Abstract
Tumor necrosis factor alpha (TNF-α) and glucocorticoids are widely recognized as mutually antagonistic regulators of adaptive immunity and inflammation. Surprisingly, we show here that they cooperatively regulate components of innate immunity. The Toll-like receptor 2 (TLR2) gene encodes a transmembrane receptor critical for triggering innate immunity. Although TLR2 mRNA and protein are induced by inflammatory molecules such as TNF-α, we show that TLR2 is also induced by the anti-inflammatory glucocorticoids in cells where they also regulate MKP-1 mRNA and protein levels. TNF-α and glucocorticoids cooperate to regulate the TLR2 promoter, through the involvement of a 3′ NF-κB site, a STAT-binding element, and a 3′ glucocorticoid response element (GRE). Molecular studies show that the IκBα superrepressor or a STAT dominant negative element prevented TNF-α and dexamethasone stimulation of TLR2 promoter. Similarly, an AF-1 deletion mutant of glucocorticoid receptor or ablation of a putative GRE notably reduced the cooperative regulation of TLR2. Using chromatin immunoprecipitation assays, we demonstrate that all three transcription factors interact with both endogenous and transfected TLR2 promoters after stimulation by TNF-α and dexamethasone. Together, these studies define novel signaling mechanism for these three transcription factors, with a profound impact on discrimination of innate and adaptive immune responses.
The host response to microbial pathogens is mediated by both innate and adaptive immune systems, with macrophages, neutrophils and natural killer cells providing the rapid responses to invading pathogens. Recently, Toll-like receptors (TLRs) on these and other cell types have also been shown to play an essential role in triggering the innate immune response by recognizing pathogen-associated molecular patterns and stimulating the activity of host immune cells against several microbial products (23). In mammals, these pattern recognition receptors (PRR) are also expressed in dendritic cells, mucosal epithelial cells, and dermal endothelial cells that are also involved in the first line of defense against pathogens. To date, 10 distinct receptors have been reported which belong to the TLR family (5, 13, 23, 27, 35). These receptors are responsible for recognizing and triggering a response to microbial products such as lipopolysaccharide (LPS), peptidoglycan, flagellin, and bacterial CpGDNA motifs.
One of the members of the TLRs, TLR2, has been shown to act as a PRR for diverse bacteria and their products, particularly gram-positive bacteria, peptidoglycan, and bacterial lipopeptides (1, 8, 34). In contrast, TLR4 has also been identified as the receptor for LPS, a major constituent of gram-negative bacteria. LPS may also regulate TLR2 expression either directly and/or through LPS-activated TLR4 (7, 19, 20), although knockout-mouse experiments suggest that TLR2 is not essential for LPS signaling (34). The cascade underlying TLR2-induced signaling is similar to that observed for other inflammatory molecules such as interleukin-1 receptor (IL-1R). On TLR activation by gram-positive bacteria, the cytoplasmic adaptor proteins MyD88 and TIRAP are recruited to the receptor complex (36). A serine/threonine kinase, IRAK, is subsequently recruited to the signaling complex, where it phosphorylates Tollip, which terminates TLR signaling. This signaling event is crucial for the TLR complex to interact with the downstream signaling molecule TRAF6, which subsequently activates NF-κB (36), Jun amino-terminal kinase (JNK), extracellular signal-related kinase (ERK), and p38 kinase (2).
Lung epithelial cells actively secrete and respond to inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) (25), and TNF-α is also produced when the TLR2 and TLR4 signaling pathways are activated (14). Recent evidence suggest that TNF-α regulates TLR2 through a classic NF-κB pathway in which TNF-α triggers IκB phosphorylation, ubiquitination, and proteosomal degradation, enabling NF-κB nuclear translocation and subsequent binding to specific genomic response elements. Once bound to DNA, NF-κB activates the transcription of proinflammatory genes such as the gene encoding IL-8 (25). TNF-α has also recently been reported to up-regulate the expression of the TLR2 receptor in macrophages (19). Together, these results provide evidence that proinflammatory cytokines may amplify both inflammatory and immune responses.
