Abstract
The peroxisome proliferator-activated receptor gamma (PPARγ) regulates adipogenesis, lipid metabolism, and glucose homeostasis, and roles have emerged for this receptor in the pathogenesis and treatment of diabetes, atherosclerosis, and cancer. We report here that induction of the PPARγ activator and adipogenesis forced by overexpression of adipogenic regulatory proteins is blocked upon expression of dominant-negative BRG1 or hBRM, the ATPase subunits of distinct SWI/SNF chromatin-remodeling enzymes. We demonstrate that histone hyperacetylation and the binding of C/EBP activators, polymerase II (Pol II), and general transcription factors (GTFs) initially occurred at the inducible PPARγ2 promoter in the absence of SWI/SNF function. However, the polymerase and GTFs were subsequently lost from the promoter in cells expressing dominant-negative SWI/SNF, explaining the inhibition of PPARγ2 expression. To corroborate these data, we analyzed interactions at the PPARγ2 promoter in differentiating preadipocytes. Changes in promoter structure, histone hyperacetylation, and binding of C/EBP activators, Pol II, and most GTFs preceded the interaction of SWI/SNF enzymes with the PPARγ2 promoter. However, transcription of the PPARγ2 gene occurred only upon subsequent association of SWI/SNF and TFIIH with the promoter. Thus, induction of the PPARγ nuclear hormone receptor during adipogenesis requires SWI/SNF enzymes to facilitate preinitiation complex function.
Differentiation of adipocytes, as with all differentiation events, involves programmatic changes in gene expression patterns. Genes specifically expressed in adipocytes must be activated; the concerted action of several transcriptional regulators, including C/EBPα, C/EBPβ, and the nuclear hormone receptor peroxisome proliferator-activated receptor gamma (PPARγ), controls these activation events via direct interaction with PPARγ and C/EBP binding sites in adipocyte-specific gene-regulatory sequences (reviewed in references 14, 33, and 36). Each of these regulators is expressed with different kinetics during adipocyte differentiation in culture, yet forced overexpression of any is sufficient to initiate adipogenic differentiation in fibroblast cells (17, 45, 50, 51). Function of the PPARγ regulator is especially critical, since many of the genes involved in adipogenesis, as well as glucose homeostasis, are activated by this nuclear hormone receptor. Since adipocyte-specific genes are not expressed prior to differentiation, it is likely that the regulatory sequences controlling the expression of these genes are incorporated into a repressive chromatin structure that is refractory to gene expression. Eukaryotic cells have evolved two classes of enzymes that can alter chromatin structure to control accessibility to the transcriptional machinery. These include histone-modifying enzymes, which posttranslationally modify the N-terminal and C-terminal domains (CTDs) of the individual histone proteins that comprise the nucleosome, and ATP-dependent chromatin-remodeling enzymes, which alter structure by disrupting the histone:DNA contacts of the nucleosome, thereby altering nucleosome conformation and, in some cases, altering the position of the histone octamer along the DNA (reviewed in references 28, 42, and 46).
The mammalian SWI/SNF family of ATP-dependent chromatin-remodeling enzymes includes members containing either the Brg1 or Brm ATPase. Although the mammalian SWI/SNF enzymes share most of the same subunits, multiple forms of these enzymes exist; these are distinguished by the ATPase present, the presence of unique subunits, and/or the presence of tissue-specific isoforms of common subunits (30, 39, 48, 49). In vitro analyses of hBRM- and different BRG1-containing enzymes reveal many similarities in chromatin remodeling assays (39). In vivo, however, clear differences in function likely exist. Brg1 knockout mice are embryonic lethal, and heterozygotes are predisposed to tumors (7). Brm knockout mice and cells, in contrast, show only modest proliferation differences compared to the wild type (35). Additionally, at the molecular level, chromatin immunoprecipitation (ChIP) analyses have revealed that Brg1 and Brm can be present on different promoters (18, 40), supporting the idea of differential functions.
Collectively, the literature reveals that Brg1 and/or Brm can physically interact with a number of different transcriptional regulatory proteins, and these proteins have been localized by ChIP studies to the promoter sequences of a number of inducible genes during transcriptional activation. In particular, previous work has indicated a requirement for or a contribution by SWI/SNF enzymes for activation of cellular differentiation genes. Myeloid, erythrocyte, enterocyte, muscle, and adipose cell differentiation events have been linked to the presence of functional SWI/SNF enzymes (2, 12, 19, 32, 40) and, in a more limited number of cases, to the ability of SWI/SNF enzymes to alter chromatin structure at or near inducible promoters. In the case of adipogenesis, the factor C/EBPα, already known to physically interact with the polymerase II (Pol II)-associated general transcription factors TBP and TFIIB (29), was shown to have the capability to interact with hBRM (32). Moreover, the domain mediating this interaction was critically required for the ability of C/EBPα to transdifferentiate fibroblasts into adipocyte-like cells. In vitro, the competency of PPARγ to activate in vitro transcription templates assembled into chromatin was dependent on a specific BRG1-containing SWI/SNF enzyme (22). However, the nature of the role that SWI/SNF enzymes play in facilitating adipogenic gene expression remains to be determined.
Here we explore the functional role of SWI/SNF enzymes during adipocyte differentiation by examining the activation of the PPARγ regulator itself. PPARγ mRNA is expressed from two distinct promoters that give rise to two distinct isoforms, termed PPARγ1 and PPARγ2 (52). We focused on PPARγ2 expression because in differentiating preadipocytes, PPARγ2 is highly induced and is the predominant isoform in differentiated adipocytes (38, 44), while in undifferentiated and differentiated fibroblasts, PPARγ1 expression was not observed (see below). We found that the SWI/SNF enzymes are critically required for transdifferentiation of fibroblasts along the adipogenic pathway. Temporal analyses of factor binding to the PPARγ2 promoter revealed that the BRG1-based SWI/SNF enzymes did not facilitate activator binding to the promoter but instead promoted preinitiation complex (PIC) function. Examination of PPARγ2 activation during differentiation of committed preadipocyte cells confirmed that changes in chromatin structure, activator binding, and assembly of multiple components of the PIC did not require SWI/SNF function. Nevertheless, activation of PPARγ2 transcription did not occur until SWI/SNF and TFIIH subsequently were brought to the promoter. Thus, using two different cellular models for adipocyte differentiation, we demonstrate that activation of the PPARγ regulator critically depends upon SWI/SNF enzymes, most likely by facilitating PIC formation and function.
MATERIALS AND METHODS
Plasmids.
The retrovirus encoding mouse PPARγ2 (45) and cDNAs encoding rat C/EBPα (20) and mouse C/EBPβ (8) were kindly provided by Bruce Spiegelman. C/EBPα and C/EBPβ were subcloned into pBabe-Puro (27).
Cell lines and differentiation methods.
The derivation and maintenance of the cell lines that express dominant-negative human BRG1 (B22 and B24), dominant-negative hBRM (H17), and the Tet-VP16 regulator (Tet-VP16) were described previously (11). To infect cell lines, BOSC23 cells were cultured in 100-mm-diameter dishes and transfected at 80% confluence by FUGENE (Roche) with 10 μg of pBabe-PPARγ2, pBabe-C/EBPα, pBabe-C/EBPβ, or the empty vector as described previously (31). Viral supernatants were harvested 48 h after transfection. Dishes (60 mm) of B22, B24, H17, or Tet-VP16 cells at 50% confluence were infected with virus in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum, 4 μg of polybrene/ml, and 2 μg of tetracycline/ml in a final volume of 5 ml. The corresponding cell lines were split 1:3 48 h after infection and placed under selection with 2 μg of puromycin/ml in the presence of tetracycline. Subsequently, each virally infected cell line was split 1:4 into media containing or lacking tetracycline. After 96 h the medium was changed. Plates designated to be undifferentiated received and continued to be maintained in DMEM plus 10% calf serum in the presence or absence of tetracycline, while the plates designated for differentiation received DMEM plus 10% fetal calf serum plus cocktail containing 0.5 mM methylisobutylxanthine (Sigma), 1 μM dexamethasone (Sigma), 5 μg of insulin/ml, and 10 μM troglitazone (Biomol) in the presence or absence of tetracycline for 48 h. The cells were subsequently maintained in DMEM plus 10% fetal calf serum with 5 μg of insulin/ml and refed every 2 days for up to 8 days.
