X-100, and protease inhibitors, and disrupted by sonication. Triton X-100 was added to a final concentration of 1%, and the lysate was incubated on ice for 15 min. Following centrifugation, the supernatant was incubated with glutathione-agarose beads (Sigma) for 1 h at 4°C. The beads were recovered by centrifugation, washed three times with phosphate-buffered saline, and used for in vitro binding assays.
In vitro pulldown assays.
[35S]methionine-labeled LEF1 proteins were generated with the TNT system (Promega), and 5 μg of GST fusion protein was incubated with 5 μl of labeled LEF1 in 750 μl of TNEN buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 0.1% Igepal, 5 mM EDTA, 1 mM dithiothreitol, and protease inhibitors). The beads were then washed with TNEN, proteins were resolved by SDS-PAGE, and interactions were revealed by phosphorimaging.
Generation and analysis of mice.
vestigial tail (Wnt3avt/vt) mice were obtained from the Jackson Laboratory (Bar Harbor, Maine). Cdx1 null mice were described previously (55). An allelic series of compound mutant offspring were generated from Wnt3avt/+ Cdx1+/− intercrosses. Mice were mated overnight, and females were examined the following morning for the presence of a vaginal plug; noon of the day that the plug appeared was considered E0.5. CD1 mice (Charles River) were used as wild-type controls; no overt differences in gene expression have been observed between CD1 offspring relative to wild-type offspring from the Wnt3avt or Cdx1 mutant colonies (49). In situ hybridization and skeletal preparations were performed as previously described (1, 49).
RESULTS
Cdx1 associates with its own promoter in vivo.
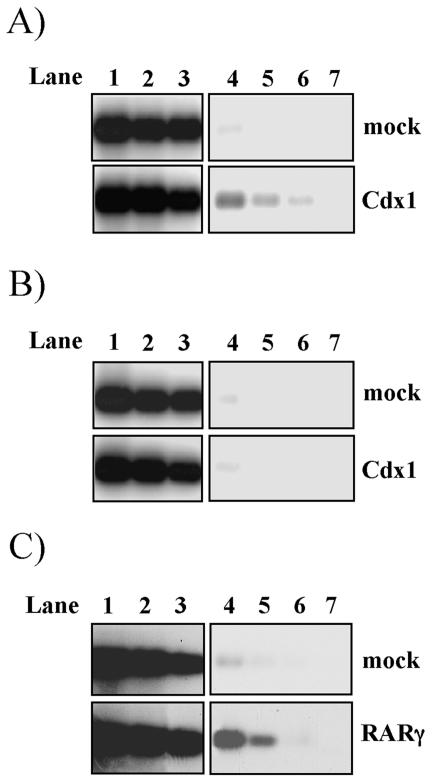
Cdx1 expression is lost in Cdx1 mutants at E8.5 (50), suggestive of an autoregulatory loop. Cdx1 can induce expression from Cdx1 promoter sequences in transfection assays. However, Cdx response elements are not evident in these sequences (16, 49), and electrophoretic mobility shift assay analysis has failed to reveal association of Cdx1 with any region of this promoter (our unpublished observation). Despite the absence of such regulatory elements, chromatin immunoprecipitation experiments demonstrated that the Cdx1 protein is present on the proximal Cdx1 promoter (Fig. 1A) but not on more distal sequences (Fig. 1B). This association was comparable to RARγ binding to this region, which likely occurs through a previously documented retinoic acid response element (28) (Fig. 1C).
FIG. 1.
Cdx1 is present on its own promoter in vivo. (A and B) Anti-Flag chromatin immunoprecipitation analysis of the endogenous Cdx1 promoter (A) and downstream sequences used as a negative control (B) on mock-transfected (upper panel) and Flag-Cdx1-transfected (lower panel) F9 cells. (C) Identical analysis of the endogenous Cdx1 promoter on mock-transfected (upper panel) and Flag-RARγ (lower panel)-transfected F9 cells. After anti-Flag immunoprecipitation of the sonicated chromatin extracts, DNA was purified and amplified by PCR with oligonucleotides spanning the region of the Cdx1 promoter encompassing the LEF/TCF response elements in panel A, the retinoic acid response element in panel C, or exon 3 in panel B. Products were resolved by agarose gel electrophoresis and assessed by Southern blot analysis with an internal oligonucleotide probe specific to the predicted amplification products. Lanes 1 to 3 (before immunoprecipitation) and 4 to 6 (after immunoprecipitation) represent PCR amplifications of serial dilutions of DNA. Lane 7 is a negative control in which DNA was not included.
Cdx1 and LEF1 act synergistically.
The above data suggest that Cdx1 is able to associate with its own promoter. While cryptic Cdx1 binding elements cannot be ruled out, one alternative possibility is that Cdx1 may interact with other relevant transcription factors, including RARs or LEF/TCF members. Transfection analysis was therefore used to examine the effect of LEF1, retinoic acid, and Cdx1, individually and in combination, on expression of a Cdx1 reporter in P19 cells.
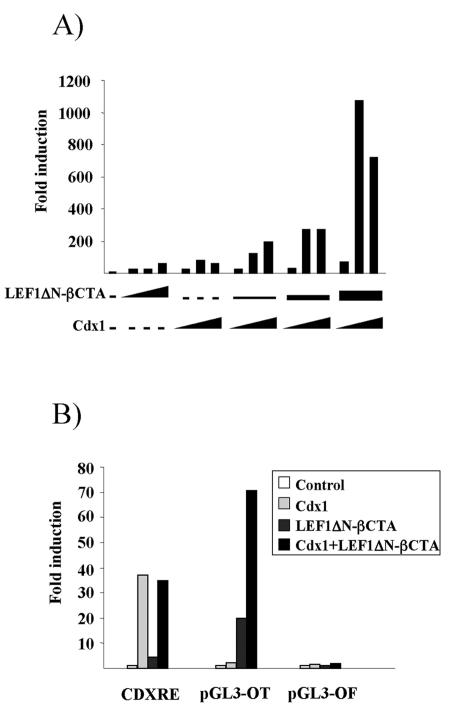
In agreement with previous findings, retinoic acid, LEF1, and Cdx1 all induced the Cdx1 reporter (Fig. 2A and data not shown) (49). Only additive interactions were seen between retinoic acid and Cdx1 in transfection analysis (data not shown). However, a strong interaction was observed between a LEF1-β-catenin fusion protein (LEF1ΔN-βCTA) (61) and Cdx1 which greatly exceeded the additive values achieved with either transcription factor alone. This outcome was not likely an artifact of the nonphysiological fusion protein, since a similar synergistic interaction was also seen between Cdx1, LEF1, and β-catenin (data not shown).
FIG. 2.
Synergy between Cdx1 and LEF1. (A) P19 cells were transfected with the 2-kb Cdx1 reporter alone or with various amounts of the expression vectors (0.05 to 0.5 μg) as indicated. Cells were harvested 48 h posttransfection, and extracts were assessed for luciferase activity. Data are expressed as induction relative to activity with the reporter vector alone. (B) P19 cells were transfected with a reporter construct harboring Cdx1 binding sites from the Hoxb8 promoter (CDXRE) or with wild-type (pGL3-OT) or mutant (pGL3-OT) LEF/TCF response elements. Transfections were done with the reporter vector alone or with the indicated expression constructs. Luciferase activity was determined as above and expressed as induction relative to activity with the reporter vector alone.
