Abstract
Ku86 plays a key role in nonhomologous end joining in organisms as evolutionarily disparate as bacteria and humans. In eukaryotic cells, Ku86 has also been implicated in the regulation of telomere length although the effect of Ku86 mutations varies considerably between species. Indeed, telomeres either shorten significantly, shorten slightly, remain unchanged, or lengthen significantly in budding yeast, fission yeast, chicken cells, or plants, respectively, that are null for Ku86 expression. Thus, it has been unclear which model system is most relevant for humans. We demonstrate here that the functional inactivation of even a single allele of Ku86 in human somatic cells results in profound telomere loss, which is accompanied by an increase in chromosomal fusions, translocations, and genomic instability. Together, these experiments demonstrate that Ku86, separate from its role in nonhomologous end joining, performs the additional function in human somatic cells of suppressing genomic instability through the regulation of telomere length.
Most human tumors display some sort of chromosomal instability, ranging from minor DNA sequence changes to GCRs (gross chromosomal rearrangements) and aneuploidy (reviewed in references 53 and 68). These genomic alterations are often the causative event(s) in the transformation of a normal cell into a neoplastic cell. This occurs when the alteration results in the activation of proto-oncogenes or the inactivation of tumor suppressor genes and/or by the acquisition of a mutator phenotype (reviewed in reference 51). There are at least three proposed pathways by which chromosomal rearrangements may originate: (i) checkpoint defects (reviewed in reference 46), (ii) stalled replication fork collapse (reviewed in reference 15), and (iii) telomere dysfunction (reviewed in reference 54). Studies with yeast have demonstrated that a deficiency in any of these three pathways enhances chromosome loss and GCRs by up to 2 to 3 orders of magnitude (reviewed in reference 46). Similarly, in higher eukaryotes, mutations in genes regulating checkpoints and the repair of stalled replication forks result in a highly elevated frequency of GCRs (reviewed in references 38 and 78). Finally, in mice and humans, evidence is accumulating that the dysfunction of telomeres may be the driving force in the generation of genomic instability, which is strongly linked to cancer predisposition (reviewed in reference 54).
Telomeres are the terminal structures of linear chromosomes. Telomeres appear to perform at least two functions: (i) they allow for the replication of the ends of chromosomes, and (ii) they stabilize chromosomes by keeping them from recombining with one another (reviewed in references 17 and 24). Telomeric DNA consists of a repetitive motif with the general form TxAyGz, which in mammals is T2A1G3. At the ends of the chromosomes, the G-rich strand is often extended over the C-rich strand for a variable number of nucleotides (reviewed in reference 7). Many of the genes involved in telomere biogenesis and stability have been identified, and their subsequent characterization has led to the identification of even more genes, leaving the field with a rich, yet complicated, picture. In yeast, for example, mutation of any of more than 25 different genes can deleteriously affect telomere length and/or structure (17). The mammalian counterparts of some of these genes have been identified, and these include the ribonucleoprotein complex consisting of TERT (telomerase reverse transcriptase) (35) and TR (telomerase RNA) (8), which is responsible for the synthesis of the T2A1G3 repeat; TRF1 and TRF2 (telomere recognition factors 1 and 2, respectively) (12, 18), which bind to the double-strand portion of the T2A1G3 repeat; and Pot1 (protection of telomeres 1) (3), which binds to the single-stranded, G-rich strand overhang. In addition, a variety of DNA repair proteins are also associated, directly or indirectly, with telomeres.
The DNA-dependent protein kinase catalytic subunit DNA-PKcs, together with the heterodimeric Ku protein (Ku86 and Ku70), comprises a complex, DNA-PK, that is critically involved in DNA DSB (double-strand break) repair and V(D)J recombination in mammalian cells (reviewed in references 41 and 50). Animals mutant at the DNA-PKcs locus are IRs (ionizing radiation sensitive), defective in DNA DSB repair, and immunodeficient. Moreover, DNA-PKcs appears to regulate telomere length (28, 36, 69) and prevent GCRs (2, 31, 32). Ku is a heterodimeric protein of 70- and 86-kDa subunits that is conserved from prokaryotes (21, 82) to humans. Ku binds in a sequence-nonspecific fashion to all double-stranded DNA ends, including 5′ and 3′ overhangs, blunt ends, duplex DNA ending in stem-loop structures, and telomeres (reviewed in references 39 and 75). Mice containing targeted disruptions of Ku86 (59, 84), Ku70 (49, 61), or DNA-PKcs (10, 30, 43, 73) are IRs and defective for DNA DSB repair and have the anticipated immune defects. In addition, inactivation of the murine Ku86 gene results in cells with severe growth retardation (60), premature senescence (79), and a marked increase in chromosomal aberrations (20, 25, 44) that show elevated telomeric fusions (2, 67). Thus, DNA-PK is an important mammalian DNA repair complex, and mutations in Ku or DNA-PKcs result in DNA DSB repair defects and immunodeficiencies.
Despite the relative uniformity of phenotypes of eukaryotic cell lines with defects in DNA-PK concerning repair and recombination, there are some glaring differences concerning telomere biology. In Saccharomyces cerevisiae, Ku86 and Ku70 mutant strains exhibit temperature-sensitive lethality and aberrant telomere shortening (11, 33, 63) or telomeric repeat maintenance (26, 29, 34, 74). Ku-null Schizosaccharomyces pombe (4, 52) and trypanosome (19) cells also show telomere shortening but no exacerbated telomere degradation and lethality. In contrast to these organisms, the majority of Ku-null chicken DT40 cells exhibit telomeres of the parental length although some telomeric expansions have been observed in independent subclones (81). In contrast to the sporadic telomeric expansions seen in the chicken—and in sharp contrast to what has been observed with yeast and trypanosomes—Ku-null Arabidopsis thaliana plants show consistent, massive telomeric expansions (65). In rodents, multiple contradictory studies have reported telomeric shortening (20), telomeric expansions—some slight (27, 67), some large (36, 69)—and/or no discernible effects (28, 31, 32) in Ku- or DNA-PKcs-null cell lines and animals. The fact that all three of the DNA-PK mutant knockout mouse lines are fertile and viable, however, suggests that if there are telomeric defects, they cannot be as severe as those caused by the loss of telomerase activity, which results in senescent and infertile animals by the sixth generation (9). Given the glaring lack of uniformity between—and even within—the various model systems, it has been unclear which model system is most applicable to humans. Moreover, because DNA-PK activity in humans appears to be essential (48), the direct effect of DNA-PK mutations on telomere function has not been reported.