3T3-L1 preadipocytes were purchased from the American Type Culture Collection, maintained in growth medium consisting of DMEM containing 10% calf serum, and induced to differentiate as described previously (50).
Oil Red O staining.
Sixty-millimeter-diameter dishes were washed twice with phosphate-buffered saline (PBS) and fixed with 10% buffered formalin for 30 min. The formalin was aspirated, and the cells were stained for 1 h in freshly diluted Oil Red O solution (Sigma), prepared by mixing six parts Oil Red O stock solution (0.5% Oil Red Oil in isopropanol) and four parts distilled water. The stain was removed, and cells were washed three times with distilled water and photographed.
RNA analysis.
RNA isolation and analysis by Northern blotting was described previously (13). Probes were derived from plasmids containing PPARγ, aP2, or adipsin cDNA (provided by B. Spiegelman) and were labeled by random priming. Washed blots were exposed to a PhosphorImager (Molecular Dynamics).
For reverse transcriptase (RT)-PCR, total RNA (3 μg) was reverse transcribed with Moloney murine leukemia virus RT (Invitrogen). cDNA was amplified by PCR with Taq polymerase (Invitrogen; 2.5 U/reaction), 0.2 mM deoxynucleoside triphosphates, 8 ng of each primer/μl, and either 1 mM (C/EBPα), 1.5 mM (PPARγ), or 2.5 mM hypoxanthine phosphoribosyltransferase (HPRT) MgCl2. The initial denaturation step was at 95°C for 5 min and was followed by 25 cycles (PPARγ and C/EBPα) or 27 cycles (HPRT). A cycle for PPARγ PCR consisted of denaturation for 30 s, annealing for 40 s at 68°C, and extension at 72°C for 30 s. For C/EBPα, it consisted of denaturation for 50 s, annealing for 55 s at 66°C, and extension at 72°C for 50 s. The final round of extension was for 5 min. The sequences of the primers were as follows: 5′-GCATGGTGCCTTCGCTGATGC-3′ and 5′-AGGCCTGTTGTAGAGCTGGGT-3′ for PPARγ2 (340-bp product), and 5′-CCGGCCGCCTTCAACGAC-3′ and 5′-CTCCTCGCGGGGCTCTTGTTT-3′ for C/EBPα (288-bp product). HPRT and PPARγ1 RT-PCRs were described previously (13, 52).
Nuclear run-on analysis was performed as described previously by Schübeler and Bode (http://juergenbode.gmxhome.de/t01176.htm) on nuclei isolated from ∼4 × 106 differentiating 3T3-L1 preadipocyte cells that were collected at days 0, 2, 4, 5, 6, and 7 after induction of differentiation. Hybond N+ membranes (Amersham) were prepared for hybridization as described previously (3). Immobilized DNAs included 0.1 μg of genomic DNA from 3T3-L1 preadipocytes, 3 μg of EcoRI-linearized pBABE and pBABE-PPARγ2, or 3 μg of PCR product corresponding to nucleotides 181 to 531 of the mouse 36B4 cDNA, to nucleotides 111 to 641 of mouse Gapdh, or to Hprt. The HPRT PCR product has been described (13).
Protein extracts and Western analysis.
Isolation of protein and Western blotting have been described (11). Antibodies utilized included C/EBPβ (Santa Cruz, sc-7962), M2 anti-FLAG (Sigma), and phosphatidylinositol (PI) 3-kinase (06-496; Upstate). Determination of the total Brg1 levels (see Fig. 4) was performed by scanning multiple film exposures and quantifying in ImageQuant (Molecular Dynamics).
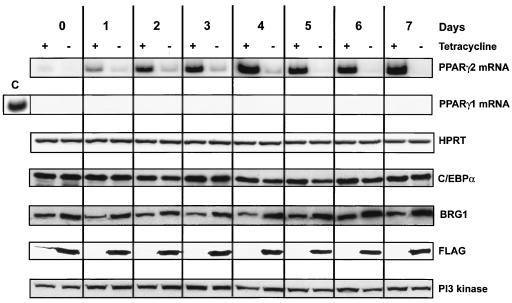
FIG. 4.
Expression levels of adipogenic regulators during differentiation of fibroblast cells along the adipogenic pathway. B22 cells were infected with a retroviral vector encoding C/EBPα, grown in the presence or absence of tetracycline, and differentiated. Expression levels of adipogenic regulators are given as a function of time of differentiation. PPARγ2 and PPARγ1 transcript levels were measured by RT-PCR. HPRT levels are shown as a control. The lane marked C is a positive control from day 7 differentiated 3T3-L1 preadipocytes. C/EBPα, total Brg1, dominant-negative, FLAG-tagged BRG1, and PI3-kinase levels were measured by Western blotting.
Accessibility assays.
Restriction enzyme and DNase I accessibility assays were performed as described previously (12), except that buffer M contained 0.15 mM spermine and 0.5 mM spermidine. DNase I was purchased from Promega. Probes P1 and P2 were PCR fragments corresponding to −1579 to −1187 and −105 to +138 from the mouse PPARγ2 promoter (52).
ChIPs.
The ChIP procedure was adopted from the Upstate protocol. Cells at the indicated time points were fixed by adding 37% formaldehyde to a final concentration of 1% and incubated at 37°C for 10 min. Cross-linking was stopped by addition of glycine to a final concentration of 0.125 M. Cells were washed twice with cold PBS and collected in 1 ml of PBS containing protease inhibitors. After centrifugation, the pellet of cells was resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) containing protease inhibitors and incubated on ice for 10 min. Cell lysates were sheared extensively by sonication (Ultrasonic processor from Cole and Parmer; 3-mm tip at 80 W) on ice to obtain fragments from 200 to 600 bp, as revealed by ethidium bromide staining of aliquots run on agarose gels. Samples were centrifuged to pellet debris, and an aliquot was taken for gel analysis and inputs. One hundred micrograms of soluble chromatin was diluted 10 times with IP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.1], 167 mM NaCl) containing protease inhibitors and precleared for 3 h at 4°C with 50% slurry of protein A, G, or L in Tris-EDTA (TE) (depending on the isotype of the antibody used) in the presence of 20 μg of sonicated salmon sperm DNA and 1 mg of bovine serum albumin/ml. After incubation, the beads were pelleted, and the supernatant was immunoprecipitated with antibodies of interest (see below) at 4°C overnight. Immune complexes were collected with 50% slurry of protein A, G, or L containing 20 μg of sonicated salmon sperm DNA and 1 mg of bovine serum albumin/ml in TE by incubation at 4°C for 1 h. Sepharose beads were washed sequentially for 5 min at 4°C with wash 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), wash 2 (wash 1 containing 500 mM NaCl), and wash 3 (0.25 M LiCl, 1% NP-40, 1% sodium deoxycolate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and finally were washed twice with TE (pH 8.0). Immune complexes were eluted from the beads with 1% SDS in TE (pH 8.0), and protein-DNA cross-links were reversed by adding 200 mM NaCl and heating at 65°C overnight. After treatment with proteinase K, the samples were purified with the QIAquick PCR purification kit (QIAGEN). One-tenth of the immunoprecipitated DNA and 1% of the input DNA were analyzed by PCR.