Proinflammatory cytokines such as TNF-α and IL-1β are well known to be produced at the site of peripheral inflammation by activated lymphocytes and macrophages. These cytokines can exert profound excitatory effects on the hypothalamic-pituitary-adrenal axis (HPA axis), leading to the production of anti-inflammatory glucocorticoids by the adrenal gland and an overall blunting of the inflammatory response (4, 37). Glucocorticoids are extensively used clinically to suppress a large variety of inflammatory and immune responses. They exert their anti-inflammatory on the adaptive immune system effects primarily by blocking the expression of proinflammatory cytokines and adhesion molecules in a glucocorticoid receptor (GR)-dependent manner. The antagonism between anti-inflammatory glucocorticoids and pro-inflammatory molecules such as TNF-α on the production of cytokines and interleukins is well established and has been previously detected in many cell types including lung cells (25). These antagonistic actions of glucocorticoids on inflammatory signaling were shown largely to involve protein-protein interactions between GR and transcription factors such as NF-κB and AP-1 (12, 22). However, simultaneous inhibitory and stimulatory effects of glucocorticoids were found in inflammatory and apoptotic gene clusters, suggesting that they can exert positive and negative effects (9).
Interestingly, glucocorticoids were recently reported to increase Haemophilus influenzae-induced expression of TLR2 mRNA and protein via a signaling pathway that involves a negative cross talk with p38 mitogen-activated protein kinase (MAPK) (29). This effect appeared to involve the up-regulation of MAPK phosphatase 1 (MKP-1), which leads to dephosphorylation of p38 (15). Dexamethasone (Dex) treatment was shown to dephosphorylate p38, resulting in the sustained expression of MKP-1 (18). However, it remains unclear if glucocorticoids also regulate TLR2 by other, more direct mechanisms such as transcriptional activation of the TLR2 gene. In this study, we provide experimental evidence for increased expression of TLR2 RNA and protein expression after treatment with both the proinflammatory molecule TNF-α and the anti-inflammatory molecule dexamethasone. This effect appears to involve unprecedented cooperative interaction between GR, NF-κB, and STAT at the level of the endogenous TLR2 gene promoter, with potential consequences for the stimulation of innate immune responses.
MATERIALS AND METHODS
Reagents and antibodies.
Recombinant human TNF-α was purchased from R&D Systems (Minneapolis, Minn.). Dexamethasone (Dex) was supplied by Steraloids (Wilton, N.H.). The EXPRE 35S35 protein 35S labeling mix (1,108 Ci/mmol) was purchased from Perkin-Elmer (Boston, Mass.). Mefipristone (RU486) was a gift from Roussel UCLAF (France). Dulbecco's modified eagle's medium lacking methionine, cysteine, and glutamate was purchased from ICN Biochemicals, Inc. (Irvine, Calif.). Oligonucleotide primers for mutagenesis and TaqMan PCR were synthesized by Oligo's Etc. (Bethel, Maine). The TaqMan probes were synthesized by Applied Biosystems (Foster City, Calif.). The hTLR2-specific antibody was kindly provided by P. Scherer (Albert Einstein Institute, New York, N.Y.) (19). Anti-Flag antibody was obtained from Sigma. The previously characterized antibody to the human GR Ab57 (6) was used in all studies. The peroxidase-labeled secondary antibodies and enhanced chemiluminescence reagents were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.).
Cells, transient transfection, and reporter gene assays.
A549 cells were grown at 37°C in a 5%-95% CO2-air atmosphere in Dulbecco's modified Eagle's medium/F12 medium supplemented with 5% fetal calf serum, 100 IU of penicillin per ml, and 100 mg of streptomycin per ml. The cell cultures were maintained in a 5% CO2 humidified incubator at 37°C and passaged every 3 to 4 days. All transfections were carried out with Fugene reagent as specified by the manufacturer (Roche, Indianapolis, Ind.). An appropriate amount of Fugene reagent (3 μl per μg of transfected plasmid) was added to Optimem (Life Sciences, Inc., St. Petersburg, Fla.) with the purified plasmid DNA and allowed to incubate for 15 min at room temperature before being added to cells in Optimem.