In the present report, we demonstrate that the inactivation of a single allele of Ku86 in human somatic cells results in dramatic telomere shortening. This telomeric shortening is accompanied by chromosomal fusions, aneuploidy, and GCRs. Thus, human Ku86 appears to be a critical suppressor of genomic instability.
MATERIALS AND METHODS
Tissue culture.
HCT116 and all of the genetically modified cell lines derived from it were cultured in McCoy's 5A medium with 10% fetal calf serum (48). For construction of complemented subclones #A6 and #A10, 25 μg of a BglI-linearized pcDNA3.1 expression plasmid (Invitrogen) containing the full-length human Ku86 gene was electroporated (240V, 975 μF) into 107 clone #70-32 cells. These cells were then plated onto two 10-cm-diameter dishes, and after 24 h, the cells were placed under selection (1 mg of G418 per ml). Ten to 14 days later, individual G418-resistant clones were isolated by toothpicking and expanded. Clones containing elevated levels of Ku86 protein were then identified by immunoblotting.
Immunoblotting.
Levels of Ku86 protein in the various cell lines were determined by Western blotting, which was performed exactly as previously described (48). Ku86 and β-actin were detected with antibodies SC-5280 (Santa Cruz) and A5441 (Sigma), respectively.
Telomeric TRF and G-strand overhang analyses.
TRF (terminal restriction fragment) and G-strand overhang assays were performed exactly as previously described (45, 77), with the restriction enzymes MboI and AluI (New England Biolabs). Oligonucleotides used for probes were obtained from Operon/Qiagen. Exonuclease I (ExoI) was purchased from Amersham.
FISH and SKY analyses.
Fluorescence in situ hybridization (FISH) analysis for the presence of telomeres was performed with metaphase-arrested cells with a protein-nucleic acid telomere-specific probe [Cy3 conjugated to (T2AG3)3] in accordance with a protocol provided by the manufacturer (DAKO) (66). Individual rearrangements in mammalian cells were analyzed by spectral karyotyping (SKY) analysis as previously described (14).
RESULTS
Functional inactivation of a single allele of Ku86 in human somatic cells results in profound telomeric shortening.
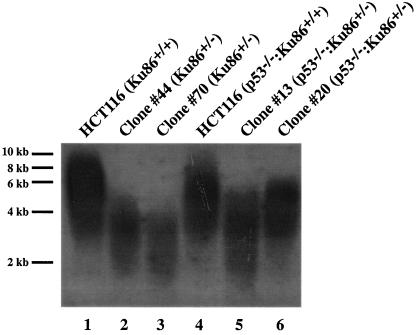
We have recently demonstrated that the functional inactivation by gene targeting of both Ku86 alleles in a diploid human somatic cancer cell line is not compatible with viability (48). Interestingly, Ku86-null cells did not die immediately but underwent 8 to 10 cell divisions before they succumbed to apoptosis. This phenotype was identical to that caused by overexpression of a dominant negative telomerase in human cancer cells (83). Moreover, two of the most distinctive phenotypes of human Ku86-heterozygous cell lines were a decrease in proliferation and a significant elevation in the spontaneous levels of the DNA damage-regulated transcription factor p53 (48). These two phenotypes have also been observed in mice in which both alleles of the murine telomeric RNA gene, TERC, have been inactivated (47). Together, these similarities suggested that at least some of the deleterious effects associated with the reduction of Ku86 expression in human somatic cells could be due to telomeric dysfunction. To experimentally test this hypothesis, we used the well-established TRF assay (57) to directly measure the telomere lengths in the telomerase-positive parental HCT116 colorectal carcinoma cell line (16), as well as in two independent Ku86-heterozygous clones, #44 and #70 (48). This assay takes advantage of the fact that the telomeric repeat region, in contrast to the adjacent unique genomic DNA, is devoid of the recognition sequences for almost all restriction enzymes. Thus, human genomic DNA that has been digested to completion with the very frequent-cutting restriction enzymes AluI and MboI will yield intact telomeric TRFs, which can be detected with a radioactive d(C3TA2)3 probe in a Southern blotting procedure. TRFs from wild-type cells were significantly longer than those observed in heterozygous cells (Fig. 1, compare lane 1 with lanes 2 and 3, respectively). The range for the TRFs from the parental line was 3 to 9 kb, with an average size of 5.6 kb (±0.8 kb, n = 11). In contrast, the range for the TRFs from Ku86-heterozygous cells was only 2 to 5 kb, with an average length of 3.1 kb (±0.1 kb, n = 5) and 2.9 kb (±0.6 kb, n = 17) for clones #44 and #70, respectively. We have constructed two additional independent Ku86-heterozygous cell lines (G. Li, G. Ghosh, and E. A. Hendrickson, unpublished data), clones #13 and #20, in an isogenic HCT116 strain that is null for p53 (13, 14). The p53-null HCT116 cells had somewhat shorter telomeres (range of 3 to 8 kb, with an average size of 5.0 kb [±0.7 kb, n = 7]) than the parental cells from which they were derived (Fig. 1, compare lane 4 with lane 1), but upon inactivation of one allele of Ku86, the telomere size was reduced even more (range of 2 to 6 kb, with average sizes of 3.8 kb [±0.5 kb; n = 6] and 4.3 kb [±0.1; n = 3] for clones #13 and #20, respectively) (Fig. 1, compare lanes 5 and 6 with lane 4). The observation that four independent Ku86-heterozygous cell lines displayed the same short-telomere phenotype strongly suggests that the phenotype was due to the reduction in Ku86 expression and was not an artifact of clonal cell line variability.