Antibodies used included Brg1, Brm, and Ini1 (11), diacetylated (K9K14) H3 (06-599; Upstate), Tetra-acetylated H4 (06-866; Upstate), Pol II CTD-Ser-5P (Covance), and the following Santa Cruz antibodies: C/EBPα (sc-9314), C/EBPβ (sc-7962), C/EBPδ (sc-151), TFIIB (sc-274), TBP (sc-273), and TFIIH (sc-293). When Pol II antibody was used, 10 mM NaF was added to all buffers. PCRs were performed with QIAGEN HotStart Taq master mix in the presence of 2 uCi of [α-32P]dATP under the following conditions: 94°C, 15 min, followed by 26 cycles (β-actin) or 27 cycles (PPARγ2) of 94°C for 30 s, and then either 65°C for 40 s (β-actin) or 49.5°C for 40 s (PPARγ2), followed by 72°C for 30 s, followed by a 72°C extension for 5 min. PCR products were resolved in 8% polyacrylamide-1× Tris-borate-EDTA gels, dried, and exposed to a PhosphoImager. Primers used were the following: β-actin, 5′ (+31)GCTTCTTTGCAGCTCCTTCGTTG-3′ and 5′(+135)TTTGCACATGCCGGAGCCGTTGT-3′; and PPARγ2 promoter, 5′(−413) TACGTTTATCGGTGTTTCAT-3′ and 5′(−247) TCTCGCCAGTGACCC-3′. Experiments utilizing Pol II or GTF antibodies were also performed using primers spanning −216 to −20 of the PPARγ2 promoter with identical results.
RESULTS
SWI/SNF enzymes are required for differentiation of fibroblasts along the adipogenic pathway.
We previously described the B22 and H17 fibroblast cell lines that utilize the tet-regulatory system to inducibly express an ATPase-deficient, FLAG-tagged human allele of BRG1 (B22) or an ATPase-deficient, FLAG-tagged human allele of BRM (H17). The parent for these cell lines, termed Tet-VP16, inducibly expresses only the tet-VP16 transactivator and thus serves as a control for our experiments. The mutant BRG1 and hBRM proteins expressed in B22 and H17 cells are competent to associate with other endogenous subunits of SWI/SNF chromatin-remodeling enzymes and can act as dominant negatives with regard to different inducible gene activation events (11, 12).
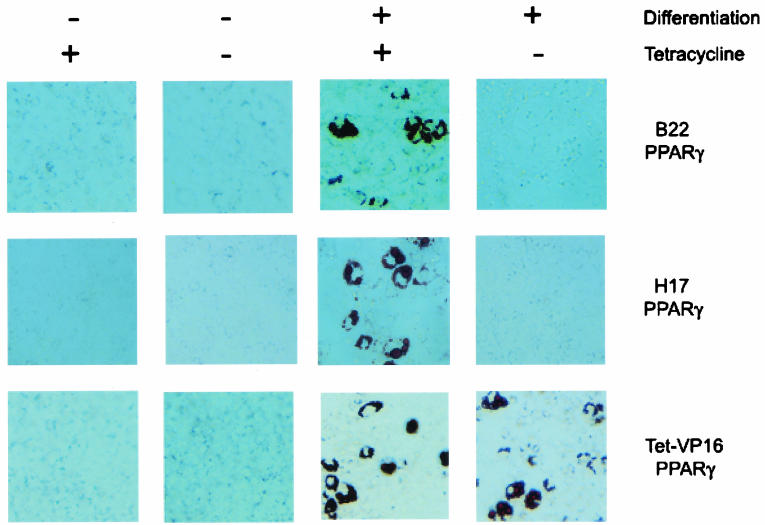
To determine the requirement for BRG1- and hBRM-based SWI/SNF enzymes in adipogenesis, we forced the differentiation of these cells in the presence (dominant negative off) or absence (dominant negative on) of tetracycline by infection with a retroviral vector encoding the PPARγ nuclear hormone receptor or the C/EBPα or C/EBPβ activator. Cells infected with empty retroviral vector served as a control. Cells then were split into medium containing or lacking tetracycline for 96 h and subsequently were cultured for 7 days in the presence or absence of a differentiation regimen that included exposure to a differentiation cocktail (see Materials and Methods) containing troglitazone, a synthetic PPARγ ligand. Adipocyte differentiation was shown by staining with Oil Red O, a lipophilic dye. In our hands, approximately 15 to 20% of the PPARγ-infected cells stained with Oil Red O, which is similar to previous data for unmodified NIH 3T3 cells infected with PPARγ (45). Tet-VP16 control cells stained positively when infected with the PPARγ retrovirus but not when infected with the empty vector (data not shown), and differentiation into adipocytes was dependent on exposure to the differentiation cocktail (Fig. 1). The presence or absence of tetracycline made no difference, as expected. In contrast, B22 and H17 cells infected with the PPARγ retrovirus and cultured in the absence of tetracycline did not differentiate (Fig. 1), suggesting that the expression of dominant-negative BRG1 or hBRM interfered with the differentiation process. The same results were obtained for cells infected with either the C/EBPα or the C/EBPβ virus (data not shown).
FIG. 1.
Expression of dominant-negative BRG1 or hBRM blocks the ability of PPARγ2 to induce adipogenesis in fibroblast cells. B22 and H17 cells, which inducibly express ATPase-deficient, dominant-negative BRG1 or hBRM, respectively, upon removal of tetracycline, or Tet-VP16 cells, which induce only the Tet-VP16 regulator, were infected with a retroviral vector encoding PPARγ2, grown in the presence or absence of tetracycline, and differentiated for 7 days in the presence or absence of differentiation cocktail (see Materials and Methods). Cells were fixed, stained with Oil Red O, and photographed.
SWI/SNF enzymes are required for the activation of adipogenic genes as well as for activation of PPARγ.
We analyzed the mRNA levels of two adipogenic marker genes, aP2 and adipsin, in cells expressing PPARγ, C/EBPα, or C/EBPβ. Cells infected with the PPARγ retrovirus and cultured in the presence of tetracycline and differentiation cocktail expressed both genes, while cells cultured in the absence of tetracycline produced dominant-negative BRG1 and inhibited the expression of both genes (Fig. 2A, right panel). Expression of PPARγ in each of the samples was monitored by RT-PCR (Fig. 2B, right panel), and induction of dominant-negative BRG1 was monitored by FLAG expression (Fig. 2C, right panel). Other experiments demonstrated that the related B24 cell line, which also inducibly expresses dominant-negative BRG1, and H17 cells, which inducibly express dominant-negative hBRM, similarly inhibited expression of adipogenic marker genes in the presence of dominant-negative protein (data not shown). Control Tet-VP16 cells infected with PPARγ-encoding virus showed accumulation of both aP2 and adipsin mRNAs, regardless of the presence or absence of tetracycline (Fig. 3, lanes 9 and 10). The results demonstrate that expression of dominant-negative BRG1 or hBRM interfered with the differentiation process by preventing expression of adipogenic genes. Since BRG1 and hBRM have been found only in cells in a large-molecular-weight complex associated with the other SWI/SNF subunits, presumably adipogenesis is also dependent on the activity of the SWI/SNF chromatin-remodeling enzymes.
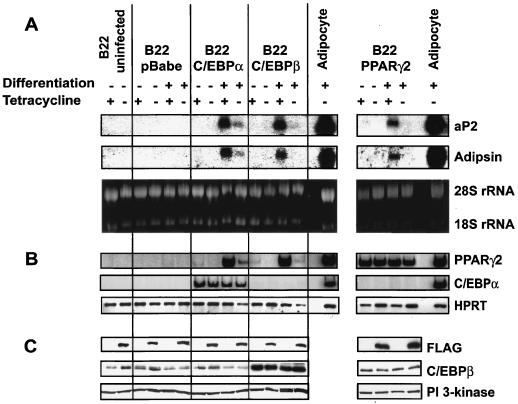
FIG. 2.
