Abstract
Background
Carcinoma of unknown primary (CUP) accounts for 3–5% of all adult solid tumors. An extensive search for the anatomic site of origin is often undertaken in an attempt to tailor systemic treatment, but the latter often has limited efficacy – especially in the setting of an initial treatment failure. Molecularly targeted therapy is an emerging approach that may offer greater efficacy and less toxicity but is most likely to be effective when pairing a tumor harboring a sensitizing genomic alteration with an agent directed at the altered gene product. We report a patient with a CUP harboring a MET amplification with a complete metabolic response to crizotinib despite also harboring a KRAS mutation.
Methods
Ge-nomic profiling was performed using a clinical next-generation-sequencing-based assay, FoundationOne®, in a CAP-accredited laboratory certified by Clinical Laboratory Improvement Amendments (Foundation Medicine, Cambridge, Mass., USA).
Results
The CUP harbored both MET amplification (16 copies) and a KRAS G12V mutation. The patient was treated with crizotinib, a MET inhibitor, and has experienced a complete normalization of tumor metabolic activity for more than 19 months. Conclusions: Genomic profiling of CUP may reveal clinically meaningful genomic alterations that can guide targeted therapy decision-making. The use of this approach should be studied prospectively as a strategy for the effective treatment of CUP patients and for avoiding resource-intensive workups to identify the tumor site of origin.
Key words: Carcinoma of unknown primary, MET amplification, KRAS mutation, Crizotinib, Next-generation sequencing
Introduction
Carcinoma of unknown primary (CUP) represents 3–5% of all malignancies, and carries a dismal prognosis. Median overall survival is less than a year (6–10 months) [1, 2], and cytotoxic therapy has limited efficacy. Molecularly targeted therapy can offer a treatment alternative that, when administered to a tumor with a sensitizing genomic alteration, is potentially more effective and less toxic than cytotoxic therapy. This is exemplified by the treatment paradigm of ERBB2-amplified breast cancer and anti-ERBB2 (HER2)-targeted therapy [3]. We report a patient with an adenocarcinoma of unknown primary in whom a genomic profiling assay identified high-level MET amplification, and who is responding to crizotinib for more than 19 months.
Patients and Methods
We reviewed the medical records of a patient who presented with a cerebral metastasis of a CUP at the community practice Maryland Hematology Oncology.
Genomic Profiling
Next-generation sequencing (NGS) was performed using the Clinical Laboratory Improvement Amendments (CLIA)-certified FoundationOne® platform (version 2012; Foundation Medicine, Cambridge, Mass., USA). FoundationOne is a targeted assay utilizing NGS in routine cancer specimens [4]. The assay simultaneously sequences the entire coding sequence of 182 cancer-related genes plus 37 introns of 14 genes frequently rearranged in cancer to a minimum coverage depth of 250X [4]. The assay detects all classes of genomic alterations (including base substitutions, insertions and deletions, copy number alterations and rearrangements) and is performed on formalin-fixed, paraffin-embedded (FFPE) surgical specimens and needle biopsies that can be as small as 0.6 mm3 [4].
Case History
The patient is a 59-year-old white female with a 60-pack-year history of smoking who presented with new-onset seizures in May 2012. MRI of the brain with gadolinium revealed a solitary 2.6 × 1.9 cm mass in the right frontal lobe with surrounding edema. Further imaging with 18F-fluorodeoxyglucose positron emission tomography (18F-FDG-PET) and computed tomography (CT) revealed a left mid-abdominal mass of 2 × 4 cm with a standardized uptake value (SUV) of 6; no other lesions nor a definitive anatomic site of origin were identified.
The brain lesion was resected, and the pathologic examination of the specimen revealed a poorly differentiated metastatic carcinoma that had positive immunohistochemical staining for CK7 and epithelial membrane antigen, weak inconclusive staining for thyroid transcription factor-1 and negative staining for S100, p63 and CK20 (fig. 1). Real-time PCR assays for specific codons (‘hotspot testing’) demonstrated a KRAS G12V mutation and wild-type EGFR, while break-apart fluorescence in situ hybridization for ALK rearrangement was negative. The patient was diagnosed with adenocarcinoma of unknown primary and treated with 4 cycles of carboplatin and docetaxel. A subsequent 18F-FDG-PET/CT in September 2012 revealed no interval change on the left mid-abdominal mass but identified a new mass measuring 4.2 × 2.2 cm, which was inseparable from small bowel loops and adjacent to the original left mid-abdominal lesion as well as suggestive of disease progression (fig. 2, table 1).