FIG. 1.
Human Ku86-deficient cell lines have shortened telomeres. Genomic DNA was purified from the indicated cell lines, digested to completion with MboI and AluI, and then subjected to terminal restriction fragment Southern blot analysis under denaturing conditions with a (C3TA2)3 5′-end-radiolabeled oligonucleotide probe. Lanes: 1, HCT116 parental cell line; 2 and 3, HCT116 Ku86-heterozygous cell lines #44 and #70, respectively; 4, HCT116 p53-null cell line; 5 and 6, HCT116 p53-null Ku86-heterozygous cell lines #13 and #20, respectively. Approximate molecular size markers are shown on the far left.
Partial functional complementation of telomere shortening by reintroduction of a Ku86 cDNA.
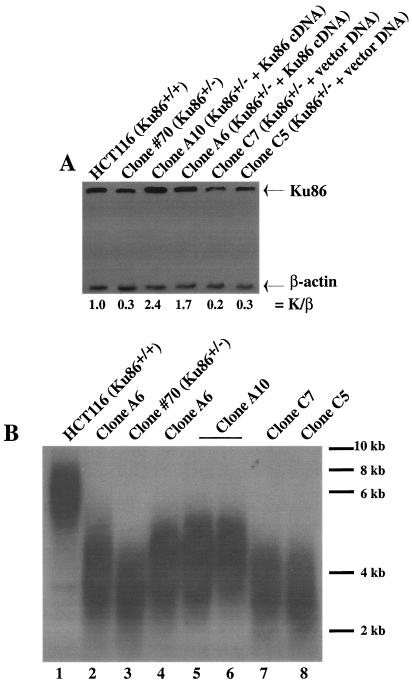
To directly test the hypothesis that a reduction in Ku86 expression was responsible for the short-telomere phenotype of these cell lines, a full-length, wild-type human Ku86 cDNA expressed from a pcDNA3.1(Neo) vector was introduced into the heterozygous clone #70 cell line. Stable subclones were selected for G418 resistance and expanded, and then cell lines with restored levels of Ku86 protein were identified by Western blot analysis. Two subclones (A6 and A10) had levels of Ku86 that were 1.7 and 2.4 times, respectively, more abundant than that of the wild type (Fig. 2A). In contrast, two clones, C7 and C5, that had only been transfected with vector DNA did not show any restoration of Ku86 expression (Fig. 2A). After ∼30 generations of cell growth, these subclones were subjected to a TRF analysis, which demonstrated that the telomeres of clones A6 and 10 (Fig. 2B, lanes 2 and 5, respectively) were partially complemented and migrated at a size intermediate between those of wild-type HCT116 (Fig. 2B, lane 1) and the heterozygous clone #70 cell line (Fig. 2B, lane 3). In contrast, there was no complementation of the telomere length in clones C7 and C5, which contained only the empty vector (Fig. 2B, lanes 7 and 8, respectively). When all of these cell lines were grown for an additional ∼30 generations, another TRF analysis was carried out. The complemented clones did not show any additional lengthening of their telomeres (Fig. 2B, lanes 4 and 6). Thus, the partially complemented phenotype was stable. These data strongly reinforced the contention that a reduction in Ku86 expression in the heterozygous cell lines was directly responsible for their short-telomere phenotype.
FIG. 2.
Introduction of a human Ku86 cDNA into Ku86-heterozygous cells results in partial restoration of telomere length. (A) Whole-cell extracts were prepared from the indicated cell lines and subjected to immunoblot analysis with commercial monoclonal antibodies directed against either Ku86 or β-actin. The signals on the autoradiogram were quantitated by densitometry, and the ratio of the level of Ku86 to β-actin (K/β) expression is shown below each lane. (B) Genomic DNA was purified from the indicated cell lines and subjected to TRF analysis by denaturing gel electrophoresis. Lanes: 1, HCT116 parental cell line; 2 and 4, clone A6 cells harvested after 30 or 60 population doublings, respectively; 3, HCT116 Ku86-heterozygous cell line #70; 5 and 6, clone A10 cells harvested after 30 or 60 population doublings, respectively; 7 and 8, clone C7 and C5 cell lines (HCT116 Ku86+/− cell lines complemented with an empty vector), respectively. Approximate molecular size markers are shown on the far right.
The G-strand overhang is longer in human Ku86-heterozygous cells.