To further understand the basis for the synergistic interaction between LEF1 and Cdx1, the ability of LEF1ΔN-βCTA and Cdx1 to regulate synthetic reporter vectors harboring either Cdx or LEF/TCF response elements was examined. The Hoxb8 promoter contains a regulatory element consisting of four functional Cdx binding sites (7). Transfection analysis revealed that Cdx1 elicited a strong transcriptional response through these elements (Fig. 2B). LEF1ΔN-βCTA alone did not activate this reporter, and cotransfection with both Cdx1 and LEF1ΔN-βCTA gave an induction comparable to that seen with Cdx1 alone. The pGL3-OT and pGL3-OF reporters contain three wild-type or mutant LEF/TCF response elements, respectively (25). As expected, LEF1ΔN-βCTA mediated a significant induction of pGL3-OT which was lost in pGL3-OF. While Cdx1 did not induce the pGL3-OT reporter, it markedly potentiated the effect of LEF1ΔN-βCTA (Fig. 2B). This interaction was abolished upon mutation of the LEF/TCF binding sites. Taken together, these data suggest that LEF1 and Cdx1 can synergize in a manner that is dependent on association of LEF1 with cognate response elements but in the absence of direct binding of Cdx1 to the DNA.
Cdx1 and LEF1 interact in vitro and in vivo.
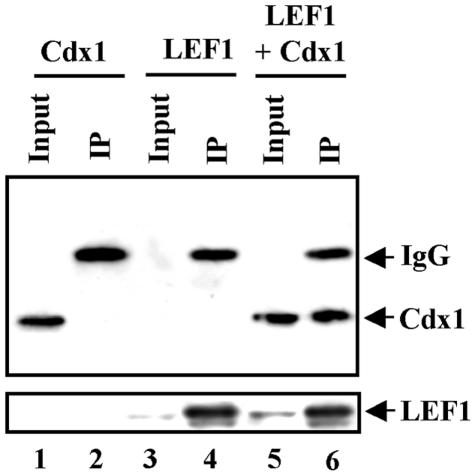
The synergy observed between LEF1 and Cdx1 in transcription assays, together with the observations from chromatin immunoprecipitation studies, suggested that these two proteins might interact physically to regulate Cdx1 expression. We initially tested this by coimmunoprecipitation experiments from Cos7 cells transfected with epitope-tagged Cdx1 and LEF1 and found that Cdx1 coimmunoprecipitated with LEF1 when the two were coexpressed (Fig. 3).
FIG. 3.
Cdx1 interacts with LEF1 in vivo. Whole-cell lysates from Cos7 cells transfected with the Cdx1-Flag and/or Lef1-HA expression vectors were immunoprecipitated with anti-HA antibody and analyzed by Western blotting with an anti-Flag antibody (top panel). Expression of LEF1 was assessed by reprobing the blot with an anti-HA antibody (bottom panel). Inputs represent 5% of the total lysate used for immunoprecipitation.
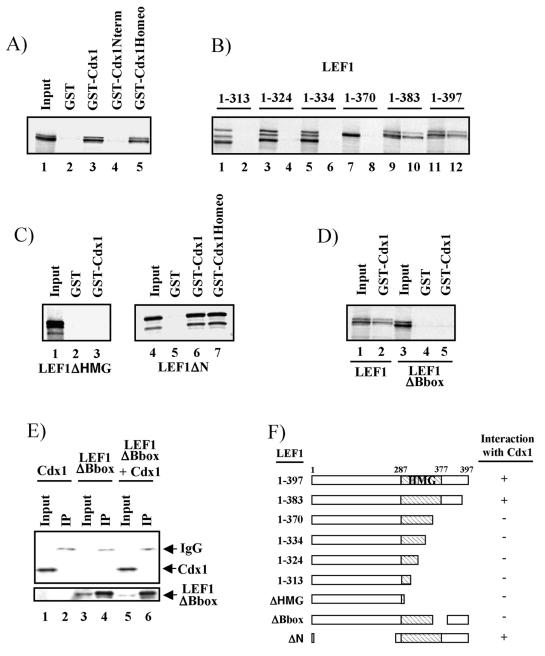
To determine if LEF1 and Cdx1 association was direct, we used GST-Cdx1 fusion proteins and in vitro-translated LEF1 to assess interaction in a cell-free system. All GST-Cdx1 fusions were stably expressed, as evaluated by Coomassie blue staining (data not shown). As shown in Fig. 4A, LEF1 interacted with GST-Cdx1 but not with GST alone. Similar assays indicated that the Cdx1 homeodomain alone was sufficient for association with LEF1, whereas N-terminal sequences failed to interact (Fig. 4A). In order to exclude the possibility that both proteins bound nonspecifically to contaminating DNA, we repeated the assay in the presence of ethidium bromide, which prevents Cdx1-DNA interactions (data not shown), and found that it did not affect the Cdx1-LEF1 association (data not shown).
FIG. 4.
Cdx1 interacts directly with LEF1. GST-Cdx1 and 35S-labeled LEF1 constructs were generated and assessed for interaction as described in Materials and Methods. (A) Both full-length Cdx1 (GST-Cdx1) and the Cdx1 homeodomain (GST-Cdx1Homeo) interact with LEF1 (lanes 3 and 5), whereas Cdx1 N-terminal sequences do not (lane 4). (B) Deletion of sequences N-terminal to position 383 of LEF1 abolished interaction with Cdx1 (compare lane 10 to lane 8; odd-numbered lanes are 20% input of the various 35S-labeled LEF1 constructs, and even-numbered lanes are from the GST-Cdx1 pulldown). (C) Deletion of the LEF1 HMG box (lane 3) abolishes interaction with Cdx1, whereas loss of N-terminal LEF1 residues has no effect on interaction with either full-length Cdx1 (lane 6) or the homeodomain sequences of Cdx1 (lane 7). (D and E) Deletion of the LEF1 B box abolishes interaction with Cdx1 in vitro (D, compare lane 5 to lane 2) and in vivo (E, lane 6). (F) Schematic representation of LEF1-Cdx1 interactions. Coimmunoprecipitations (panel E) were performed as in Fig. 2 except the ΔBbox mutant was used instead of full-length LEF1.
To further characterize the LEF1-Cdx1 association, we used deletion analysis to determine the LEF1 sequences mediating the interaction with the Cdx1 homeodomain. Progressive deletion of the C-terminal region of LEF1 completely abolished interaction with GST-Cdx1 when the integrity of the HMG box was affected (Fig. 4B; summarized in Fig. 4F). Indeed, internal deletion of the C-terminal HMG box sequences eliminated interaction with Cdx1, whereas loss of the entire N-terminal region of LEF1 had no effect on association with either the full-length protein or the Cdx1 homeodomain (Fig. 4C).