Fig. 1.
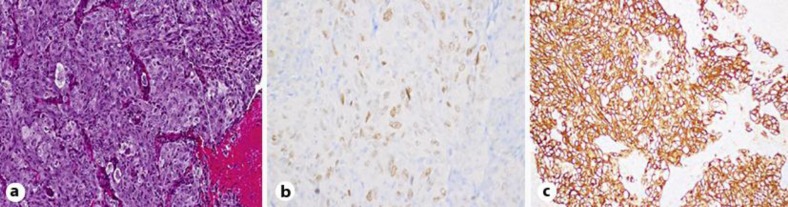
Adenocarcinoma metastatic to the brain. Resection of a cerebral metastasis sample revealed a high-grade adenocarcinoma lacking tubular lumens but containing mucin and signet ring cells (a; ×20). Immunohistochemical workup demonstrated weak inconclusive thyroid transcription factor-1 staining (b; ×40), positive staining for CK7 (c; ×20) and negative staining for S100, p63 and CK20 (data not shown).
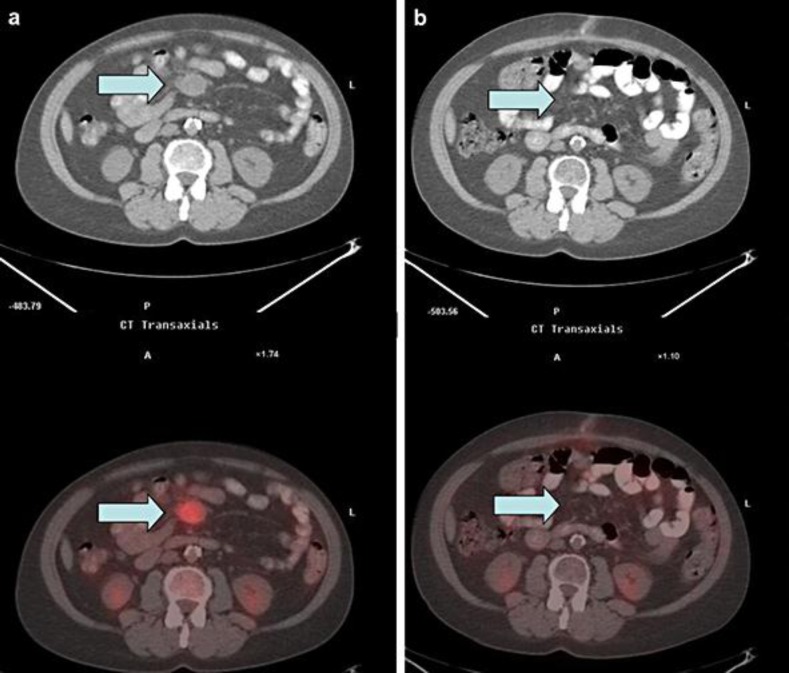
Fig. 2.
Metabolic imaging of tumor response. Low-dose CT (top) and 18F-FDG PET/CT (bottom) fusion of the transaxial left mid-abdominal mesenteric mass. a One month prior to starting treatment with crizotinib (max. SUV = 5.5; mass measures 3.6 cm by 2.8 cm AP). b Three months after starting treatment with crizotinib (max. SUV = 2.0; indicates that the minimal residual mass is metabolically inactive).
Table 1.
Imaging summary of CUP lesions before and after MET-targeted therapy
| September 2012 | January 2013 | May 2013 | January 2014 | |
|---|---|---|---|---|
| Lesion 1 | Mass in central mesentery level of the tip of the liver/right lower kidney pole 3.6 × 2.8 cm (max. SUV 5.5) | Mass has moved to the left side of midline due to peristaltic motion 1.3 × 2.4 cm (max. SUV 2); metabolic remission | No abnormal uptake in the region of the mesentery to indicate recurrent active tumor | Unchanged from previous imaging |
| Lesion 2 | New additional mass inseparable from small bowel loops 4.2 × 2.2 cm (max. SUV 1.4) | Unchanged from previous imaging | – | – |
Crizotinib treatment was initiated in October 2012.