Mutations in Ku affect the length of the G-strand overhang in yeast (6, 33) and plants (64). To assess the length of the G-strand overhang in human Ku86-heterozygous cells, we used a TRF analysis combined with nondenaturing gel electrophoresis (57). In a native (nondenaturing) gel, the single-stranded G-strand overhang is the only substrate capable of hybridizing to the probe (Fig. 3A, −ExoI lanes). This interpretation was confirmed by prior treatment of the samples with ExoI (57), a single-stranded exonuclease, which removed the overhangs and completely eliminated all of the hybridization signal (Fig. 3A, +ExoI lanes). The total amount of telomeric DNA in each lane was quantitated by subsequently denaturing the gel and rehybridizing it with the probe (Fig. 3B). The ratio of the signal obtained from the native gel (i.e., a function of the length of the G-strand overhang only) to that obtained from the denatured gel (i.e., a function of the total telomere length) was arbitrarily set to 1.0 for the HCT116 cell line. Since the overall telomere length in the heterozygous cells is shorter than that of wild-type cells, their G-strand overhang-to-total ratio was normalized to account for this difference. Even with this normalization, however, the G-strand overhang ratio for the four Ku86-heterozygous clones, which ranged from an average of 1.49 to 2.95, was always greater than that for the wild type (Table 1). When a similar analysis was carried out with Ku86 cDNA-complemented clones #A6 and #A10, their G-strand overhang ratios were slightly lower than that of the wild type (Table 1). Together, these experiments suggest that a deficiency in Ku86, while causing overall telomere shortening, results in more telomeres with G-strand overhangs and/or telomeres with slightly longer G-strand overhangs. Moreover, the overexpression of a Ku86 cDNA in Ku86-heterozygous cells completely complemented this defect.
FIG. 3.
The G-strand overhang is elongated in Ku86-heterozygous cells. (A) Genomic DNA was purified from the indicated cell lines and either left untreated (−) or digested overnight (+) with ExoI. The DNA was subsequently purified, digested to completion with MboI and AluI, and then subjected to terminal restriction fragment Southern blot analysis under nondenaturing (Native) conditions with a (C3TA2)3 5′-end-radiolabeled oligonucleotide probe. (B) The gels shown in panel A were denatured and rehybridized with the identical probe (Denatured). Exp., experiment.
TABLE 1.
The G-strand overhang is elongated in Ku86+/− cellsa
| Cell line | G overhang ratio | SDb | nc |
|---|---|---|---|
| HCT116 | 1.00 | NAd | 8 |
| Clone #44 | 1.49 | NA | 1 |
| Clone #70-32 | 1.51 | ±0.30 | 4 |
| Clone #13 | 1.93 | ±0.34 | 2 |
| Clone #20 | 2.95 | NA | 1 |
| Clone #A6 | 0.78 | ±0.36 | 3 |
| Clone #A10 | 0.90 | ±0.31 | 3 |
Gels such as those shown in Fig. 3 were quantitated with a phosphoimager, and the ratio of the signal from the native gel in comparison to that from the denatured gel was arbitrarily set to 1.00 for the parental HCT116 cell line. Clones #44, #70-32, #13, and #20 are all Ku86+/− cell lines, and clones #A6 and #A10 are Ku86+/− cell lines complemented with a Ku86 cDNA (see the text for a fuller description). All cell lines were normalized relative to HCT116 for total telomere length.
SD, standard deviation.
n, number of independent experimental data points.
NA, not applicable.
Confirmation of telomere dysfunction in Ku86-heterozygous cells by FISH analyses.
To independently confirm the telomere defect observed by TRF analysis, parental HCT116 cells and the two independent p53-positive, Ku86-heterozygous cell lines were arrested in metaphase, hybridized with a protein-nucleic acid telomere probe, and analyzed by FISH. All of the 1,840 telomeres examined in the parental HCT116 cells hybridized to the probe (Fig. 4A and Table 2). In striking contrast, 61 of 4,232 telomeres examined in Ku86-heterozygous cells were completely devoid of any telomeric signal (Fig. 4B and Table 1). Thus, 1.4% of the telomeres in Ku86-heterozygous cells were either entirely missing or so short that they could no longer be detected by this probe. Moreover, the FISH analysis uncovered additional defects in the Ku86-heterozygous cells. In 6 of the 23 metaphases examined, two examples of telomere fusions (Fig. 4C) and four examples of ring chromosomes that completely lacked telomeric signals (Fig. 4D) were observed. In contrast, no such abnormalities were detected in the parental HCT116 cell line (Fig. 4A and Table 2). Thus, Ku86-heterozygous cells had shortened telomeres and this appeared to promote chromosomal instability.
FIG. 4.
Human Ku86-heterozygous cells contain chromosomes with telomere abnormalities. FISH analysis with a telomere-specific Cy3-(C3TA2)3 protein-nucleic acid probe of either wild-type cells (A) or three independent Ku86-heterozygous cells (B to D). Telomeres are seen as red dots, and the metaphase chromosomes are stained blue. In panel A, every chromosome contains four discrete spots of hybridization (two at each end). In panel B, the arrows point to chromosomes where no discernible hybridization was detected. In panel C, an example of two chromosomes that have fused is shown and the position of the internal telomere signal is designated by the arrow. In panel D, an example of a ring chromosome lacking any telomeric DNA is shown by the arrow.
TABLE 2.
Telomere abnormalities in Ku86 heterozygous cells
| Type of telomere abnormality | HCT116 | HCT116 Ku86+/− |
|---|---|---|
| Telomere loss | 0/1,840 | 61/4,232 |
| Chromosome fusion | 0/10 | 6/23a |
Confirmation of genomic instability in Ku86-heterozygous cells by SKY analyses.