Expression of dominant-negative BRG1 blocks the ability of PPARγ2, C/EBPα, or C/EBPβ to induce adipogenesis in fibroblast cells. B22 cells were infected with retroviral vectors encoding PPARγ2, C/EBPα, or C/EBPβ, grown in the presence or absence of tetracycline, and differentiated for 7 days in the presence or absence of differentiation cocktail. Samples in lanes marked Adipocyte were taken from 3T3-L1 preadipocytes differentiated for 7 days. (A) Northern blot showing levels of aP2 and adipsin mRNA. Ethidium bromide staining of rRNA is shown as a control. (B) RT-PCR showing levels of PPARγ2 and C/EBPα mRNA. HPRT mRNA levels are shown as a control. (C) Western blot showing levels of FLAG-tagged dominant-negative BRG1, C/EBPβ, and PI3-kinase.
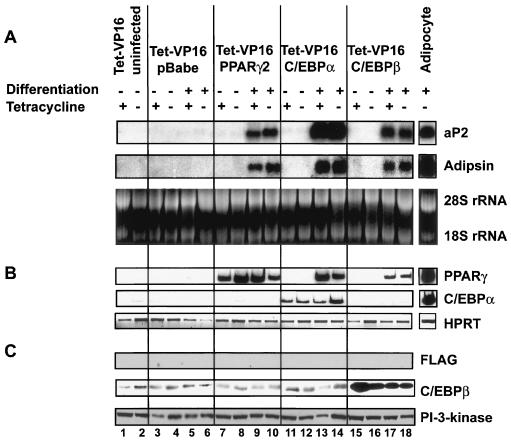
FIG. 3.
PPARγ2, C/EBPα, and C/EBPβ induce adipogenesis in Tet-VP16 control fibroblasts. Tet-VP16 cells were infected and manipulated as described for Fig. 2. (A) Northern blot showing levels of aP2 and adipsin mRNA. Ethidium bromide staining of rRNA is shown as a control. (B) RT-PCR showing levels of PPARγ2 and C/EBPα mRNA. HPRT mRNA is shown as a control. (C) Western blot showing levels of FLAG-tagged dominant-negative BRG1, C/EBPβ, and PI3-kinase.
Adipogenesis also can be induced in culture via ectopic expression of the cellular transcription factor C/EBPα or C/EBPβ (17, 50, 51). When B22 cells were infected with retrovirus expressing either C/EBPα or C/EBPβ, a similar block to aP2 and adipsin gene expression was observed in the presence of dominant-negative BRG1, though in cells expressing C/EBPα, the aP2 and adipsin mRNA levels were reduced 85 to 90% and were not completely absent (Fig. 2A, left panel). Expression levels of C/EBPα (Fig. 2B, left panel), C/EBPβ, and dominant-negative BRG1 (Fig. 2C, left panel) in these samples were monitored by RT-PCR or Western blotting. Similar results were obtained in H17 cells expressing dominant-negative hBRM (data not shown). As anticipated, Tet-VP16 cells infected with C/EBPα- or C/EBPβ-encoding virus expressed both aP2 and adipsin mRNAs in both the presence and absence of tetracycline (Fig. 3, lanes 13 to 14 and 17 to 18). Thus, SWI/SNF chromatin-remodeling enzymes also are required for activation of the adipogenic gene program by C/EBPα or C/EBPβ.
Interestingly, cells induced to differentiate via expression of C/EBPα or C/EBPβ activated PPARγ expression in a SWI/SNF-dependent manner (Fig. 2B, left panel). This suggests that during differentiation promoted by C/EBPα or C/EBPβ, induction of PPARγ—one of the earliest steps in differentiation—is dependent on the activity of BRG1 or BRM. This explains the significant decrease in aP2 and adipsin expression in these cells. However, the data presented here also indicate that SWI/SNF enzymes play a broader role in adipogenesis than just promoting induction of PPARγ, because when the induction of PPARγ is bypassed by providing PPARγ via retroviral infection, expression of the downstream adipogenic marker genes remained SWI/SNF dependent (Fig. 2, right panels). Thus, the data indicate that SWI/SNF enzymes are required for both early and late gene activation events during adipocyte differentiation.
Kinetics of PPARγ2 expression in differentiating cells.
To further analyze the molecular events controlling activation of the adipogenic pathway, we initiated differentiation by infecting cells with C/EBPα, since we could observe that nearly 70% of cells differentiated for 1 week would accumulate lipid droplets (data not shown). RT-PCR analysis of PPARγ2 mRNA levels in cells grown in the presence of tetracycline showed a detectable accumulation on day 1 of differentiation, with robust levels present from days 2 to 7 (Fig. 4). Low levels of PPARγ2 transcripts were observed in cells grown in the absence of tetracycline. We could not detect PPARγ1 mRNA in these cells. Western analyses confirmed that C/EBPα was present at equivalent levels in each sample (Fig. 4). Analysis of FLAG-tagged-protein levels indicated that expression of the mutant BRG1 protein occurred in all samples grown in the absence of tetracycline. Reprobing of this blot for total Brg1 levels revealed only a 1.5- to 2.0-fold increase in levels of Brg1 in the cells expressing the dominant-negative BRG1. This suggests that the levels of mutant Brg1 are only a fraction of the total Brg1 present in the cells, yet they still function as dominant-negative proteins. Alternatively, expression of the Brg1 mutant reduces expression from the endogenous locus, perhaps for the purpose of maintaining a specific overall level of Brg1 protein. In either case, the results indicate that high levels of Brg1 overexpression are not occurring in the differentiating cells.
Histone hyperacetylation and binding of C/EBP factors to the PPARγ2 promoter are independent of Brg1-based SWI/SNF activity.
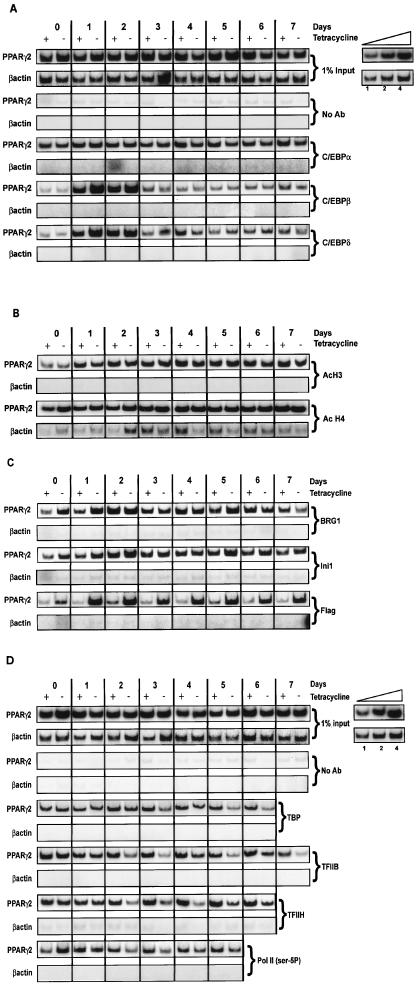
The mouse PPARγ2 promoter has been defined but not well characterized functionally. Of interest, however, is the presence of multiple C/EBP binding sites in the promoter, including two half-consensus sites around −325 relative to the mRNA start site that contribute to activation of PPARγ2 reporter genes (16). To temporally analyze factor interactions at the endogenous PPARγ2 promoter, we performed ChIP experiments and amplified either PPARγ2 promoter sequences or β-actin sequences as a control. Because we forced differentiation by ectopically expressing C/EBPα before inducing differentiation, we were not surprised to see interaction of C/EBPα with the PPARγ2 promoter at the initiation of the differentiation process and throughout the time course. This observation likely reflects and explains the capacity of C/EBPα to initiate the adipogenic gene expression program in nonadipogenic cells. The presence of dominant-negative BRG1 had no or little effect on C/EBPα interactions (Fig. 5A). In differentiating adipocytes, there is a temporal order to expression of the C/EBP family members, where C/EBPβ and C/EBPδ are rapidly induced and then are shut off over the first few days of differentiation. Subsequently, C/EBPα is induced at about day 2 to 3 and is maintained during differentiation (reviewed in reference 21). We therefore examined interactions of C/EBPβ and C/EBPδ with the PPARγ2 promoter. Somewhat surprisingly, both C/EBPβ and C/EBPδ showed a robust induction of occupancy of the promoter at days 1 to 2 of differentiation, despite the presence of ectopic C/EBPα and regardless of the presence of dominant-negative BRG1 (Fig. 5A). Thus, there appears to be a preference during the early stages of differentiation for interaction with C/EBPβ and C/EBPδ. In addition, binding of these factors does not require functional SWI/SNF enzymes.