Previous work suggested that a 9-amino-acid sequence, the B box, located within the C-terminal region of the HMG domain is involved both in nuclear localization of LEF1 and in contributing to DNA binding (48). To assess the potential involvement of the B box in Cdx1 interaction, we generated a LEF1 mutant lacking the B box and assessed its ability to associate with Cdx1. As shown in Fig. 4D, GST-Cdx1 interacted with full-length LEF1 but not with the B box mutant. Similar results were also seen in vivo, where a LEF1ΔBbox mutant failed to interact with Cdx1 (Fig. 4E). Together, these data demonstrate that the Cdx1 homeodomain binds to LEF1 B box sequences.
Transactivation domain of Cdx1 confers transcriptional activity on LEF1 HMG box sequences.
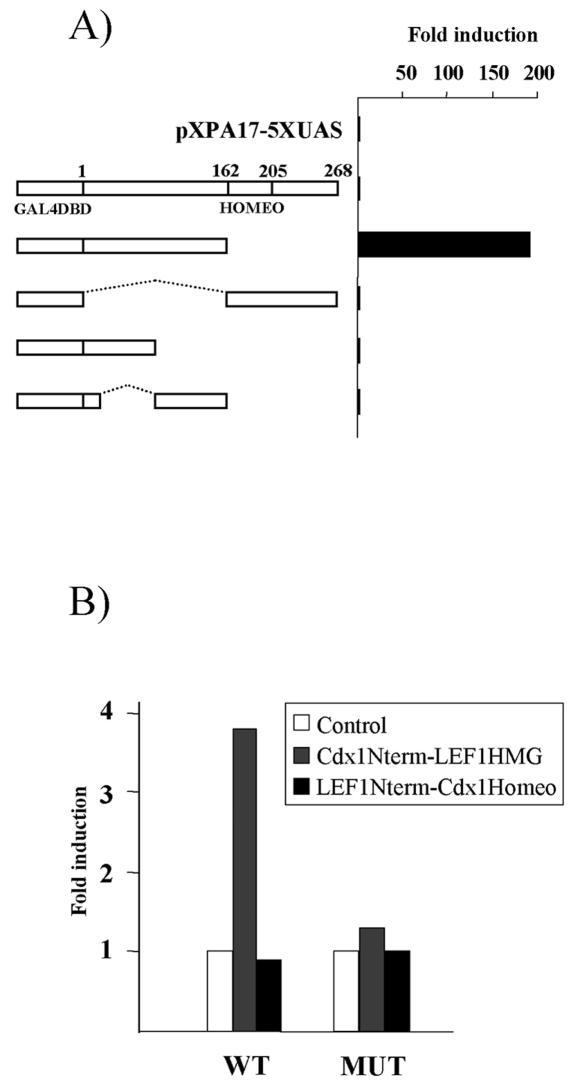
The mechanism by which Cdx1 activates transcription is poorly understood. We used a mammalian one-hybrid approach to identify transactivation domains, using Cdx1-GAL4 DNA binding domain chimeras and assaying their ability to activate transcription from GAL4 binding elements. Western blot analysis indicated that all fusion proteins were stably expressed (data not shown). Unexpectedly, the chimera containing full-length Cdx1 did not exhibit significant transcriptional activity (Fig. 5A), perhaps due to steric hindrances, which may prohibit proper DNA binding and/or coactivator recruitment. In marked contrast, deletion of the C-terminal sequences resulted in significant reporter activation. However, additional dissection of this region resulted in loss of transactivation potential (Fig. 5A and data not shown). Interestingly, the transactivation domains of Cdx2 (57, 58) and Xcad3 (the Xenopus homologue of Cdx4) (30) map to a similar region despite the relatively weak conservation between these sequences (22).
FIG. 5.
Transcriptional activation domain of Cdx1 activates the Cdx1 promoter. (A) The N-terminal domain of Cdx1 contains a transactivation function. P19 cells were transfected with a reporter vector containing GAL4 binding elements (pXPA17-5XUAS), alone or with the indicated GAL4 DNA binding domain-Cdx1 fusion. Luciferase activity was measured 48 h posttransfection and expressed as induction relative to activity with the reporter vector alone. (B) The N-terminal domain of Cdx1 can activate transcription from the Cdx1 promoter when fused to the LEF1 HMG box. P19 cells were transfected with the 180-bp proximal Cdx1 reporter construct (WT) or an identical construct lacking the LEF/TCF response elements (MUT). Transfections were performed either with the reporter vector alone or with the indicated fusion construct. Luciferase activity and results were determined as above.
We subsequently determined whether recruitment of the Cdx1 transactivation domain to the Cdx1 LEF/TCF response elements could regulate expression. To assess this, we generated chimeras consisting of either the N-terminal Cdx1 sequences and the HMG box of LEF1 or the LEF1 N-terminal region fused to the Cdx1 homeodomain and compared their ability to induce transcription of a reporter containing 180 bp of the proximal Cdx1 promoter, a region which harbors two LEF/TCF response elements (38, 49). As shown in Fig. 5B, only the Cdx1 N terminus-LEF1 HMG box fusion protein transactivated this reporter, and this outcome required the LEF/TCF response elements. This observation is consistent with a model in which LEF1 serves to recruit Cdx1 to the Cdx1 promoter, where it contributes to transactivation. It is notable that this chimera elicited a considerably weaker induction than that seen with Cdx1-LEF1 and β-catenin. This may be due to a key role for β-catenin in regulating transcription from the Cdx1 promoter, consistent with our finding that all three proteins coimmunoprecipitate (data not shown).
Genetic evidence for Cdx1-LEF/TCF interactions.
To determine the in vivo relevance of Cdx1-LEF1 interactions, we assessed the skeletal morphology of an allelic series of Cdx1 Wnt3avt compound mutants. The Wnt3avt mouse, a Wnt3a hypomorph, was chosen for this analysis because Wnt3a null mutants have severe caudal defects which preclude assessment of the direct impact of Wnt signaling on Cdx1 expression; a similar caveat applies to LEF1 TCF1 double null mutants (21, 56).
The murine vertebral column is composed of seven cervical (C1 to C7), 13 thoracic (T1 to T13), six lumbar (L1 to L6), three or four sacral (S1 to S4), and 31 caudal vertebrae. The first cervical vertebra (C1 or atlas) exhibits thick neural arches and possesses a ventrally located tubercle, the anterior arch of the atlas (Fig. 6A). The neural arches of C2 are intermediate to those of C1 and more posterior cervical vertebrae. C3, C4, and C5 are virtually identical to one another, while C6 is distinguished by the ventrally protruding anterior tuberculi. The thoracic vertebrae are characterized by the presence of ribs, the first seven of which (T1 to T7) are attached to the sternum.