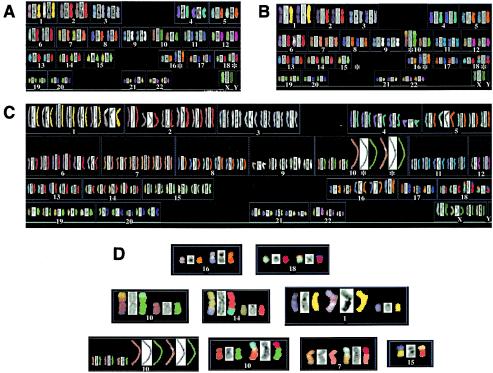
To independently confirm the genomic instability suggested by the FISH analyses, we next performed SKY analyses (5). The HCT116 cell line has previously been analyzed by SKY by two independent laboratories, and three identical karyotypic anomalies have been detected: (i) a translocation involving chromosome 16 [der(16)t(8;16)], (ii) a translocation involving chromosome 18 [der(18)t(17;18)], and (iii) a variable number of cells retaining the Y chromosome (1, 56). Similarly, we detected precisely the same three karyotypic abnormalities (Fig. 5A). Importantly, in 17 metaphases examined, no (0 of 17, 0%) additional chromosomal aberrations were detected (Table 3). Thus, the HCT116 cell line appears to be karyotypically very stable. In stark contrast, we detected seven additional gross chromosomal abnormalities in 28 metaphases (7 of 28, 25%) from the HCT116 Ku86-heterozygous cell lines (Table 3). These included various instances of aneuploidy (Fig. 5C), translocations involving whole chromosomes (Fig. 5B) or parts of chromosomes (Fig. 5D), fragmented chromosomes (Fig. 5D), and amplifications (Fig. 5D and Table 3). These data confirmed the contention that the loss of a single Ku86 allele in human cells results in a highly elevated frequency of GCRs.
FIG. 5.
Human Ku86-heterozygous cells contain chromosomal abnormalities. SKY analysis of either wild-type cells (A) or two independent Ku86-heterozygous cell lines (B and C). Each chromosome is represented in sets of three corresponding to the actual image, the 4′,6′-diamidino-2-phenylindole (DAPI)-stained image, and the computer-generated, false-color image, respectively. In panel A, the two derivative chromosomes (16 and 18) common to the parental cell line are marked with an asterisk. Panel B shows an example of a Ku86-heterozygous cell that contains—in addition to the parental translocations—a translocation of chromosome 15 to chromosome 10, each of which is also marked with an asterisk. Panel C shows an example of a Ku86-heterozygous cell that has become aneuploid and has sustained an amplification of chromosome 10. Panel D shows individual examples of the parental translocations, as well as some of the abnormalities detected in Ku86-heterozygous cells (see also Table 3).
TABLE 3.
Genomic instability of Ku86 heterozygous cells
| Incidence of GCRs | HCT116a | HCT116 Ku86+/−b |
|---|---|---|
| All cells | der(16)t(8;16) | der(16)t(8;16) |
| der(18)t(17;18) | der(18)t(17;18) | |
| Only Ku86+/− cells | der(10)t(10;15) | |
| der(14)t(14;14;4) | ||
| del(1) | ||
| dup(10) | ||
| der(10)t(10;18) | ||
| der(7)t(7;18) | ||
| der(15)t(15;21) | ||
| 2 cases of polyploidy |
The incidence of GCRs was 0 of 17 (0%).
The incidence of GCRs was 7 of 28 (25%).
DISCUSSION
Telomere biology appears to vary radically between different species.
Modern biology is replete with examples of genes and processes that are highly conserved throughout evolution. Indeed, the very existence of telomeres appears to be nature's consistent answer to the problem of chromosome end maintenance. Yet, despite the near universal presence of telomeres in eukaryotic organisms, it is a clear understatement to suggest that the way that different species maintain telomeres is idiosyncratic (reviewed in reference 23). The confusing diversity of mechanisms used by various organisms is perhaps best exemplified by mutations in Ku, which, while being involved in telomere maintenance in most of the species examined, can give rise to dramatically shortened telomeres (in yeast) (11, 33, 63), unaltered telomeres (in chicken cells) (81), or massively expanded telomeres (in plants) (65) when mutated. It is particularly noteworthy that there appear to be significant differences in telomere biology between humans and mice. (i) Human telomeres are shorter, whereas murine telomeres are substantially longer. (ii) Human somatic cells generally do not express telomerase, whereas mouse somatic cells often do. (iii) Humans can regulate telomere damage via two redundant pathways (p53 and p16/Rb), whereas mice regulate telomere damage only through p53 (reviewed in references 17 and 24). The best model for studying telomere structure and damage regulation in human cells was via the overexpression of a dominant-negative form of TRF2 that resulted in telomere uncapping, telomeric fusions, and cell death (77). Human model systems using loss-of-function approaches have not been described.
Haploinsufficiency of Ku86 results in short telomeres.
Here we have demonstrated that the functional inactivation via gene targeting of a single allele of human Ku86 results in profound telomere shortening. One possibility is that Ku might regulate TRF1 and/or TRF2. Ku is known to physically interact with TRF1 (40) and TRF2 (71). Moreover, overexpression of TRF1 (76) or TRF2 (70) results in telomeric shortening in human cells. Thus, if Ku negatively regulates TRF1 or TRF2, then a Ku86-heterozygous cell could be functionally equivalent to a TRF1- or TRF2-overexpressing cell. This hypothesis, while attractive, would not explain the increase in the frequency or length of the G-strand overhangs (Fig. 3 and Table 1). Thus, we favor a model in which Ku acts as a physical barrier to nucleases, probably by binding to the double-stranded-to-single-stranded transition—a structure to which Ku is known to tightly bind (75)—present at the telomeric end. The absence of Ku at this transition could generally reduce protection of the telomere from nucleases and could also account for the increase in the frequency or length of the G-strand overhangs (Fig. 3) by allowing preferential resection of the C-rich strand (42). Restoration of the G-strand overhang to the wild-type length upon reintroduction of a Ku86 cDNA (Table 1) strongly supports this hypothesis. In Ku86-null yeast strains, the elongation of the G-strand overhang can be upwards of an order of magnitude greater (6, 33, 72). In Ku86-heterozygous HCT116 cells, the effect we observed, 1.5- to 3-fold, was much smaller. While this difference may be simply due to the comparison between null and heterozygous cell lines, respectively, it is also possible that human cells possess alternative or additional end protection activities.