FIG. 5.
ChIP analysis of interactions on the PPARγ2 promoter as a function of time of differentiation. B22 cells were infected with a retroviral vector encoding C/EBPα, grown in the presence or absence of tetracycline, and differentiated. One percent of input is shown for each experiment. Portions of the PPARγ2 promoter or the β-actin 5′ untranslated region and coding sequence were amplified from each sample. (A) Levels of interactions with C/EBPα, -β, and -δ. (B) Levels of interactions with tetra-acetylated H4 and K9-, K14-diacetylated H3. (C) Levels of interactions with total Brg1, dominant-negative, FLAG-tagged BRG1, and Ini1. (D) Levels of interactions with TBP, TFIIB, TFIIH p89, and Pol II phosphorylated on Ser-5 of the CTD. Inset: linearity controls for PCR amplifications. Each ChIP was repeated in two to five independent experiments. The data shown in parts A, B, and C derive from a single differentiation experiment. ChIPs in part D derive from a different differentiation experiment.
There are a multitude of histone modifications and other ATP-dependent chromatin-remodeling activities that may mediate C/EBP factor binding in the presence of mutant SWI/SNF enzymes. We examined diacetylation of histone H3 on lysines 9 and 14 as well as tetra-acetylation of H4 (Fig. 5B). Levels of tetra-acetylated H4 were high at the beginning of the differentiation process and remained constant throughout. In contrast, there was induction of diacetylated H3 at day 1, concurrent with occupancy of the promoter by the C/EBPβ and C/EBPδ factors. Acetylation of H3 and H4 was unaffected by the presence of dominant-negative BRG1.
C/EBP factors initiate recruitment of SWI/SNF and Pol II-associated GTFs to the promoter.
Previous work has demonstrated that both C/EBPα and C/EBPβ can physically interact with the SWI/SNF component hBRM in cells overexpressing both proteins (19, 32). Additionally, C/EBPα interacts with the Pol II-associated general transcription factors (GTFs) TFIIB and TBP (29). These data have been used to argue that the C/EBP factors can recruit SWI/SNF and GTFs to facilitate transcription. ChIP experiments with the FLAG-tagged ATPase-deficient BRG1 produced in B22 cells revealed that the mutant ATPase, when expressed, was present on the PPARγ2 promoter as early as day 0 and maximally from day 1 through day 7 of differentiation (Fig. 5C). Thus, the presence of C/EBPα may be recruiting SWI/SNF to the promoter at or before the onset of differentiation, possibly reflecting the ability of these proteins to physically interact. ChIP analysis of Brg1 and the SWI/SNF component Ini1 revealed kinetics of promoter occupancy that were similar to each other and to FLAG-tagged mutant BRG1. Promoter occupancy in cells not expressing the dominant-negative BRG1 (plus tetracycline) differed in that interaction of SWI/SNF components peaked at day 2, which coincides with full induction of PPARγ2 transcription (Fig. 4). The observed interaction of the mutant BRG1 with the promoter on day 0 may be due to the modest overexpression of total Brg1 in the cells expressing mutant BRG1 (Fig. 4).
Analyses of phosphorylated RNA Pol II and its associated GTFs, TBP, TFIIB, and TFIIH, indicated that each of these factors is present on the PPARγ2 promoter on day 0, and occupancy in the presence of the dominant-negative SWI/SNF complex is essentially unaffected on days 0 and 1 (Fig. 5D). Thus, SWI/SNF function is not required to assemble these factors onto the promoter in the presence of ectopic C/EBPα. However, the presence of the C/EBP factors, Brg1-based SWI/SNF enzyme, and these GTFs on the promoter are not sufficient to initiate transcription at day 0. Maximal induction of PPARγ2 transcription in the presence of functional SWI/SNF enzymes occurs on day 2 (Fig. 4). In contrast, starting on day 2 and continuing throughout the rest of the time course, the association of TBP, TFIIB, Pol II, and especially TFIIH is compromised in cells expressing the dominant-negative BRG1 (Fig. 5D). Western blot analysis of these GTFs and Pol II shows that the levels of these factors are not altered by differentiation or by expression of the dominant-negative BRG1 (data not shown). Thus, on or about day 2 of differentiation, SWI/SNF function is necessary to maintain the PIC on the PPARγ2 promoter and to promote transcription.
Examination of PPARγ2 promoter accessibility and activation in differentiating 3T3-L1 preadipocytes.
Although forced differentiation of fibroblasts has been utilized for many years as a model system and has been essential in the identification and characterization of adipogenic regulatory proteins, we were concerned that the events leading to activation of the PPARγ2 promoter under conditions where the C/EBPα activator was prematurely expressed might not precisely reflect the events that occurred during differentiation of a committed preadipocyte cell. We therefore examined PPARγ2 activation in differentiating 3T3-L1 preadipocytes.
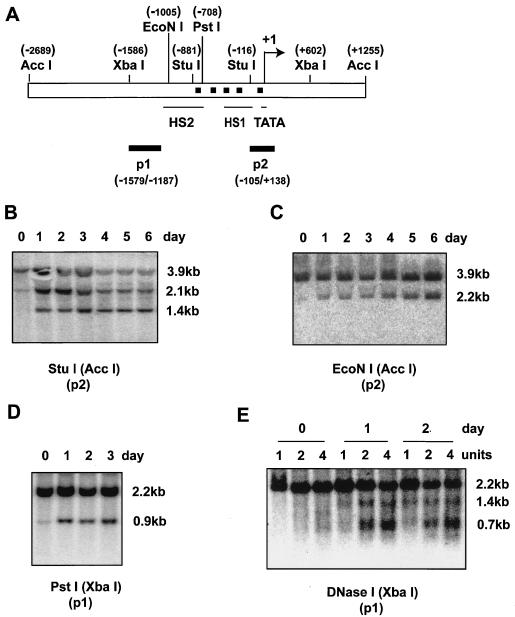
Initially, we examined changes in PPARγ2 promoter structure as reflected by increases in nuclease accessibility. A schematic of the promoter is presented in Fig. 6A. Examination of two StuI sites, an EcoNI site, and a PstI site located between −100 and −1000 on the PPARγ2 promoter revealed a dramatic increase in accessibility in this region on day 1 of the differentiation process (Figs. 6B to D), reflecting changes in promoter structure. A previous report (34) indicated that two DNase I hypersensitive sites exist on the PPARγ2 promoter in undifferentiated 3T3-L1 cells (Fig. 6A). Our analysis indicated these DNase I hypersensitive sites were difficult to detect in undifferentiated, day 0 cells. However, by day 1, they were clearly visible, indicating that that these hypersensitive sites are induced by differentiation (Fig. 6E). Together, the results clearly demonstrate a change in PPARγ2 promoter structure on day 1 of 3T3-L1 differentiation, prior to induction of PPARγ2 gene expression.
FIG. 6.