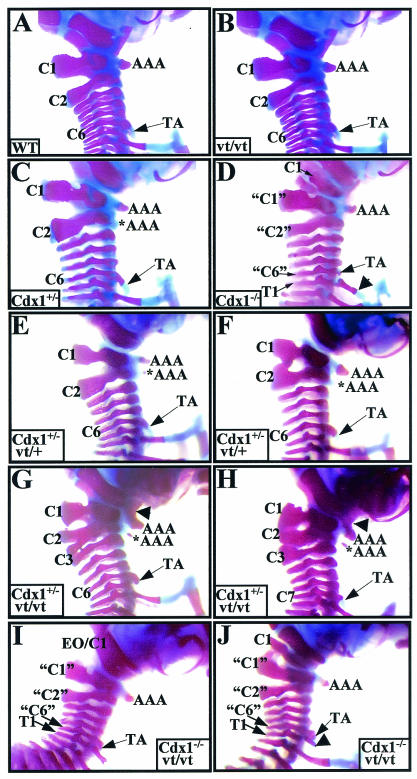
FIG. 6.
Skeletal analysis of Cdx1 Wnt3avt compound mutants. The cervical region of whole-mount skeletal preparations from (A) wild-type, (B) Wnt3avt/vt, (C) Cdx1+/−, (D) Cdx1−/−, (E and F) Cdx1+/− Wnt3avt/+, (G and H) Cdx1+/− Wnt3avt/vt and (I and J) Cdx1−/− Wnt3avt/vt offspring. Note the normal cervical region for both wild-type (A) and Wnt3avt/vt (B) mice. (C, E, and F) Cdx1+/− and Cdx1+/− Wnt3avt/+ mice exhibit similar phenotypes, notably an ectopic anterior arch of the atlas (*AAA), a broader C2 neural arch, and a C1-C2 fusion (F). (D, I, and J). Cdx1 null and Cdx1−/− Wnt3avt/vt mice exhibit identical vertebral defects, notably malformation and malposition of C1 and anterior transformation of C2, C3, and C7, denoted “C1,” “C2,” and “C6,” respectively. The arrowhead in panel J indicates a partial rib associated with presumptive T1 which does not reach the sternum. (G and H) Cdx1+/− Wnt3avt/vt offspring exhibit an exacerbated phenotype compared to either single mutant (compare panels G and H to panels B and C). In particular, note the fusion of the anterior arch of the atlas with the basioccipital bone (arrowhead) and the presence of an ectopic anterior arch of the atlas. Note also the malformations and fusions of neural arches C1 to C3 and the presence of TA on C7 instead of C6 (H). Abbreviations: AAA, anterior arch of the atlas; *AAA, ectopic anterior arch of the atlas; TA, tuberculum anterior; EO, exoccipital; C, cervical vertebrae; T, thoracic vertebrae. Quotation marks indicate presumptive anterior transformations.
All combinations of Cdx1 Wnt3avt compound mutants were viable and fertile. Wnt3avt/vt offspring exhibited a foreshortened tail, as previously described (29, 56), and this phenotype was not exacerbated by subsequent loss of Cdx1 (data not shown). We did not detect any vertebral defects in the Wnt3avt/vt background, although a low incidence of a subtle C2-C1 malformation has been reported by others (29) (Table 1 and Fig. 6B). As described previously (1), Cdx1+/− offspring exhibited certain vertebral defects; C2 often presented characteristics indicative of a C1 morphology, including an anterior arch of the atlas-like structure and/or thicker neural arches (Table 1 and Fig. 6C). The neural arches of C1, C2, and C3 were frequently malformed (Table 1 and Fig. 6C), and basioccipital-anterior arch of the atlas fusions were observed at a lower frequency.
TABLE 1.
Vertebral phenotypes of compound Wnt3avt Cdx1 mutantsa
| Phenotypea | No. (%) with genotype:
|
||||||
|---|---|---|---|---|---|---|---|
| Wild type (n = 14) | Wnt3avt/vt or Wnt3avt/+ (n = 20) | Cdx1+/− (n = 18) | Cdx1−/− (n = 29)b | Wnt3avt/vt Cdx1+/− (n = 20) | Wnt3avt/vt Cdx1+/− (n = 21) | Wnt3avt/vt Cdx1−/− (n = 20) | |
| Basioccipital fusion to AAA | 2 (11) | 29 (100) | 2 (10) | 9 (43) | 20 (100) | ||
| Vertebra 1 | |||||||
| Malformed NA | 3 (17) | 6 (30) | 5 (24) | ||||
| Fusion to occipitals | 29 (100) | 18 (90) | |||||
| Vertebra 2 | |||||||
| C1 identity (AAA and/or thick NA) | 10 (56) | 29 (100) | 12 (60) | 20 (96) | 20 (100) | ||
| Malformed NA | 2 (11) | 8 (40) | 18 (86) | ||||
| Vertebra 3 | |||||||
| C2 identity | 29 (100) | 4 (19) | 20 (100) | ||||
| Malformed NA | 1 (6) | 4 (14) | 5 (24) | 1 (5) | |||
| NA fusions | |||||||
| V1-V2 | 1 (6) | 6 (30) | 3 (14) | ||||
| V2-V3 | 1 (6) | 4 (19) | |||||
| TA on vertebra 7 instead of vertebra 6 | 28 (97) | 1 (5) | 4 (19) | 20 (100) | |||
| Vertebra 8 | |||||||
| C7 identity | 11 (38) | 2 (10) | |||||
| Incomplete ribs | 18 (62) | 11 (55) | |||||
AAA, anterior arch of the atlas; NA, neural arch; TA, anterior tuberculi.
From Allan et al. (1).
Cdx1−/− offspring also exhibited vertebral defects consistent with prior work (Table 1, Fig. 6D) (1, 55). Close apposition or fusion of C1 to the basioccipital bone, accompanied by a reduction of C1 neural arches and loss of the anterior arch of the atlas, was observed in all Cdx1 mutants. This occurred concomitant with C2 to C1, C3 to C2, and C6 to C5 transformations, determined by morphological criteria as previously described (1, 55). Most Cdx1−/− skeletons also exhibited partial ribs on the eighth vertebra (i.e., presumptive T1). A significant percentage of the skeletons also exhibited a T1 to C7 phenotype.
The Cdx1+/− Wnt3avt/+ mice presented phenotypes approximating that of Cdx1 heterozygote mutants, albeit with a higher incidence of malformed neural arches and C2-C1 fusion (Table 1 and Fig. 6E and F). The Cdx1−/− Wnt3avt/vt double mutants also closely phenocopied Cdx1 single null offspring (Table 1 and Fig. 6I and J). This result is in accordance with previous observations that put Wnt3a signaling upstream of Cdx1 (49).
In marked contrast to the double null mutant phenotype, Cdx1+/− Wnt3avt/vt offspring exhibited a profound synergistic interaction relative to the defects observed in either individual background (Table 1 and Fig. 6G and H). Fusion between the anterior arch of the atlas and the basioccipital bone, observed at a low incidence in Cdx1 heterozygotes and never in Wnt3avt/vt offspring, was found in approximately half of the Cdx1+/− Wnt3avt/vt compound mutants. The incidence of malformation of the neural arches of C1, C2, and C3 and C2 to a C1 transformation was also markedly increased. Transformation of C3 to a C2 identity occurred in about 20% of Cdx1+/− Wnt3avt/vt offspring but was never seen in the Cdx1+/− or Wnt3avt/vt background.