Genomic Profiling
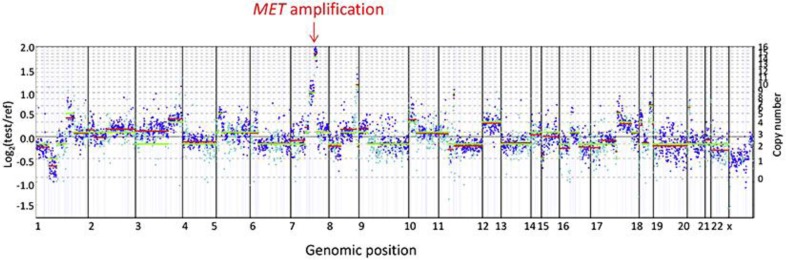
The FFPE-resected brain specimen was submitted to a CAP-accredited, CLIA-certified laboratory (Foundation Medicine) for genomic profiling by a clinical NGS-based assay [4]. Extracted tumor DNA underwent hybrid capture for the entire coding regions from 182 cancer-related genes plus 36 introns of 14 genes frequently rearranged in cancer and was sequenced to a median depth of 1,392X. The sequence was assessed for base substitutions, insertions and deletions, select rearrangements, and copy number changes. The tumor harbored amplification of MET (16 copies; fig. 3), CCND1 (8 copies) and MYC (9 copies) as well as KRAS G12V, TP53 R273L and CARD11 N184S alterations with estimated mutant allele frequencies of 27, 46 and 71, respectively. A previous report described a MET-amplified non-small cell carcinoma (NSCLC) patient who responded to crizotinib, a commercially available inhibitor of MET [5]. On this basis, our patient began treatment with crizotinib (250 mg orally twice daily). Subsequent scans at 3-month intervals demonstrated an ongoing durable response exceeding 19 months with the fused PET-CT revealing complete resolution of the FDG-avid tumor in the abdominal mass (fig. 2, table 1).
Fig. 3.
Genomic profiling identified amplification of MET. Genomic profiling by a NGS-based assay was performed on the resected brain specimen (FFPE tissue) to a coverage depth of 1,392X, revealing 6 distinct genomic alterations including MET amplification. MET copy number (16 copies) was determined by modeling copy variation and aneuploidy across the genome and was compared to process-matched normal controls.
Discussion
CUP is an advanced malignancy, which by definition presents with metastatic disease and accounts for 3–5% of all solid tumors [1]. As treatment, cytotoxic chemotherapy is administered based on a presumed or ‘most likely’ site of origin, which often follows a resource-intensive search for the anatomic site of origin. In our patient, a somatic MET amplification was identified by genomic profiling, and the patient experienced a durable response to crizotinib, all without knowledge of the site of tumor origin. Similarly, another CUP patient was identified as harboring a somatic EGFR L858R alteration via hotspot testing and responded to gefitinib, an EGFR-targeted therapy [4]. However, for our patient, broad-based genomic profiling, which analyzed more than 200 genes somatically altered in human cancer and identified all four classes of genomic alterations, was necessary to detect the clinically meaningful MET amplification. Such an unbiased approach of characterizing the same set of oncogenes and tumor suppressor genes in all solid tumors, given the unpredictable nature of genomic alterations that may be present in any given tumor, has been shown to identify genomic alterations associated with FDA-approved targeted therapies or clinical trials in more than 70–80% of the patients [6, 7].
Other groups have utilized molecular approaches to identify the site of origin and then selected tissue-appropriate chemotherapy regimens, which increased median survival to 12.5 months relative to a historical median survival of 6–10 months for CUP patients [5]. In contrast, the genomic profiling utilized in our case was intended to identify clinically meaningful somatic genomic alterations that suggest potential responsiveness to targeted therapy rather the site of origin. For this patient, inferring the site of origin from the MET-amplified genomic profile was unlikely to provide a definitive answer, as MET amplification has been reported in numerous tumor types, including glioblastoma multiforme, gastroesophageal carcinoma, lung adenocarcinomas, and lung squamous cell carcinomas, with all of these tumors responding to crizotinib [5, 8, 9, 10]. KRAS is also mutated in many tumor types, including pancreatic ductal adenocarcinoma, NSCLC, and colorectal carcinoma, and thus this alteration does not suggest a site of origin [11].