Changes in PPARγ2 promoter accessibility occur on day 1 of differentiation. 3T3-L1 preadipocytes were induced to differentiate, and nuclei were harvested at the indicated times for nuclease accessibility experiments. (A) Schematic of the PPARγ2 promoter. The locations of relevant restriction enzyme sites are indicated, as are the previously reported DNase I hypersensitive sites (HS1 and HS2) (34). Black squares represent potential C/EBP binding sites. Black bars P1 and P2 indicate the fragments used as Southern blot probes. (B to D) Restriction enzyme accessibility of EcoNI, StuI, and PstI. The enzymes used for flanking digests are indicated in parentheses. (E) Accessibility of DNase I.
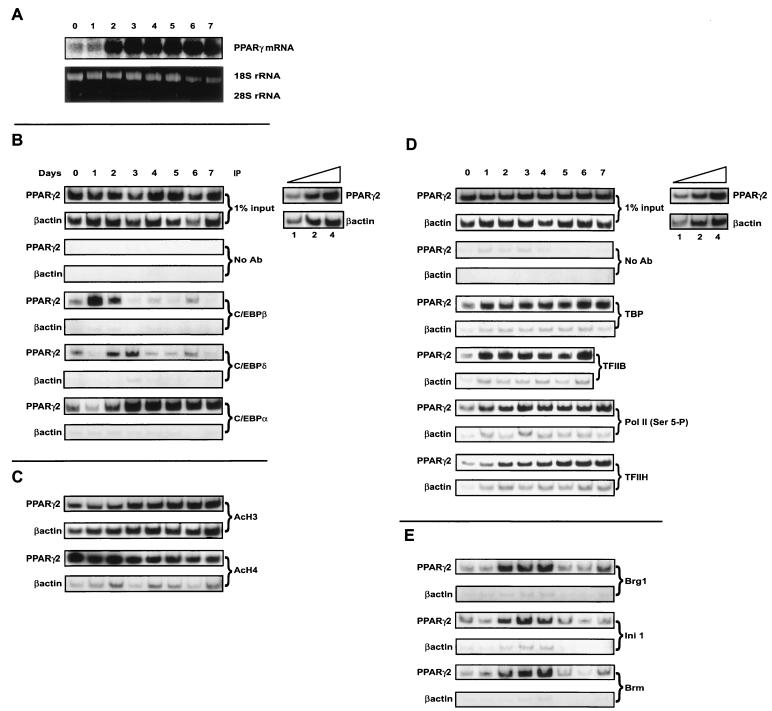
Northern analysis of PPARγ transcript levels revealed a robust induction of expression on day 2 of 3T3-L1 preadipocyte differentiation (Fig. 7A), in agreement with prior studies (45). Thus, induction of the PPARγ2 is temporally distinct from changes in promoter accessibility.
FIG. 7.
(A) PPARγ expression levels in differentiating 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were induced to differentiate. PPARγ expression levels were analyzed by Northern blotting. Ethidium bromide-stained 28S and 18S rRNA is shown as a control. (B to E) ChIP analysis of interactions on the PPARγ2 promoter as a function of time of differentiation of 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were induced to differentiate and processed for ChIP assays at the indicated times. Portions of the PPARγ2 promoter or the β-actin gene were amplified from each sample. (B) Levels of interactions with C/EBPα, -β, and -δ. (C) Levels of interactions with tetra-acetylated H4 and K9-, K14-diacetylated H3. (D) Levels of interactions with TBP, TFIIB, TFIIH p89, and Pol II phosphorylated on Ser-5 of the CTD. (E) Levels of interactions with Brg1, Brm, and Ini1. Inset: linearity controls for PCR amplifications. Each ChIP was repeated in three to five independent experiments. The data shown in parts B, C, and E derive from a single differentiation experiment. ChIPs in part D derive from a different differentiation experiment.
Binding of C/EBP factors, hyperacetylated histones, Pol II, and most GTFs precedes the onset of PPARγ2 induction.
ChIP experiments to identify interactions of the C/EBP family of factors with the PPARγ2 promoter showed an induction of C/EBPβ and C/EBPδ binding on days 1 and 2 of differentiation, respectively (Fig. 7B), largely consistent with the expression patterns of these C/EBPs in differentiating 3T3-L1 cells and with the ChIP results from the forced fibroblast system (Fig. 5A). Thus, interaction of C/EBPβ correlated temporally with changes in nuclease accessibility on the promoter. Over days 2 and 3, C/EBPβ and -δ disappeared from the promoter, concurrent with the appearance of C/EBPα (Fig. 7B). Thus, in differentiating 3T3-L1 cells, the C/EBP binding sites in the PPARγ2 promoter undergo a transition of factor occupancy from C/EBPβ to C/EBPα.
As in the C/EBPα forced fibroblast differentiation experiments, ChIP analysis of tetra-acetylated H4 indicated that levels were high prior to differentiation (Fig. 7C). In contrast to the forced fibroblasts, levels of diacetylated H3 were present on the PPARγ2 promoter at day 0 but increased on day 3 (Fig. 7C). This increase clearly occurred after the onset of PPARγ2 expression but correlated with the transition from binding of C/EBPβ and -δ to C/EBPα. Whether the change in diacetylated H3 levels is a cause or effect of the transition to C/EBPα binding has not been determined. Diacetylated H3 was also present at relatively high levels on the control β-actin sequences.
Examination of phosphorylated Pol II and associated GTFs by ChIP showed that Pol II, TBP, and TFIIB became associated with the promoter on day 1 of differentiation, prior to expression of PPARγ2 (Fig. 7D). Collectively, these data agree with the results obtained with the fibroblast differentiation model and indicate that much of the transcriptional machinery is present on the promoter prior to the onset of gene activation.
PPARγ2 expression is coincident with the binding of TFIIH and SWI/SNF components to the promoter.
In contrast to the other factors examined, the increase in association of TFIIH did not occur until day 2 of 3T3-L1 differentiation (Fig. 7D). Similarly, association of the SWI/SNF components Brg1, Brm, and Ini1 occurred on day 2 (Fig. 7E), coincident with the onset of PPARγ2 transcription. Unlike the case with Pol II and all of the GTFs examined, the interaction of SWI/SNF subunits was transient, with only background levels present on the promoter after day 4. The Ini1 subunit is common to all SWI/SNF enzymes examined, but the Brg1 and Brm ATPases form distinct SWI/SNF enzymes, indicating that at least two distinct forms of the enzyme are present at the PPARγ2 promoter at the onset of transcription. The data indicate that histone hyperacetylation, changes in promoter structure, C/EBP activator binding, and association of phosphorylated Pol II and multiple GTFs with the PPARγ2 promoter occur over the first 24 h of adipocyte differentiation but that these events are not sufficient to initiate transcription. PPARγ2 expression is facilitated by association of TFIIH and SWI/SNF enzymes on day 2 of differentiation.
A decrease in the rate of PPARγ transcription correlates with the dissociation of SWI/SNF enzyme components from the PPARγ2 promoter.
The ChIP experiments shown in Fig. 7E clearly demonstrate that SWI/SNF components were no longer stably associated with the PPARγ2 promoter after day 4 of differentiation, even though the GTFs and C/EBPα remained. Analysis of stable mRNA levels indicated that PPARγ mRNA was abundant on days 5 to 7 (Fig. 7A). Two possible explanations for these data exist. The promoter may achieve a stable structure that is permissive for continued transcription in the absence of SWI/SNF, or the transcription of PPARγ2 decreases or stops after day 4 and the PPARγ message observed on days 5 to 7 represents stable mRNAs produced on day 4 or earlier.