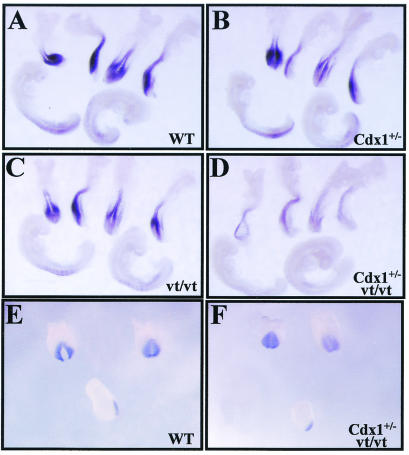
As Cdx1+/− Wnt3avt/+ mutants phenocopy Cdx1+/− mice and Wnt3avt/vt offspring exhibit a wild-type axial skeleton, the increased penetrance of the defects seen in Cdx1+/− Wnt3avt/vt offspring cannot be explained by a simple additive effect of Cdx1 and Wnt3a on vertebral patterning but rather by a synergistic interaction. Such a mechanism is in agreement with the physical interaction between Cdx1 and LEF1 on the Cdx1 promoter. To further investigate this possibility, Cdx1 expression was assessed in the relevant mutant backgrounds by whole-mount in situ hybridization of E8.5 and E9.5 embryos. Despite the high incidence of homeotic transformations in Cdx1+/− mice, Cdx1 expression was not appreciably altered (Fig. 7A and B). This is consistent with the prior finding that both wild-type and mutant Cdx1 transcripts are equally stable and therefore that the homeosis observed in Cdx1+/− offspring is due to reduction in Cdx1 protein per se (49). Cdx1 expression was marginally downregulated in the tailbud of Wnt3avt/vt embryos, as previously described (Fig. 7C) (49), although this reduction is apparently not sufficient to affect vertebral patterning. Finally, a strong reduction of Cdx1 expression was observed in Cdx1+/− Wnt3avt/vt embryos (Fig. 7D), in close agreement with the synergistic effect of these mutant alleles on vertebral morphogenesis. We also found that this effect was stage specific, as no differences were detected at E7.5 (Fig. 7E and F), which is in accordance with previous findings demonstrating that Cdx1 autoregulation commences around E8.5 (49). These observations are consistent with a mechanism for Cdx1 autoregulation involving an interaction between Cdx1 and LEF/TCF on the Cdx1 promoter.
FIG. 7.
Stage-specific attenuation of Cdx1 expression in Cdx1+/− Wnt3avt/vt compound mutants. Cdx1 expression was assessed by whole-mount in situ hybridization in E8.5 (A to D) or E7.5 (E and F) embryos. (A and E) Wild-type control. (B) Cdx1+/−. (C) Wnt3avt/vt. (D and F) Cdx1+/− Wnt3avt/vt. Note the nearly complete loss of Cdx1 expression at E8.5 in Cdx1+/− Wnt3avt/vt embryos relative to that in the cognate mutant backgrounds, while expression is unaffected at E7.5.
Cdx1 interacts with Sox2.
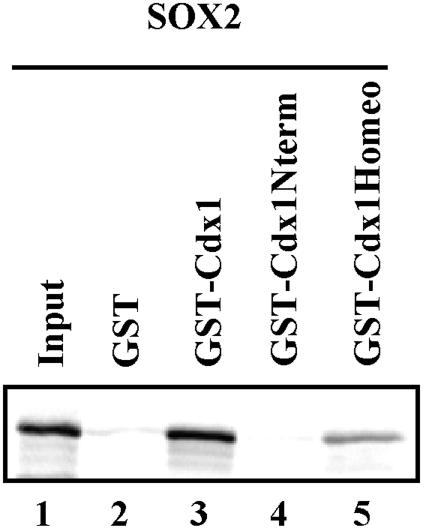
The HMG box is a conserved sequence shared by many transcription factors. We therefore asked whether Cdx1-HMG box interaction could reflect a more general paradigm. Using in vitro interaction assays, we found that Cdx1 and the homeodomain sequences alone could interact with Sox2, a member of the Sry-related HMG box family of transcription factors (12) (Fig. 8). Conversely, Sox2 failed to interact with Cdx1 N-terminal sequences. This association appeared to be specific, since no interaction was observed under identical conditions with Sox9 (data not shown), suggesting that Cdx1 may interact with a subset of HMG box-containing proteins.
FIG. 8.
Cdx1 homeodomain interacts with Sox2 in vitro. 35S-labeled Sox2 was incubated with GST alone or GST-Cdx1 (full length, N terminus, or homeodomain only). Sox2 specifically interacted with both full-length Cdx1 (lane 3) and the Cdx1 homeodomain (lane 5).
DISCUSSION
Our prior work suggested that Cdx1 expression is dependent on some form of autoregulation (49). Since we could not demonstrate direct Cdx binding on Cdx1-responsive promoter sequences, we postulated that the autoregulation either involves an intermediate which is itself a Cdx1 target or is dependent on a cofactor(s) which acts in concert with Cdx1 to regulate Cdx1 expression. In the present study, we present evidence supporting the latter mechanism of action. We found that Cdx1 can be detected on its own promoter in vivo and can synergize with LEF1 in inducing transcription from Cdx1 sequences containing LEF/TCF response elements. We also demonstrated that Cdx1 and LEF1 physically interact. The importance of this interaction in vivo is suggested by the synergistic effect of Cdx1 and Wnt3avt mutant alleles on Cdx1 expression and vertebral patterning. As Cdx1 can also interact with Sox2, these data further suggest that Cdx-HMG interactions may be involved in the transcription of additional target genes.
Cdx1 interacts with LEF1 to mediate Cdx1 expression in presomitic mesoderm.
We and others (16, 49) have been unable to find evidence for direct binding of Cdx proteins to Cdx1 promoter sequences. However, in the present study, chromatin immunoprecipitation studies indicated that Cdx1 was indeed present on its own promoter in a manner similar to RARγ, a direct regulator of Cdx1 expression (28). Moreover, we found synergistic interactions between Cdx1 and LEF1 in transfection assays, suggesting that Cdx1 autoregulation might be mediated via a LEF1-dependent mechanism. Consistent with this, we found that Cdx1 and LEF1 interact physically and mapped the Cdx1-interacting domain to the B box of LEF1.
The B box sequence of all LEF/TCF proteins is highly conserved, suggesting that other members of this family can substitute for LEF1 in Cdx1 association and autoregulation. In this regard, TCF4 can mediate Wnt induction of Cdx1 expression in several cell types (16, 26, 38). However, as regards vertebral patterning, LEF1 and TCF1 are coexpressed with Cdx1 in the posterior embryo, with transcripts for all three genes found with a caudal-high distribution in the posterior embryo (41, 46). In agreement with this, Cdx1 is a Wnt3a target, and it has been shown that LEF1 and TCF1 are critical transducers of the Wnt3a signal in the caudal embryo (29, 30). Analysis of Wnt3avt Cdx1 compound mutants, discussed below, also supports a role for LEF1 in Cdx1-dependent vertebral patterning.