Complementation experiments suggest the existence of different mechanisms of Ku86-dependent telomere maintenance.
Reintroduction of a Ku86 cDNA into Ku86-heterozygous cells resulted in significant, albeit partial, complementation of the overall telomere length (Fig. 2B). If Ku86 only functioned in end protection as postulated above, there was no a priori reason to expect that the telomeres should be re-extended at all by reintroduction of Ku86. The fact that the telomeres did elongate suggests that Ku86 may also participate in telomere elongation. One possibility is that Ku86 is required for telomerase activity and that the short-telomere phenotype is an indirect effect of reduced telomerase activity. This is consistent with a report that Ku physically interacts with telomerase and that telomerase activity is slightly reduced in Ku86-heterozygous cells (16). A second possibility is that Ku86 is required for hTR biogenesis. Biochemical and genetic data demonstrate that—in yeast—Ku86 can bind to the RNA component of telomerase (6, 62, 72). If human Ku86 possesses a similar hTR binding activity, it is likely that telomere length maintenance would be aberrantly effected in Ku86-heterozygous cell lines. A third possibility is that Ku86 may be required for recruiting telomerase to the telomere. In its role as a DNA DSB repair protein, Ku normally binds to a broken double-stranded end and recruits DNA-PKcs (50). At a telomere, Ku may perform an analogous role by recruiting telomerase. Whichever, if any, of these three hypotheses is correct, it is clear that Ku86 is not essential for these functions since only partial complementation of the short-telomere phenotype was observed upon restoration of Ku86 expression (Fig. 2B). Last, it should be emphasized that in human cells, Ku86 is often associated with the DNA-PK complex. Thus, the short-telomere phenotype could also be due to the reduced DNA-PK activity in these cells (48). A model consistent with all of our results is that the Ku heterodimer alone normally provides end-blocking activity but that Ku86, as part of the DNA-PK complex, may also be required for telomere elongation. Many aspects of this model can ultimately be addressed by the construction of conditionally null Ku86 and DNA-PKcs cell lines in isogenic backgrounds.
Ku86 protects cells from genomic instability.
The telomeres observed in a Ku86-heterozygous cell, while shortened, appear relatively stable, as some of these cell lines have been grown in continuous culture for more than a year. One possibility is that the residual levels of Ku are sufficient to keep the telomeres in this short-but-stable configuration. This would be consistent with the observations that further reductions in Ku86 levels by gene targeting (48) or RNA interference (data not shown; I. Jaco, P. Munoz, and M. A. Blasco, personal communication) result in cell death. Alternatively, although Ku may be important for proper telomere length maintenance, a Ku-independent mechanism may exist to protect or maintain short telomeres as a last defense against genomic instability. Presumably, when this final barrier, perhaps mediated through TRF2 (45), is overcome, the cells begin a GCR process that is either lethal or oncogenic or leads to senescence. This model is consistent with observations in the mouse that suggest that it is not the average telomere length that regulates genomic stability but the frequency of chromosomes containing very short or no telomeres (27, 37). Similarly, while our TRF analysis (Fig. 1) showed that the average telomere length of all chromosomes in a Ku86-heterozygous cell is reduced, the FISH experiments (Fig. 4) demonstrated that only 1.4% of the chromosomes contained no detectable telomere sequences (Table 2). Indeed, this value may actually be an overestimation of the number of chromosomes lacking telomeres since some chromosomes with very short, albeit functional, telomeres may have escaped detection. If 1.4 out of 100 chromosomes were completely lacking telomeres and each telomereless chromosome led to a detectable GCR, then maximally one out of every two cells would be expected to contain a GCR. By FISH (Fig. 4 and Table 2) and SKY (Fig. 5 and Table 3) analyses, we observed that 26 and 25%, respectively, of the human Ku86+/− cells had a GCR, which is in fairly good agreement with the expected frequency. Consistent with this model is the observation that the p53-null Ku86+/+ cell line had, on average, slightly shorter telomeres than the parental control cell line (Fig. 1). However, by FISH analysis, we did not observe any telomere loss (data not shown) and this cell line is not prone to GCRs (data not shown; 14). In summary, Ku86 levels appear to critically regulate telomere shortening, which, in turn, is a key step in the production of the nonfunctional telomeric ends that generate GCRs and cellular catastrophe.
Where are all of the human Ku mutants?
In the preceding decade, mutations of many DNA repair genes have been linked to human pathologies including, predominately, cancer predisposition (38). This is not the case for Ku and DNA-PK. To our knowledge, not a single case study of a Ku- or DNA-PKcs-null or heterozygote patient has ever been reported. The demonstration that the functional inactivation of both alleles of Ku86 in a somatic cell line is lethal provides a partial explanation for this discrepancy (48). If Ku86 is essential in somatic cells, it is likely that human Ku86-null individuals are inviable. Our current demonstration that Ku86-heterozygous cells have profound telomere defects and genomic instability suggests that even haploinsufficiency of Ku86 in humans may be lethal. Alternatively, while the genomic instability of the heterozygous cell lines is significant, their radiosensitivity is rather slight (48). Thus, the target group of radiosensitive, immunodeficient patients among whom Ku and DNA-PK mutant individuals have been sought (see, e.g., reference 22) may, in retrospect, not be the group in which they are most likely to be found. Our data suggest that if Ku and DNA-PKcs patients do exist, (i) they will only be heterozygous or contain hypomorphic alleles, (ii) their chromosomes will have shortened telomeres, and (iii) they will likely present with some clinical feature of genomic instability in a haploinsufficient state. DKC (dyskeratosis congenita) is a rare inherited disorder that encompasses all three of these phenotypes. In particular, while DKC patients usually die an early death from bone marrow failure, they are also afflicted with an increased risk of cancer (reviewed in reference 55). The disease is defined by multiple complementation groups, and two of the relevant genes have been cloned and identified. One is the RNA component of telomerase, hTR (80), while the other is in dsykerin (58). Dyskerin is a nucleolar protein that binds to snoRNAs (small nucleolar RNAs) and appears to be critical for proper formation of the telomerase ribonuclear protein particle. DKC patients with mutations in either hTR or dsykerin have lower levels of hTR, produce lower levels of telomerase activity, and have shortened telomeres (55). Intriguingly, some of the DKC-hTR patients are heterozygotes and transmit the disease in what appears to be an autosomal dominant fashion that is due to haploinsufficiency (80). Last, in yeast, Ku86 has been shown to bind to the telomeric RNA (6, 62, 72). Together, these observations suggest that patients with Ku or DNA-PK mutations may present with the clinical features that are encompassed by DKC. Future research should clarify this issue.