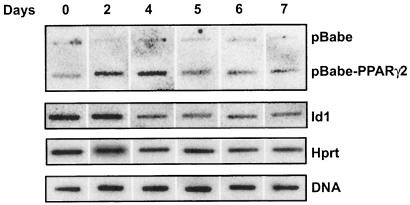
To distinguish between these possibilities, we performed a time course of nuclear run-on experiments. Linearized plasmid DNA containing the PPARγ2 cDNA or empty vector was immobilized on membranes and hybridized to radiolabeled run-on transcripts produced by nuclei isolated from differentiating 3T3-L1 cells on the days indicated. The data demonstrated induction of PPARγ transcription on day 2, continued transcription on day 4, but little transcription on days 5 to 7 (Fig. 8). Thus, the rate of PPARγ transcription decreased between days 4 and 5 and correlated with the loss of SWI/SNF enzyme components on the PPARγ2 promoter (Fig. 7E). In contrast, the rate of Id1 transcription decreased as a function of differentiation (Fig. 8), in agreement with previously published results from the differentiation of human preadipocyte cells (26). We utilized nonspecific hybridization to plasmid sequences and hybridization to 3T3-L1 genomic DNA as controls, as has been described (10, 24, 41, 47), since we were unable to identify other genes that gave a constant rate of transcription over the differentiation time course. The rates for Hprt, 36B4, and Gapdh all decreased over the 7-day time course (Fig. 8 and data not shown).
FIG. 8.
The rate of PPARγ transcription decreases after day 4 of 3T3-L1 differentiation. Nuclear run-on assays were performed on nuclei isolated from differentiating 3T3-L1 cells on the days indicated. Run-on transcripts were hybridized to 3T3-L1 genomic DNA, linearized pBABE vector, linearized pBABE containing the PPARγ2 cDNA, or PCR products corresponding to Id1 or Hprt cDNAs.
We note that the immobilized template contained the entire PPARγ2 cDNA; thus, PPARγ1 transcription would also have been detected. We were unable to detect run-on signal when only the short, 121-bp portion of the cDNA unique to PPARγ2 (52) was used for the hybridizations (data not shown). However, PPARγ mRNA at day 5 of 3T3-L1 differentiation and beyond is predominantly expressed from the PPARγ2 promoter (38, 44). Thus, the data most likely reflect a decrease in the rate of PPARγ2 or in the rates of both PPARγ2 and PPARγ1 transcription.
DISCUSSION
PPARγ is involved in adipocyte differentiation, insulin sensitivity and diabetes, atherosclerosis, and the control of cell proliferation in some cancer cells (reviewed in references 5 and 37). Consequently, its function has been the subject of intense investigation. Relatively little, however, is known about the mechanisms controlling its expression. Here we utilized two different cellular models for adipocyte differentiation to temporally describe the molecular interactions that occur at the promoter of the inducible PPARγ2 gene during adipocyte differentiation, with particular emphasis on the requirement for SWI/SNF chromatin-remodeling enzymes. Through use of a differentiation system driven by introduction of the adipogenic regulator, C/EBPα, we demonstrate a requirement for SWI/SNF enzymes in the activation of the PPARγ regulator as well as in the activation of adipogenic marker genes expressed later during differentiation. Moreover, these experiments revealed that this requirement for SWI/SNF enzymes was relatively late in the cascade of events leading to PPARγ2 activation. Activator binding, Pol II and associated GTF interactions at the promoter, and histone H3 and H4 acetylation occurred prior to and independently of SWI/SNF function. Instead, the data revealed a role for SWI/SNF enzymes in the function of the PIC components at the promoter at the time of transcriptional activation.
Because of the inherent differences between forcing differentiation of fibroblasts into the adipocyte lineage and genuine preadipocyte differentiation, we analyzed differentiation of 3T3-L1 preadipocytes and confirmed both the general order of events that occur during PPARγ2 activation and a role for SWI/SNF enzymes in facilitating PIC function. We expect that the differences exhibited by the two systems reflect the ability of the C/EBPα activator to recruit GTFs and SWI/SNF enzymes (32) prematurely at the initiation of the forced differentiation program. Despite the differences, it is important to note that the data from both systems are consistent with a need for SWI/SNF enzymes to promote the function of the PIC.
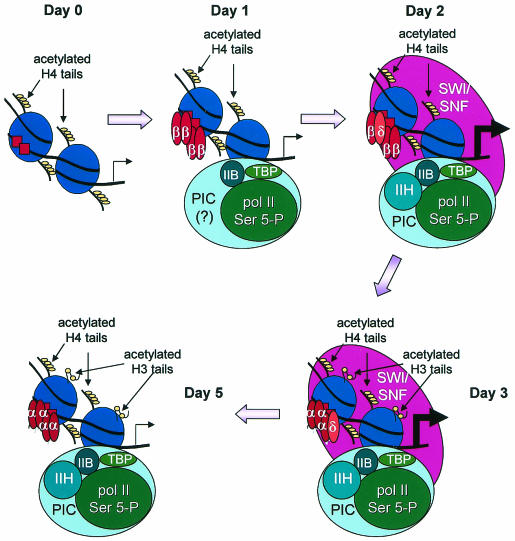
The order of events occurring in differentiating 3T3-L1 cells is diagrammed schematically in Fig. 9. Acetylation of H4 occurs before the onset of differentiation, followed by concurrent changes in promoter accessibility, binding of the C/EBPβ and -δ activators, and assembly of Pol II and most of the GTFs on day 1 of differentiation. Subsequently, on day 2, SWI/SNF enzymes and TFIIH associate with the promoter, indicating that the SWI/SNF enzymes likely facilitate completion of the preinitiation complex, thereby permitting PPARγ2 transcription to commence. On day 3, there is both an increase in the levels of H3 acetylation and a transition from binding of C/EBPβ and -δ to binding of C/EBPα. Which event, if either, is causal remains to be determined. Following day 4, the SWI/SNF enzymes disappear from the promoter and the rate of PPARγ transcription drops, indicating that the presence of SWI/SNF is required for continued transcription.
FIG. 9.
Schematic model of the temporal changes in factor interactions at the PPARγ2 promoter during 3T3-L1 preadipocyte differentiation. Nucleosome positions are presented for illustrative purposes only.
The data from differentiating 3T3-L1 preadipocytes indicate that both BRG1 and BRM are present on the PPARγ2 promoter, suggesting that both complexes are contributing to function. Alternatively, the two complexes could be redundant in function, and the presence of either SWI/SNF enzyme might be sufficient. Our studies using cell lines that inducibly express dominant-negative BRG1 or BRM also suggest that both ATPases are required for PPARγ2 activation and adipocyte differentiation. However, because the BRG1 and BRM SWI/SNF complexes share multiple subunits, it is possible that expression of one mutant ATPase deleteriously affects both complexes by sequestering subunits from the other, endogenous ATPase. Thus, we cannot rigorously state at present whether BRG1, BRM, or both are required for PPARγ2 activation and adipocyte differentiation.
One of the interesting results from our studies is the demonstration that the C/EBP binding sites undergo a transition during the time course of differentiation from binding C/EBPβ and -δ to binding C/EBPα. Although the kinetics of expression for these factors has long supported this idea, this is the first documentation that such a transition occurs at a promoter expressed during adipogenesis. These results differ from previously published work that showed that C/EBPα and -δ, but not C/EBPβ, could bind to the C/EBP sites in the PPARγ2 promoter in a gel shift study and could activate a transiently transfected PPARγ2 reporter plasmid (16). The differences between the studies may be attributed to the likelihood that the chromatin structure at the genomic locus differs from that on a transfected template and undergoes changes during the differentiation process that affect factor interactions.
Previous reports documenting the potential of C/EBPα and -β to physically interact with BRM in cells transfected with both the C/EBP isoform and BRM suggested that these factors may recruit SWI/SNF enzymes (19, 32). Our temporal analysis of factor interactions on the PPARγ2 promoter during C/EBPα-driven differentiation strongly suggests that targeting of SWI/SNF by C/EBPα can occur. However, C/EBPα is prematurely present on the promoter in this differentiation system; the temporal differences in the appearance of the different C/EBP factors on the PPARγ2 promoter in the differentiating 3T3-L1 adipocytes suggest that in a more natural differentiation context, C/EBPβ and -δ may target SWI/SNF enzymes, which then later recruit C/EBPα.