The durable response to crizotinib in this MET-amplified, KRAS-mutated patient is intriguing as the latter is a contraindication for EGFR-targeted therapy in colorectal carcinoma. Recent work has suggested that KRAS, EGFR and ALK alterations are largely mutually exclusive in NSCLC [12]. Moreover, no concurrent KRAS mutations are known to exist in MET-amplified patients who have responded to crizotinib [5, 9], but only limited molecular testing was performed on these tumors.
Dual MET-amplified, KRAS-mutated tumors have been identified at a low frequency in the TCGA database (February 2014) as 1 of 230 NSCLC harboring concurrent MET amplification and KRAS G12V and 1 of 219 stomach adenocarcinoma harboring MET amplification and KRAS G12D. Retrospective evaluation of KRAS alterations in responders to MET inhibitors may help understand whether co-existence of a KRAS alteration paradoxically enhances sensitivity to MET inhibitors as some evidence suggests [13].
Conclusion
Comprehensive genomic profiling led to the detection of an unexpected genomic alteration and successful institution of an available targeted therapy for our patient with previously treated CUP, while avoiding a resource-intensive investigation for the anatomic site of origin. Moreover, despite KRAS mutation being a contraindication for the treatment with RTK-targeted therapy in some carcinomas, our MET-amplified, KRAS-mutated CUP patient responded to crizotinib. Genomic profiling that identifies all four classes of genomic alterations may be of value in treatment decision-making for CUP patients and warrants further study.
Disclosure Statement
N.P., S.A., D.D., S.S., G.A.P., D.M., J.S.R., D.L. and V.A.M. report to be employed by and have equity interest in Foundation Medicine, Inc. J.O., K.W. P.J.S., M.P. and N.K. have nothing to disclose.
References
- 1.Massard C, Loriot Y, Fizazi K. Carcinomas of an unknown primary origin – diagnosis and treatment. Nat Rev Clin Oncol. 2011;8:701–710. doi: 10.1038/nrclinonc.2011.158. [DOI] [PubMed] [Google Scholar]
- 2.Tan DSW, Montoya J, Ng QS, et al. Molecular profiling for druggable genetic abnormalities in carcinoma of unknown primary. J Clin Oncol. 2013;31:e237–e239. doi: 10.1200/JCO.2012.44.3937. [DOI] [PubMed] [Google Scholar]
- 3.Ross JS. Breast cancer biomarkers and HER2 testing after 10 years of anti-HER2 therapy. Drug News Perspect. 2009;22:93–106. doi: 10.1358/dnp.2009.22.2.1334452. [DOI] [PubMed] [Google Scholar]
- 4.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ou SHI, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011;6:942–946. doi: 10.1097/JTO.0b013e31821528d3. [DOI] [PubMed] [Google Scholar]
- 6.Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecol Oncol. 2011;130:554–559. doi: 10.1016/j.ygyno.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Ross JS, Wang K, Al-Rohil RN, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27:271–280. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 8.Schwab R, Petak I, Kollar M, et al. Major partial response to crizotinib, a dual MET/ALK inhibitor, in a squamous cell lung (SCC) carcinoma patient with de novo c-MET amplification in the absence of ALK rearrangement. Lung Cancer. 2014;83:109–111. doi: 10.1016/j.lungcan.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–4810. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi AS, Batchelor TT, Kwak EL, et al. Rapid radiographic and clinical improvement after treatment of a MET-amplified recurrent glioblastoma with a mesenchymal-epithelial transition inhibitor. J Clin Oncol. 2012;30:e30–e33. doi: 10.1200/JCO.2011.38.4586. [DOI] [PubMed] [Google Scholar]
- 11.Neuzillet C, Tijeras-Raballand A, de Mestier L, et al. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141:160–171. doi: 10.1016/j.pharmthera.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Gainor JF, Varghese AM, Ou S-HI, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sequist LV, von Pawel J, Garmey EG, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]