For promoter activity experiments, 2 μg of the mTLR2 luciferase construct (24) in combination with 10 ng of phRLSV40 (Renilla reporter vector) was transfected into A549 cells. At 24 h after transfection, the cells were stimulated with TNF-α or Dex for 18 h. They were then lysed in passive lysis buffer (Promega Corp., Madison, Wis.), and the lysates were used to determine luciferase activity in the dual luciferase reporter assay system (Promega). The luciferase activity was measured using the 96-well plate format with an MLX automated microtiter plate luminometer from Dynex. Luciferase activity was then normalized to the Renilla activity and/or the protein content in each sample and to the control. All of the luciferase assays reported here represent three separate experiments assayed in duplicate. Representative results are shown for each experiment.
TLR2 promoter mutagenesis and plasmids.
Site-directed mutagenesis was used to generate a specific mutation in the potential NF-κB site at −160 in the mTLR2 promoter, using the pGL3-297 construct as the template (24). The native sequence of the potential NF-κB site in the sense strand is 5′-GGGAATTCCC-3′, and the mutated sequence is 5′-GGcccATTCC-3′ (mutated bases in lowercase type). Cloning was used to separate the NF-κB site and the STAT-binding site in the TLR2 promoter deletion construct pGL3-297. A 300- or 600-bp DNA fragment amplified from the origin-E1 region of pGL3 basic was inserted into a newly generated PstI site at −245, generating the constructs pGL3-297 insert300 and pGL3-297 insert600. To remove the STAT-binding site, we generated a deletion construct (pGL3-297 wtΔ285-269) by introducing a PstI site at position −284. Through PstI digestion, we removed the STAT transcription consensus-binding site. To decrease the spacing between the NF-κB and STAT-binding site, a PstI site was introduced at position −180. Through PstI digestion, a new deletion mutant was generated (pGL3-297 wtΔ269-208). The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to accomplish this mutagenesis on the pGL3-297 TLR2 promoter construct. Direct sequencing analysis was carried out on all mutant constructs to verify the DNA sequence. Recombinant products containing one of the above mutations were used to replace the native sequence in the pGL3-297 construct of mTLR2. All the transcription binding-site mutations in the native pGL3-297 were analyzed with the software TFSearch: Searching Transcription Factor Binding Sites (version 1.3), (Parallel Application TRC Laboratory, RWCP, Tokyo, Japan) or MatInspector software (26) to confirm the presence or absence of the transcription factor-binding sites and any other potential motif such as glucocorticoid response elements (GREs). Using a threshold score of 85.0, no GRE was found in the native or wild-type pGL3-297 with the TFSearch software. However, a half GRE was identified at −127 when MatInspector software was used. Site-directed mutagenesis was used to generate a specific mutation in the potential GRE. The native sequence of the half GRE site in the sense strand is 5′-AGTTCT-3′, and the mutated sequence is 5′-ACCTCT-3′.
A dominant negative STAT5b construct (Y699F), which cannot homodimerize or heterodimerize with wild-type STATs, was kindly provided by J. Rosen (Baylor College of Medicine, Houston, Tex.) (16). A constitutively active IκBα (superrepressor), provided by A. Baldwin (30), was also cotransfected with the TLR2 promoter deletion mutants as indicated. For TLR2 protein expression experiments, a Flag-tagged TLR2 construct (provided by A. Mantovani, Instituto di Ricerche M. Negri, Milan, Italy) (36) was used at 1 μg/ml for transient expression in A549 cells. After 24 h, transiently transfected cells were harvested and processed for immunoprecipitation studies, using an anti-Flag antibody or the anti-hTLR2 (raised against the COOH terminus; provided by P. Scherer, Albert Einstein Institute) (19) to pull down transiently expressed hTLR2-Flag or the endogenous TLR2.
Real-time PCR (TaqMan PCR).