Changes in nuclease accessibility and the binding of C/EBP factors, Pol II, and many of the GTFs on day 1 prior to the appearance of SWI/SNF enzymes on the PPARγ2 promoter indicate that other factors must control the initial accessibility of these promoter sequences. Changes in H4 acetylation did not correlate with initial factor binding to the promoter. Changes in other histone modifications at the PPARγ2 promoter have not been tested but potentially could mediate factor accessibility. Alternatively, a different ATP-dependent remodeling enzyme(s) could alter chromatin structure and promote activator binding prior to SWI/SNF function. This hypothesis is supported by in vitro studies showing that ISWI containing chromatin-remodeling enzymes facilitated stable interaction of RARα:RXR on chromatin templates prior to SWI/SNF enzyme-mediated stimulation of transcription (15). Finally, transcriptional regulators present on the promoter prior to differentiation, such as GATA-2 and -3 (43) and KLF2 (4), might influence chromatin structure in a manner that promotes the transition to an actively transcribing gene.
Our analysis of GTF interactions on the PPARγ2 promoter also revealed that serine 5-phosphorylated Pol II is present at the promoter before TFIIH. TFIIH contains a kinase activity that is capable of phosphorylating the Pol II CTD; however, the temporal order of factor appearance at the PPARγ2 promoter suggests that either a different kinase is responsible for the CTD phosphorylation or TFIIH mediates CTD phosphorylation independently of promoter binding. Additionally, the presence of Pol II phosphorylated at serine 5 of the CTD raises the possibility that the polymerase may be transcriptionally engaged at day 1 and that the inclusion of SWI/SNF enzymes and TFIIH on day 2 promotes release of the polymerase and/or elongation. Multiple other genes are regulated at the level of transcriptional elongation, including hsp70, where elongation can be stimulated by SWI/SNF enzymes in vitro and in vivo (6, 9).
The concurrent entry of SWI/SNF enzymes and TFIIH onto the PPARγ2 promoter in differentiating 3T3-L1 cells suggests that SWI/SNF facilitates the interaction of TFIIH with the rest of the preinitiation complex. Such a role for SWI/SNF enzymes has not previously been documented. However, the data presented here agree with and extend findings from temporal analyses of other mammalian promoters that show the following: (i) some form of histone hyperacetylation precedes association of SWI/SNF with the promoter, and (ii) SWI/SNF enzymes work late in the activation of many genes, typically after some, if not most, of the components driving transcription have associated with the promoter (1, 23, 25, 40). Thus, the data we present on PPARγ2 activation during adipocyte differentiation support a general model where SWI/SNF enzymes function subsequent to activator binding by completing or stabilizing preinitiation complex formation and/or by promoting promoter clearance and elongation.
Acknowledgments
We are grateful to B. Spiegelman and S. Sif for providing reagents and to P. Pekala, R. Kingston, C. Peterson, and members of the Imbalzano and Peterson labs for suggestions, advice, and discussion.
This work was supported by the UMMS Diabetes Endocrine Research Center, by a Scholar Award from the Leukemia and Lymphoma Society, and by an NIH grant to A.N.I.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Banerjee, S. S., M. W. Feinberg, M. Watanabe, S. Gray, R. L. Haspel, D. J. Denkinger, R. Kawahara, H. Hauner, and M. K. Jain. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 278:2581-2584. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J., and D. E. Moller. 2002. The mechanisms of action of PPARs. Annu. Rev. Med. 53:409-435. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S. A., A. N. Imbalzano, and R. E. Kingston. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479-1490. [DOI] [PubMed] [Google Scholar]
- 7.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 9.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelius, P., M. Marlowe, M. D. Lee, and P. H. Pekala. 1990. The growth factor-like effects of tumor necrosis factor-alpha. Stimulation of glucose transport activity and induction of glucose transporter and immediate early gene expression in 3T3-L1 preadipocytes. J. Biol. Chem. 265:20506-20516. [PubMed] [Google Scholar]
- 11.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 13.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 14.Debril, M. B., J. P. Renaud, L. Fajas, and J. Auwerx. 2001. The pleiotropic functions of peroxisome proliferator-activated receptor gamma. J. Mol. Med. 79:30-47. [DOI] [PubMed] [Google Scholar]
- 15.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-Driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell. 6:1049-1058. [DOI] [PubMed] [Google Scholar]
- 16.Elberg, G., J. M. Gimble, and S. Y. Tsai. 2000. Modulation of the murine peroxisome proliferator-activated receptor gamma 2 promoter activity by CCAAT/enhancer-binding proteins. J. Biol. Chem. 275:27815-27822. [DOI] [PubMed] [Google Scholar]
- 17.Freytag, S. O., D. L. Paielli, and J. D. Gilbert. 1994. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 8:1654-1663. [DOI] [PubMed] [Google Scholar]
- 18.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 19.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 20.Landschulz, W. H., P. F. Johnson, E. Y. Adashi, B. J. Graves, and S. L. McKnight. 1988. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2:786-800. [DOI] [PubMed] [Google Scholar]
- 21.Lane, M. D., Q. Q. Tang, and M. S. Jiang. 1999. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266:677-683. [DOI] [PubMed] [Google Scholar]
- 22.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 24.Long, S. D., and P. H. Pekala. 1996. Lipid mediators of insulin resistance: ceramide signalling down-regulates GLUT4 gene transcription in 3T3-L1 adipocytes. Biochem. J. 319:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens, J. H., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moldes, M., F. Lasnier, B. Feve, J. Pairault, and P. Djian. 1997. Id3 prevents differentiation of preadipose cells. Mol. Cell. Biol. 17:1796-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 29.Nerlov, C., and E. B. Ziff. 1995. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 14:4318-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olave, I., W. Wang, Y. Xue, A. Kuo, and G. R. Crabtree. 2002. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16:2509-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangwala, S. M., and M. A. Lazar. 2000. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 20:535-559. [DOI] [PubMed] [Google Scholar]
- 34.Ren, D., T. N. Collingwood, E. J. Rebar, A. P. Wolffe, and H. S. Camp. 2002. PPARγ knockdown by engineered transcription factors: exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev. 16:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 37.Rosen, E. D., and B. M. Spiegelman. 2001. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 276:37731-37734. [DOI] [PubMed] [Google Scholar]
- 38.Saladin, R., L. Fajas, S. Dana, Y. D. Halvorsen, J. Auwerx, and M. Briggs. 1999. Differential regulation of peroxisome proliferator activated receptor gγ1 (PPARγ1) and PPARγ2 messenger RNA expression in the early stages of adipogenesis. Cell Growth Differ. 10:43-48. [PubMed] [Google Scholar]
- 39.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 41.Stephens, J. M., and P. H. Pekala. 1991. Transcriptional repression of the GLUT4 and C/EBP genes in 3T3-L1 adipocytes by tumor necrosis factor-alpha. J. Biol. Chem. 266:21839-21845. [PubMed] [Google Scholar]
- 42.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 43.Tong, Q., G. Dalgin, H. Xu, C. N. Ting, J. M. Leiden, and G. S. Hotamisligil. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290:134-138. [DOI] [PubMed] [Google Scholar]
- 44.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224-1234. [DOI] [PubMed] [Google Scholar]
- 45.Tontonoz, P., E. Hu, and B. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 46.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 47.Waite, K. J., Z. E. Floyd, P. Arbour-Reily, and J. M. Stephens. 2001. Interferon-gamma-induced regulation of peroxisome proliferator-activated receptor gamma and STATs in adipocytes. J. Biol. Chem. 276:7062-7068. [DOI] [PubMed] [Google Scholar]
- 48.Wang, W., J. Côte, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 50.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 51.Yeh, W. C., Z. Cao, M. Classon, and S. L. McKnight. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, Y., C. Qi, J. R. Korenberg, X. N. Chen, D. Noya, M. S. Rao, and J. K. Reddy. 1995. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. USA 92:7921-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]