Although the above observations suggest that LEF1 and TCF1 are the principal mediators of Cdx1 expression in the caudal embryo, Cdx1 is coexpressed with these and other LEF/TCF family members in a number of other tissues, including the limb bud, intestinal epithelium, and, to a lesser extent, central nervous system (10, 36, 41, 53). While a role for Cdx1 in these tissues has not been described, perhaps due to functional redundancy with Cdx2 (60), this expression pattern may reflect an autoregulatory loop similar to that proposed for Cdx1 in the caudal embryo.
DNA-binding-independent role for Cdx1?
The Cdx1-LEF1 interaction is not the first example of homeodomain-HMG box interactions. Indeed, the nonhistone chromatin-associated protein HMG1 is able to interact with numerous Hox proteins (64). However, HMG1 null mutants do not exhibit a phenotype related to Hox loss of function (6), suggesting that this may be a spurious or vestigial interaction. A number of Pou homeodomain and Sox-HMG box proteins have also been shown to associate (40, 54), although these interactions are typically accompanied by direct DNA binding of each factor to cognate response elements. An interaction between the paired-like homeodomain protein Alx4 and LEF1 has also been reported (4). In this case, the N-terminal region of Alx4, not its homeodomain, has been shown to interact with the LEF1 HMG box. Moreover, as with Pou-Sox interactions, both Alx4 and LEF1 bind to cognate response elements, with Alx4 DNA binding being enhanced by interaction with LEF1; similar observations have been made with regard to HMG1-Hox-DNA interactions (4, 64).
The above data are indicative of HMG-homeodomain interactions in which both partners directly bind to DNA, and in at least some cases, this binding is modulated by protein-protein interaction. We have been unable to detect direct Cdx1 binding with any Cdx1 promoter sequences. It is conceivable that a cryptic Cdx binding site exists that functions, for example, in the context of chromatin. Indeed, such a chromatin-dependent event could explain our inability to demonstrate the presence of LEF1-Cdx1 complexes on DNA in electrophoretic mobility shift assays (our unpublished observation). In this regard, chromatin has been shown to have profound effects on LEF1 interaction with β-catenin (59).
While we cannot unequivocally discount direct Cdx1-DNA interactions as a mechanism contributing to Cdx1 autoregulation, our data support a model in which Cdx1 exerts effects on transcription through an interaction with LEF1. In addition to genetic analysis of Wnt3avt Cdx1 compound mutants and physical interaction data, this possibility is also supported by the finding that Cdx1 potentiated LEF1-induced expression from the pGL3-OT reporter, a Wnt signaling reporter (25) which does not respond to Cdx1 alone. It is also interesting that LEF1 has been suggested to participate in an autoregulatory loop for the melanocyte-specific microphthalmia-associated transcription factor (MITF-M) (51) in a manner reminiscent of our proposed mechanism for Cdx1 autoregulation.
In the canonical Wnt pathway, LEF1 serves to recruit β-catenin, which plays a pivotal role in transcriptional activation (reviewed in reference 42). In this regard, Alx4-LEF1 functional interaction is β-catenin dependent, whereas MITF-M-LEF1 is not (4, 51). As Cdx1 harbors a transcription activation domain, it is conceivable that its interaction with LEF1 could obviate the necessity for β-catenin for transcriptional regulation. However, the synergistic interaction between the Cdx1 mutant and Wnt3avt alleles suggests that, on the Cdx1 promoter, the Cdx1-LEF1 interaction is β-catenin dependent. Moreover, immunoprecipitation assays revealed that Cdx1 associates with a LEF1-β-catenin complex (our unpublished observation), and therefore Cdx1 binding to LEF1 does not preclude LEF1-β-catenin interaction. Nonetheless, we cannot exclude the possibility that some genes may be regulated by a Cdx1-LEF1 complex in a β-catenin-independent manner.
Cdx-Wnt interactions in vertebral patterning.
Our analysis of Wnt3avt Cdx1 compound mutants supports a role for LEF1-Cdx1 interactions in Cdx1 autoregulation. While the convergent effect of loss of Cdx1 and Wnt3a function on Cdx1 expression could be mediated through two independent processes, our present data suggest that Cdx1-LEF1 interaction, operating through proximal LEF/TCF response elements (38, 49), mediates this interaction. Indeed, the ability of Cdx1 to augment LEF1-dependent transcription was seen on the natural Cdx1 promoter as well as the synthetic Wnt-responsive pGL3-OT reporter, which does not respond to Cdx1 alone.
In Cdx1 Wnt3avt mutants, attenuation of Cdx1 and Wnt signaling would lead to a reduction, below a critical level, in Cdx1-LEF/TCF complexes on the Cdx1 promoter. This hypothetical outcome is clearly reflected by the nearly complete loss of expression of Cdx1 in the Cdx1+/− Wnt3avt/vt compound mutants at E8.5. The residual Cdx1 expression in this background was likely due to the nature of the Wnt3avt allele, which is a weak hypomorph (24). This model is also supported by relative effects of these mutant backgrounds on vertebral patterning. Indeed, the Cdx+/− Wnt3avt/vt alleles exhibited synergistic interaction that was more pronounced from the second cervical vertebra and more caudal, whereas there was little increase in the defects of more-rostral elements. This is consistent with a role for Wnt3a signaling impacting on Cdx1 shortly after its initial onset of expression (and normal patterning of rostral vertebrae) with subsequent failure of autoregulation leading to defects of more posterior vertebrae.
HMG box proteins as Cdx partners: a general paradigm?
Cdx-HMG box interactions might contribute to regulatory interactions in addition to Cdx1 autoregulation. As Cdx homeodomains and LEF/TCF HMG boxes are highly conserved, it is conceivable that other such interactions do indeed occur. In this regard, a Cdx2-Tcf4 interaction might explain the inhibitory effect of Cdx2 on the Cdx1 promoter in colon cancer cells (16). Indeed, this outcome requires the Cdx2 homeodomain and occurs in the absence of demonstrated Cdx2 binding sites on the Cdx1 promoter (16). LEF1 and TCF1 as well as Cdx2 are also involved in events related to posterior specification (8, 21, 60), suggesting an additional pathway(s) in which these transcription factors may interact.
In an attempt to identify other potentially relevant HMG box-Cdx1 interactions, we investigated the ability of Sry HMG box-related protein (Sox) family members to interact with Cdx1 and found that Cdx1 could associate with Sox2 in vitro. In contrast to Sox2, Sox9 did not interact with Cdx1. In this regard, vertebrate Sox1, Sox2, and Sox3 form a subdivision of the Sox family, the B1 group, based on HMG box sequence similarity (12, 52). As our current findings suggest a role for HMG box sequences in association with Cdx1, these data indicate that the Cdx1-Sox2 interaction is likely reliant on specific HMG box sequences. In addition, Cdx members exhibit highly conserved homeodomains, and Cdx1 and -2 have overlapping functions in anterior-posterior patterning (60). It is therefore tempting to speculate that all Cdx members have the potential to interact with members of the B1 group of Sox proteins and that Cdx-HMG box interactions may be involved in a number of ontogenic programs.