RNA used for TaqMan PCR was prepared from cells using the Absolutely RNA RT-PCR miniprep kit (Stratagene) followed by PCR with the core reagent kit (Perkin-Elmer). The TaqMan hTLR2 and cyclophilin B probe-primer combinations used in these studies were identified using the Primer Express software package (Applied Biosystems, Foster City, Calif.). A 10 μM stock solution of each forward and reverse primer as well as the probe was prepared, and 1 μl was used per 50 μl of reaction mixture. AmpliTaq Gold polymerase and universal reaction buffer with 5.5 mM MgCl2 (Applied Biosystems) was used for master mix preparation. Predeveloped TaqMan assay reagents and Universal PCR master mix were used for IL-8 and MKP-1 (Applied Biosystems). Threshold cycle numbers (Ct) were determined with Sequence Detector software (version 1.6; Applied Biosystems) and transformed using the ΔCt or ΔΔCt method as described by the manufacturer. CyclophilinB was used as a control gene for calibration. Data were expressed as fold induction or repression by normalizing the data to the control condition for each transcript in each of three experiments run in duplicate. Statistical significance was determined by the Tukey-Kramer pair comparison analysis method.
Radiolabeled immunoprecipitation and immunoblots.
Since endogenous TLR2 and MKP1 expression in A549 cells is normally very low, immunoprecipitation was used to detect the TLR2and MKP1 protein. Briefly, for TLR2 expression, cells were treated with TNF-α, Dex, or a combination of the two for 3 h and were further incubated with EXPRE 35S35 protein labeling mix for 1 h. After the incubation period, the cells were detached from the flasks using EDTA solution (2.6 mM MgCl2, 1.5 mM KH2PO4, 136 mM NaCl, 0.5 mM EDTA, 8.1 mM Na2HPO4), pelleted, and then resuspended in low-detergent buffer (LDB) (20 mM Tris-Cl [pH 7.5], 2 mM EDTA, 150 mM NaCl, 0.5% Triton ×-100, 0.5% protease inhibitors, 5% phosphatase inhibitors) and homogenized using a Tissuemizer. The total amount of protein was measured using a protein assay reagent (Bio-Rad Laboratories, Inc., Hercules, Calif.) as specified by the manufacturer, and equivalent amounts of total protein were used for immunoprecipitation. Normal mouse immunoglobulin G was initially used to reduce nonspecific binding, and total homogenate was incubated for 15 min at 4°C with end-over-end rotation. Protein A-Sepharose was then used to remove the normal mouse immunoglobulin G, and the cleared supernatant was incubated with the hTLR2-specific antibody. Finally, the antigen-antibody complex was pulled down with a second exposure to protein A-Sepharose, and the pellet was washed with LDB. Sample buffer containing sodium dodecyl sulfate and β-mercaptoethanol was used to elute the immunoprecipitated protein, and samples were run on a sodium dodecyl sulfate-8% polyacrylamide gel and then transferred to nitrocellulose membranes. Radiolabeled TLR2 was detected by overnight exposure of the nitrocellulose membranes to chemoluminescent film at −80°C. The amount of MKP-1 protein was determined by immunoprecipitation without metabolic labeling, as described in the legend to Fig. 7. Densitometric analysis of immunoreactive bands was performed with the NIH-Image software.
FIG. 7.