Acknowledgments
We thank Richard Wang and Titia de Lange (The Rockefeller University) for the TRF protocols and Bert Vogelstein (The Johns Hopkins University) for the HCT116 p53-null cell line. Jamie Borton provided some preliminary data for the TRF studies. We are indebted to Judith Berman and Anja-Katrin Bielinsky (University of Minnesota) and David Bodine and Jennifer Puck (National Institutes of Health) for helpful discussions and comments on the manuscript. We are grateful to Maria Blasco (Spanish National Cancer Center) for communicating results prior to publication.
REFERENCES
- 1.Abdel-Rahman, W. M., K. Katsura, W. Rens, P. A. Gorman, D. Sheer, D. Bicknell, W. F. Bodmer, M. J. Arends, A. H. Wyllie, and P. A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA 98:2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, P., and T. R. Cech. 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 11:3265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayani, J. M., and J. A. Squire. 2002. Applications of SKY in cancer cytogenetics. Cancer Investig. 20:373-386. [DOI] [PubMed] [Google Scholar]
- 6.Bertuch, A. A., and V. Lundblad. 2003. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol. Cell. Biol. 23:8202-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 8.Blasco, M. A., W. Funk, B. Villeponteau, and C. W. Greider. 1995. Functional characterization and developmental regulation of mouse telomerase RNA. Science 269:1267-1270. [DOI] [PubMed] [Google Scholar]
- 9.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 10.Bogue, M., C. Jhappan, and D. B. Roth. 1998. Analysis of variable (diversity) joining recombination in DNA-dependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc. Natl. Acad. Sci. USA 95:15559-15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 13.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 14.Bunz, F., C. Fauth, M. R. Speicher, A. Dutriaux, J. M. Sedivy, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2002. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 62:1129-1133. [PubMed] [Google Scholar]
- 15.Carr, A. M. 2002. Checking that replication breakdown is not terminal. Science 297:557-558. [DOI] [PubMed] [Google Scholar]
- 16.Chai, W., L. P. Ford, L. Lenertz, W. E. Wright, and J. W. Shay. 2002. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277:47242-47247. [DOI] [PubMed] [Google Scholar]
- 17.Chan, S. W., and E. H. Blackburn. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21:553-563. [DOI] [PubMed] [Google Scholar]
- 18.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 19.Conway, C., R. McCulloch, M. L. Ginger, N. P. Robinson, A. Browitt, and J. D. Barry. 2002. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 277:21269-21277. [DOI] [PubMed] [Google Scholar]
- 20.d'Adda di Fagagna, F., M. P. Hande, W.-M. Tong, D. B. Roth, P. M. Lansdorp, Z.-Q. Wang, and S. P. Jackson. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11:1192-1196. [DOI] [PubMed] [Google Scholar]
- 21.d'Adda di Fagagna, F., G. R. Weller, A. J. Doherty, and S. P. Jackson. 2003. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 4:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai, Y., B. Kysela, L. A. Hanakahi, K. Manolis, E. Riballo, M. Stumm, T. O. Harville, S. C. West, M. A. Oettinger, and P. A. Jeggo. 2003. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc. Natl. Acad. Sci. USA 100:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lange, T. 2001. Cell biology. Telomere capping—one strand fits all. Science 292:1075-1076. [DOI] [PubMed] [Google Scholar]
- 24.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 25.Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig, E. E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Driller, L., R. J. Wellinger, M. Larrivee, E. Kremmer, S. Jaklin, and H. M. Feldmann. 2000. A short C-terminal domain of Yku70p is essential for telomere maintenance. J. Biol. Chem. 275:24921-24927. [DOI] [PubMed] [Google Scholar]
- 27.Espejel, S., S. Franco, S. Rodriguez-Perales, S. D. Bouffler, J. C. Cigudosa, and M. A. Blasco. 2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21:2207-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espejel, S., S. Franco, A. Sgura, D. Gae, S. M. Bailey, G. E. Taccioli, and M. A. Blasco. 2002. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 21:6275-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fellerhoff, B., F. Eckardt-Schupp, and A. A. Friedl. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics 154:1039-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D. T. Weaver, and F. W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for Ku in V(D)J recombination. Immunity 9:367-376. [DOI] [PubMed] [Google Scholar]
- 31.Gilley, D., H. Tanaka, M. P. Hande, A. Kurimasa, G. C. Li, M. Oshimura, and D. J. Chen. 2001. DNA-PKcs is critical for telomere capping. Proc. Natl. Acad. Sci. USA 98:15084-15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goytisolo, F. A., E. Samper, S. Edmonson, G. E. Taccioli, and M. A. Blasco. 2001. The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gravel, S., M. Larrivee, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed] [Google Scholar]
- 34.Gravel, S., and R. J. Wellinger. 2002. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22:2182-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 36.Hande, P., P. Slijepcevic, A. Silver, S. Bouffler, P. van Buul, P. Bryant, and P. Lansdorp. 1999. Elongated telomeres in scid mice. Genomics 56:221-223. [DOI] [PubMed] [Google Scholar]
- 37.Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 38.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 39.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 40.Hsu, H. L., D. Gilley, S. A. Galande, M. P. Hande, B. Allen, S. H. Kim, G. C. Li, J. Campisi, T. Kohwi-Shigematsu, and D. J. Chen. 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 42.Jacob, N. K., K. E. Kirk, and C. M. Price. 2003. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell 11:1021-1032. [DOI] [PubMed] [Google Scholar]