In this regard, Sox2 is expressed in a partially overlapping manner with Cdx1 in the neurectoderm and with the chicken Cdx2 homologue CdxA in the developing gut (31, 32, 41, 63). While there is presently no indication that Sox2 has a role in gut development, studies in Xenopus laevis suggest a function for both Sox2 and Xcad3 (the Xenopus Cdx4 homologue) in the neurectoderm (30, 35, 47). Analysis of Cdx1-Cdx2 compound mutant mice and recent studies in the chicken embryo also suggest a role for Cdx in regulating Hox gene expression in the neurectoderm (3, 60). Given that Sox2 is an important regulator of Hoxb1 expression in the hindbrain (15), it is tempting to hypothesize that Cdx-Sox2 interactions may play critical roles in development of the central nervous system.
Acknowledgments
We thank L. Attisano, A. Hecht, D. Rimm, B. Vogelstein, and M. Featherstone for kind gifts of several of the plasmids used in this study and Nathalie Bouchard for technical assistance.
This work was supported by grants to D.L. from the Canadian Institutes for Health Research. N.P. is a recipient of a fellowship from CIHR, and M.H. was supported by a studentship from the Medical Research Council of Canada. D.L. is a Senior Scholar of the Fonds de la Recherches en Santé de Québec.
REFERENCES
- 1.Allan, D., M. Houle, N. Bouchard, B. I. Meyer, P. Gruss, and D. Lohnes. 2001. RAR gamma and Cdx1 interactions in vertebral patterning. Dev. Biol. 240:46-60. [DOI] [PubMed] [Google Scholar]
- 2.Beck, F., T. Erler, A. Russell, and R. James. 1995. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204:219-227. [DOI] [PubMed] [Google Scholar]
- 3.Bel-Vialar, S., N. Itasaki, and R. Krumlauf. 2002. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129:5103-5115. [DOI] [PubMed] [Google Scholar]
- 4.Boras, K., and P. A. Hamel. 2002. Alx4 binding to LEF-1 regulates N-CAM promoter activity. J. Biol. Chem. 277:1120-1127. [DOI] [PubMed] [Google Scholar]
- 5.Burke, A. C., C. E. Nelson, B. A. Morgan, and C. Tabin. 1995. Hox genes and the evolution of vertebrate axial morphology. Development 121:333-346. [DOI] [PubMed] [Google Scholar]
- 6.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, S. Ferrari, and M. E. Bianchi. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276-280. [DOI] [PubMed] [Google Scholar]
- 7.Charité, J., W. de Graaff, D. Consten, M. J. Reijnen, J. Korving, and J. Deschamps. 1998. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development 125:4349-4358. [DOI] [PubMed] [Google Scholar]
- 8.Chawengsaksophak, K., R. James, V. E. Hammond, F. Kontgen, and F. Beck. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386:84-87. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F., J. Greer, and M. R. Capecchi. 1998. Analysis of Hoxa7/Hoxb7 mutants suggests periodicity in the generation of the different sets of vertebrae. Mech. Dev. 77:49-57. [DOI] [PubMed] [Google Scholar]
- 10.Cho, E. A., and G. R. Dressler. 1998. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech. Dev. 77:9-18. [DOI] [PubMed] [Google Scholar]
- 11.Christ, B., and C. P. Ordahl. 1995. Early stages of chick somite development. Anat. Embryol. 191:381-396. [DOI] [PubMed] [Google Scholar]
- 12.Collignon, J., S. Sockanathan, A. Hacker, M. Cohen-Tannoudji, D. Norris, S. Rastan, M. Stevanovic, P. N. Goodfellow, and R. Lovell-Badge. 1996. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development 122:509-520. [DOI] [PubMed] [Google Scholar]
- 13.Condie, B. G., and M. R. Capecchi. 1993. Mice homozygous for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and the axis. Development 119:579-595. [DOI] [PubMed] [Google Scholar]
- 14.Deschamps, J., and M. Wijgerde. 1993. Two phases in the establishment of HOX expression domains. Dev. Biol. 156:473-480. [DOI] [PubMed] [Google Scholar]
- 15.Di Rocco, G., A. Gavalas, H. Popperl, R. Krumlauf, F. Mavilio, and V. Zappavigna. 2001. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 276:20506-20515. [DOI] [PubMed] [Google Scholar]
- 16.Domon-Dell, C., and J. N. Freund. 2002. Stimulation of Cdx1 by oncogenic beta-catenin/Tcf4 in colon cancer cells; opposite effect of the CDX2 homeoprotein. FEBS Lett. 518:83-87. [DOI] [PubMed] [Google Scholar]
- 17.Duboule, D. 1998. Vertebrate hox gene regulation: clustering and/or colinearity? Curr. Opin. Genet. Dev. 8:514-518. [DOI] [PubMed] [Google Scholar]
- 18.Duboule, D., and P. Dollé. 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favier, B., F. M. Rijli, C. Fromental-Ramain, V. Fraulob, P. Chambon, and P. Dollé. 1996. Functional cooperation between the nonparalogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development 122:449-460. [DOI] [PubMed] [Google Scholar]
- 20.Ferrier, D. E. K., and P. W. H. Holland. 2001. Ancient origin of the Hox gene cluster. Nat. Rev. Genet. 2:33-38. [DOI] [PubMed] [Google Scholar]
- 21.Galceran, J., I. Farinas, M. J. Depew, H. Clevers, and R. Grosschedl. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/−Tcf1−/− mice. Genes Dev. 13:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamer, L. W., and C. V. Wright. 1993. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech. Dev. 43:71-81. [DOI] [PubMed] [Google Scholar]
- 23.Gaunt, S. J. 1994. Conservation in the Hox code during morphological evolution. Int. J. Dev. Biol. 38:549-552. [PubMed] [Google Scholar]
- 24.Greco, T. L., S. Takada, M. M. Newhouse, J. A. McMahon, A. P. McMahon, and S. A. Camper. 1996. Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 10:313-324. [DOI] [PubMed] [Google Scholar]
- 25.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 26.Hecht, A., and M. P. Stemmler. 2003. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 278:3776-3785. [DOI] [PubMed] [Google Scholar]
- 27.Horan, G. S., R. Ramirez-Solis, M. S. Featherstone, D. J. Wolgemuth, A. Bradley, and R. R. Behringer. 1995. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 9:1667-1677. [DOI] [PubMed] [Google Scholar]
- 28.Houle, M., P. Prinos, A. Iulianella, N. Bouchard, and D. Lohnes. 2000. Retinoic acid regulation of Cdx1: an indirect mechanism for retinoids and vertebral specification. Mol. Cell. Biol. 20:6579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeya, M., and S. Takada. 2001. Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103:27-33. [DOI] [PubMed] [Google Scholar]
- 30.Isaacs, H. V., M. E. Pownall, and J. M. Slack. 1998. Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 17:3413-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii, Y., K. Fukuda, H. Saiga, S. Matsushita, and S. Yasugi. 1997. Early specification of intestinal epithelium in the chicken embryo: a study on the localization and regulation of CdxA expression. Dev. Growth Differ. 39:643-653. [DOI] [PubMed] [Google Scholar]