Dex induces MKP-1. (A) MKP-1 mRNA was measured by real-time quantitative reverse transcription-PCR in A549 cells after 8 h of treatment with the indicated concentrations of Dex. Asterisks denote a significant increase between untreated and Dex-treated cultures as determined by Tukey-Kramer pair comparison analysis (P < 0.05). (B) Kinetics of MKP-1 induction by Dex. The MKP-1 mRNA level was measured by real-time quantitative reverse transcription-PCR. A549 cells were treated with 100 nM Dex and harvested at the indicated time points (0, 1, 2, 4, 6, 8, and 24 h). Asterisks denote a significant increase between untreated and Dex-treated cultures as determined by Tukey-Kramer pair comparison analysis (P < 0.05). (C) The GR antagonist, RU486, counteracts the Dex-induced up-regulation of the MKP-1 mRNA level. A549 cells were stimulated with Dex for 8 h, and the MKP-1 mRNA levels were measured by real-time quantitative reverse transcription-PCR. RU486 counteracts the enhancing effect of Dex on MKP-1 mRNA levels. For each well, MKP-1 expression was normalized to cyclophilin B, and the fold induction for each experiment was determined by dividing the normalized expression from treated wells by that from control (untreated) wells. The asterisk denotes a statistically significant increase between the indicated Dex-treated wells and all other treatments as determined by Tukey-Kramer pair comparison analysis (P < 0.05). Values are the mean ± SE of three experiments. (D) MKP-1 protein levels in A549 cells after Dex treatment. MKP-1 protein expression was detected after Dex treatment. Western blot (IB) analysis was performed to confirm the Dex-induced effect. A549 cells were harvested after 8 h of treatment and immunoprecipitated (IP) with anti-MKP-1 antibody. The graph shows MKP-1 protein expression from cells treated with Dex and/or RU486, normalized to the control immunoprecipitated protein (vehicle treated cells). Analysis of the immunoreactive bands with NIH-Image software reproduced the Dex-induced increase in the level of MKP-1. Values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test).
ChIP assays.
The chromatin immunoprecipitation (ChIP) assay was performed using the ChIP assay kit from Upstate Biotechnologies (Lake Placid, N.Y.). Cells (2 × 106 to 3 × 106) were plated on T75 flasks After 24 h, the cells were treated with TNF-α and/or Dex for 1 h. They were fixed with 1% formaldehyde and lysed, the DNA was fragmented by sonication, and 10 μl of the chromatin solution was saved as input. A 1-μg amount of anti-GR antibody Ab57, anti-p65, anti-STAT5, or rabbit immunoglobulin G was added to tubes containing 900 μl of chromatin solution. After incubation, the antibody complexes were captured with protein A-agarose beads and subjected to serial washes. The initial content of GR was assessed in the GR-immunoprecipitated fraction by Western blotting. The chromatin fraction was further extracted and reverse cross-linked at 65°C in the presence of 200 mM NaCl. The DNA was then purified using chloroform-isoamyl alcohol. In another set of experiments, cultured cells under similar conditions to those detailed above were transiently transfected using the pGL3-297wt TLR2 promoter or the pGL3-297/NFm or pGL3-297Δ285-269 mutants. Transfected cells received further identical treatment, and after 1 h the cells were subjected to a overexpressed ChIP assay using 1 μg of anti-GR antibody Ab57. For all the ChIP protocols, immunoprecipitated DNA (5 μl) and input DNA (5 μl) were subjected to 30 cycles of PCR with Taq Gold polymerase and primer pairs that amplify a 279-bp region spanning the NF-κB- and STAT-binding sites (−297 to −18) of the proximal promoter of the TLR2 the (forward primers to detect endogeous and overexpressed TLR2 promoter mutants were TLR2f1 [5′-cat tca gcc atc att gtccag gc-3′] and RV3 [5′-cta gca aaa tag gct gtc cc-3′] from pGL3 basic, respectively; the reverse primer for both endogenous and overexpressed promoter was NF3r [5′-cag ttc tgt ttt gcc tgc cc-3′]). The input PCR product amplified by the primer set specific for the TLR2 promoter was the same in the presence and absence of TNF-α and/or Dex treatment. PCR products were then run on an agarose gel, stained with ethidium bromide, and photographed.
Statistics.
All pair-treated groups were compared by the Tukey-Kramer test. Significant differences have a P of <0.05. Analysis was carried out with JMP software (Statistics Made Visual, SAS Institute Inc., Cary, N.C.).
RESULTS
TNF-α and Dex regulate IL-8 and TLR2 mRNA in A549 cells.