- 43.Jhappan, C., H. C. Morse, R. D. Fleischmann, M. M. Gottesman, and G. Merlino. 1997. DNA-PKcs: a T-cell tumor suppressor encoded at the mouse scid locus. Nat. Genet. 17:483-486. [DOI] [PubMed] [Google Scholar]
- 44.Karanjawala, Z. E., U. Grawunder, C. L. Hsieh, and M. R. Lieber. 1999. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9:1501-1504. [DOI] [PubMed] [Google Scholar]
- 45.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295:2446-2449. [DOI] [PubMed] [Google Scholar]
- 46.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 47.Leri, A., S. Franco, A. Zacheo, L. Barlucchi, S. Chimenti, F. Limana, B. Nadal-Ginard, J. Kajstura, P. Anversa, and M. A. Blasco. 2003. Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. EMBO J. 22:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, G., C. Nelsen, and E. A. Hendrickson. 2002. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. USA 99:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, G. C., H. Ouyang, X. Li, H. Nagasawa, J. B. Little, D. J. Chen, C. C. Ling, Z. Fuks, and C. Cordon-Cardo. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2:1-8. [DOI] [PubMed] [Google Scholar]
- 50.Lieber, M. R., Y. Ma, U. Pannicke, and K. Scharz. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Mol. Cell. Biol. Rev. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 51.Loeb, L. A., K. R. Loeb, and J. P. Anderson. 2003. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA 100:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marx, J. 2002. Debate surges over the origins of genomic defects in cancer. Science 297:544-546. [DOI] [PubMed] [Google Scholar]
- 54.Maser, R. S., and R. A. DePinho. 2002. Connecting chromosomes, crisis, and cancer. Science 297:565-569. [DOI] [PubMed] [Google Scholar]
- 55.Mason, P. J. 2003. Stem cells, telomerase and dyskeratosis congenita. Bioessays 25:126-133. [DOI] [PubMed] [Google Scholar]
- 56.Masramon, L., M. Ribas, P. Cifuentes, R. Arribas, F. Garcia, J. Egozcue, M. A. Peinado, and R. Miro. 2000. Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet. Cytogenet. 121:17-21. [DOI] [PubMed] [Google Scholar]
- 57.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 59.Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol, M. C. Nussenzweig, and G. C. Li. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382:551-555. [DOI] [PubMed] [Google Scholar]
- 60.Nussenzweig, A., K. Sokol, P. Burgman, L. Li, and G. C. Li. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival and development. Proc. Natl. Acad. Sci. USA 94:13588-13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang, H., A. Nussenzweig, A. Kurimasa, V. C. Soares, X. Li, C. Cordon-Cardo, W. H. Li, N. Cheong, M. Nussenzweig, G. Iliakis, D. Chen, and G. Li. 1997. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination In vivo. J. Exp. Med. 15:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 63.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 64.Riha, K., and D. E. Shippen. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riha, K., J. M. Watson, J. Parkey, and D. E. Shippen. 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romanov, S. R., B. K. Kozakiewicz, C. R. Holst, M. R. Stampfer, L. M. Haupt, and T. D. Tlsty. 2001. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409:633-637. [DOI] [PubMed] [Google Scholar]
- 67.Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. W. van Buul, and M. A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schar, P. 2001. Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell 104:329-332. [DOI] [PubMed] [Google Scholar]
- 69.Slijepcevic, P., M. P. Hande, S. D. Bouffler, P. Lansdorp, and P. E. Bryant. 1997. Telomere length, chromatin structure and chromosome fusigenic potential. Chromosoma 106:413-421. [DOI] [PubMed] [Google Scholar]
- 70.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song, K., D. Jung, Y. Jung, S. G. Lee, and I. Lee. 2000. Interaction of human Ku70 with TRF2. FEBS Lett. 481:81-85. [DOI] [PubMed] [Google Scholar]
- 72.Stellwagen, A. E., Z. W. Haimberger, J. R. Veatch, and D. E. Gottschling. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17:2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taccioli, G. E., A. G. Amatucci, H. J. Beamish, D. Gell, X. H. Xiang, M. I. Torres Arzayus, A. Priestley, S. P. Jackson, A. M. Rothstein, P. A. Jeggo, and V. L. M. Herrera. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9:355-366. [DOI] [PubMed] [Google Scholar]
- 74.Teo, S. H., and S. P. Jackson. 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35:1-33. [DOI] [PubMed] [Google Scholar]
- 76.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 77.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 78.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 79.Vogel, H., D.-S. Lim, G. Karsenty, M. Finegold, and P. Hasty. 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96:10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vulliamy, T., A. Marrone, F. Goldman, A. Dearlove, M. Bessler, P. J. Mason, and I. Dokal. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413:432-435. [DOI] [PubMed] [Google Scholar]
- 81.Wei, C., R. Skopp, M. Takata, S. Takeda, and C. M. Price. 2002. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 30:2862-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, X., V. Mar, W. Zhou, L. Harrington, and M. O. Robinson. 1999. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 13:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu, C., M. A. Bogue, D.-S. Lim, P. Hasty, and D. B. Roth. 1996. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86:379-389. [DOI] [PubMed] [Google Scholar]