- 32.Ishii, Y., M. Rex, P. J. Scotting, and S. Yasugi. 1998. Region-specific expression of chicken Sox2 in the developing gut and lung epithelium: regulation by epithelial-mesenchymal interactions. Dev. Dyn. 213:464-475. [DOI] [PubMed] [Google Scholar]
- 33.Kessel, M., and P. Gruss. 1991. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67:89-104. [DOI] [PubMed] [Google Scholar]
- 34.Kieny, M., A. Mauger, and P. Sengel. 1972. Early regionalization of somitic mesoderm as studied by the development of axial skeleton of the chick embryo. Dev. Biol. 28:142-161. [DOI] [PubMed] [Google Scholar]
- 35.Kishi, M., K. Mizuseki, N. Sasai, H. Yamazaki, K. Shiota, S. Nakanishi, and Y. Sasai. 2000. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development 127:791-800. [DOI] [PubMed] [Google Scholar]
- 36.Korinek, V., N. Barker, K. Willert, M. Molenaar, J. Roose, G. Wagenaar, M. Markman, W. Lamers, O. Destree, and H. Clevers. 1998. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 18:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecuyer, E., S. Herblot, M. Saint-Denis, R. Martin, C. G. Begley, C. Porcher, S. H. Orkin, and T. Hoang. 2002. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood 100:2430-2440. [DOI] [PubMed] [Google Scholar]
- 38.Lickert, H., C. Domon, G. Huls, C. Wehrle, I. Duluc, H. Clevers, B. I. Meyer, J. N. Freund, and R. Kemler. 2000. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development 127:3805-3813. [DOI] [PubMed] [Google Scholar]
- 39.Lickert, H., and R. Kemler. 2002. Functional analysis of cis-regulatory elements controlling initiation and maintenance of early Cdx1 gene expression in the mouse. Dev. Dyn. 225:216-220. [DOI] [PubMed] [Google Scholar]
- 40.Ma, Y., K. Certel, Y. Gao, E. Niemitz, J. Mosher, A. Mukherjee, M. Mutsuddi, N. Huseinovic, S. T. Crews, W. A. Johnson, and J. R. Nambu. 2000. Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J. Neurosci. 20:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer, B. I., and P. Gruss. 1993. Mouse Cdx-1 expression during gastrulation. Development 117:191-203. [DOI] [PubMed] [Google Scholar]
- 42.Moon, R. T., B. Bowerman, M. Boutros, and N. Perrimon. 2002. The promise and perils of Wnt signaling through beta-catenin. Science 296:1644-1646. [DOI] [PubMed] [Google Scholar]
- 43.Nordeen, S. K. 1988. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6:454-458. [PubMed] [Google Scholar]
- 44.Nowicki, J. L., and A. C. Burke. 2000. Hox genes and morphological identity: axial versus lateral patterning in the vertebrate mesoderm. Development 127:4265-4275. [DOI] [PubMed] [Google Scholar]
- 45.Oosterveen, T., K. Niederreither, P. Dolle, P. Chambon, F. Meijlink, and J. Deschamps. 2003. Retinoids regulate the anterior expression boundaries of 5′ Hoxb genes in posterior hindbrain. EMBO J. 22:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oosterwegel, M., M. van De Wetering, J. Timmerman, A. Kruisbeek, O. Destree, F. Meijlink, and H. Clevers. 1993. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development 118:439-448. [DOI] [PubMed] [Google Scholar]
- 47.Pownall, M. E., A. S. Tucker, J. M. Slack, and H. V. Isaacs. 1996. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development 122:3881-3892. [DOI] [PubMed] [Google Scholar]
- 48.Prieve, M. G., K. L. Guttridge, J. E. Munguia, and M. L. Waterman. 1996. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J. Biol. Chem. 271:7654-7658. [DOI] [PubMed] [Google Scholar]
- 49.Prinos, P., S. Joseph, K. Oh, B. I. Meyer, P. Gruss, and D. Lohnes. 2001. Multiple pathways governing Cdx1 expression during murine development. Dev. Biol. 239:257-269. [DOI] [PubMed] [Google Scholar]
- 50.Roelen, B. A., W. de Graaff, S. Forlani, and J. Deschamps. 2002. Hox cluster polarity in early transcriptional availability: a high order regulatory level of clustered Hox genes in the mouse. Mech. Dev. 119:81. [DOI] [PubMed] [Google Scholar]
- 51.Saito, H., K. Yasumoto, K. Takeda, K. Takahashi, A. Fukuzaki, S. Orikasa, and S. Shibahara. 2002. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J. Biol. Chem. 277:28787-28794. [DOI] [PubMed] [Google Scholar]
- 52.Schepers, G. E., R. D. Teasdale, and P. Koopman. 2002. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell. 3:167-170. [DOI] [PubMed] [Google Scholar]
- 53.Silberg, D. G., G. P. Swain, E. R. Suh, and P. G. Traber. 2000. Cdx1 and cdx2 expression during intestinal development. Gastroenterology 119:961-971. [DOI] [PubMed] [Google Scholar]
- 54.Soriano, N. S., and S. Russell. 1998. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development 125:3989-3996. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian, V., B. I. Meyer, and P. Gruss. 1995. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83:641-653. [DOI] [PubMed] [Google Scholar]
- 56.Takada, S., K. L. Stark, M. J. Shea, G. Vassileva, J. A. McMahon, and A. P. McMahon. 1994. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8:174-189. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, J. K., T. Levy, E. R. Suh, and P. G. Traber. 1997. Activation of enhancer elements by the homeobox gene Cdx2 is cell line specific. Nucleic Acids Res. 25:2293-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trinh, K. Y., T. Jin, and D. J. Drucker. 1999. Identification of domains mediating transcriptional activation and cytoplasmic export in the caudal homeobox protein Cdx-3. J. Biol. Chem. 274:6011-6019. [DOI] [PubMed] [Google Scholar]
- 59.Tutter, A. V., C. J. Fryer, and K. A. Jones. 2001. Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro. Genes Dev. 15:3342-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Akker, E., S. Forlani, K. Chawengsaksophak, W. de Graaff, F. Beck, B. I. Meyer, and J. Deschamps. 2002. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129:2181-2193. [DOI] [PubMed] [Google Scholar]
- 61.Vleminckx, K., R. Kemler, and A. Hecht. 1999. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech. Dev. 81:65-74. [DOI] [PubMed] [Google Scholar]
- 62.Wodarz, A., and R. Nusse. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14:59-88. [DOI] [PubMed] [Google Scholar]
- 63.Wood, H. B., and V. Episkopou. 1999. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pregastrulation to early somite stages. Mech. Dev. 86:197-201. [DOI] [PubMed] [Google Scholar]
- 64.Zappavigna, V., L. Falciola, M. Helmer-Citterich, F. Mavilio, and M. E. Bianchi. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981-4991. [PMC free article] [PubMed] [Google Scholar]