Lung epithelial cells are a classic target for both pro- and anti-inflammatory molecules, and A549 cells have previously been shown to be responsive to both TNF-α and glucocorticoids, with a typical pro- and anti-inflammatory response (25). To validate this model system, we studied the effect of TNF-α treatment on IL-8 mRNA expression following TNF-α exposure, using real-time PCR analysis (Fig. 1A). TNF-α treatment for 8 h with increasing concentrations from 0.1 to 100 ng/ml led to the predicted increase in IL-8 mRNA level (Fig. 1A, left panel). Similarly, Dex treatment of A549 cells repressed the expression of endogenous IL-8 mRNA, albeit slightly and in a manner consistent with the reported anti-inflammatory actions of glucocorticoids (right panel). This effect is probably being mediated via the classical GR, since concentrations of glucocorticoid as low as 1 nM were effective in repressing IL-8 mRNA levels.
FIG. 1.
TNF-α and Dex induce TLR2 mRNA in A549 cells. (A) The expression of IL-8 mRNA in TNF-α-treated and untreated human lung A549 cells was measured by real-time quantitative PCR. IL-8 mRNA was used to confirm the pro- and anti-inflammatory effect of TNF-α and Dex, respectively. (B) TNF-α or Dex significantly increases TLR2 mRNA levels. TLR2 mRNA levels after TNF-α or Dex treatment were determined by real-time quantitative reverse transcription-PCR. (C) Cyclophilin B (cyclo) was used as a control for the amount of RNA used in each reaction and for each treatment. All sample analyses were carried out in duplicate after 8 h of treatment. Values are the mean ± standard error (SE) of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test).
Using the same RNA samples, the expression of TLR2, a member of the TLR family, was also analyzed. As shown for IL-8 mRNA levels, 8 h of treatment with TNF-α at concentrations from 0.1 to 100 ng/ml resulted in a dose-dependent increase in TLR2 mRNA expression (Fig. 1B). Concentrations of TNF-α as low as 0.1 ng/ml were effective in increasing the TLR2 mRNA level, and a plateau in the response was observed at 10 ng/ml (Fig. 1B, left panel). Interestingly, and contrary to our expectation, the anti-inflammatory steroid Dex also increased TLR2 mRNA expression in a dose-dependent manner, although to a lesser extent than was observed for TNF-α (right panel). After 8 h of Dex treatment, the maximum effect on TLR2 mRNA levels was reached with a Dex concentration of 100 nM. The decrease in TLR2 mRNA levels at 1,000 nM Dex is probably the result of homologous down-regulation of GR and desensitization of the cells to hormone, which is known to occur under these conditions. A constitutively expressed transcript, cyclophilin B, was also evaluated in parallel for each sample, and no significant changes were observed for this gene in A549 cells treated with either TNF-α or Dex (Fig. 1C). Together, these results show the predicted opposing effect between TNF-α and Dex on IL-8 expression, but an unprecedented up-regulation of TLR2 mRNA was seen after treatment of the cells with both the pro- and anti-inflammatory agents.
A cooperative TNF-α and Dex effect on TLR2 mRNA and protein.
Using these initial studies as a basis for further experiments, we next examined the kinetics of TNF-α and Dex treatment on the regulation of IL-8 and TLR2 mRNA levels. A concentration of 10 ng of TNF-α per ml induced a rapid increase in IL-8 mRNA level, causing a 100-fold increase within 30 min. The IL-8 mRNA levels increased to 600-fold after 2 h and decreased to 80-fold thereafter (Fig. 2A, upper graph). The kinetics of the effects of glucocorticoid on IL-8 mRNA levels was also studied, and a slight inhibition was seen in a manner consistent with the data observed in Fig. 1 (Fig. 2A, upper graph). These data are consistent with the known pro- and anti-inflammatory actions of TNF-α and Dex, respectively. Combined addition of TNF-α and Dex to A549 cells significantly reduced the maximal response to TNF-α alone in IL-8 mRNA by 1 h, followed by a time-dependent inhibition at all times thereafter (Fig. 2A, upper graph).
FIG. 2.
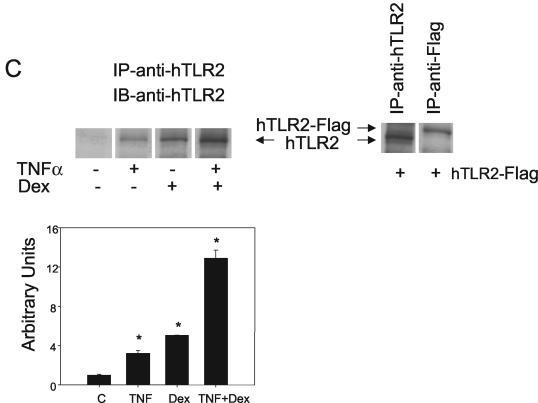
Glucocorticoids synergistically enhance TNF-α-induced TLR2 expression. (A) Glucocorticoids enhance TNF-α-induced TLR2 mRNA production. (Upper panel). IL-8 was profoundly up-regulated by TNF-α treatment and down-regulated by Dex treatment. When the two agents were added together, the TNF-α-induced increase in the amount of IL-8 mRNA was partially counteracted as determined by real-time PCR. A549 cells were harvested at six different time points (0, 0.5, 1, 2, 4, and 8 h), after addition of TNF-α and/or Dex. (Lower panel). Kinetics of the TNF-α and Dex treatments. The increase in the amount of TNF-α-induced TLR2 mRNA was sensitized by Dex. A549 cells were harvested at six different time points (0, 0.5, 1, 2, 4, and 8 h) after addition of TNF-α and/or Dex, and the TLR2 mRNA content was detected by real-time quantitative reverse transcription-PCR. (B) Mechanism of the cooperative effect of Dex on the TNF-α-induced TLR2 mRNA up-regulation. (Upper graph). The GR antagonist, RU486, counteracts the repressive effect of Dex on the TNF-α-induced up-regulation of the level of IL-8 mRNA. A549 cells were stimulated with each of the agents or the combination of all of them for 8 h, and the IL-8 mRNA levels were measured by real-time quantitative PCR in duplicate. RU486 counteracts the enhancing effect of Dex on TNF-α-induced up-regulation of TLR2 mRNA levels. A549 cells were stimulated with each of the agents or a combination of all of them for 8 h, and the TLR2 mRNA levels were measured by real-time quantitative PCR in duplicate. Values are the mean ± SE of three experiments. (C) TLR2 protein levels in A549 cells after TNF-α and Dex treatment. (Left) Major TLR2 protein expression was detected after TNF-α and Dex treatment. Western blot analysis was performed to confirm the cooperative effect between Dex and TNF-α. A549 cells were harvested after 6 h of treatment and subsequently labeled for 1 h with [35S]Cys-[35S]Met and then immunoprecipitated (IP) with anti-TLR2 antibody. (Right) Exogenous expression of TLR2-Flag comigrated with endogenously stimulated receptor with TNF-α and Dex. A549 cells were transiently transfected with the complete TLR2 cDNA tagged to a Flag epitope, labeled for 1 h with [35S]Cys-[35S]Met, and then immunoprecipitated with anti-TLR2 or anti-Flag antibodies. Cells were then harvested, and Western blot (IB) analysis was performed using the anti-TLR2 antibody. (Graph) TLR2 protein expression from cells treated with TNF-α and/or Dex was normalized to the control immunoprecipitated receptor (vehicle-treated cells). Analysis of the immunoreactive bands with the NIH-Image software reproduced the cooperative effect of Dex on the TNF-α-induced increase in the level of TLR2. Values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test).
The kinetics of the effects of TNF-α and glucocorticoids on TLR2 mRNA levels was also evaluated. When 10 ng of TNF-α was applied to the A549 cells, a 30-fold increase in the TLR2 mRNA level occurred within 2 h (Fig. 2A, lower graph), which further increased to 60-fold after 4 to 8 h. Dex at 10 ng/ml induced a slight increase in TLR2 mRNA levels to 10-fold after 4 to 8 h of treatment (Fig. 2A, lower graph). Surprisingly, TNF-α and Dex treatment induced a time-dependent increase of 180-fold in the TLR2 mRNA level by 4 h (Fig. 2A, lower graph). The TLR2 mRNA levels remained elevated up to 8 h of treatment, with a slight decrease thereafter. In contrast, cyclophilin B levels remained unchanged during the entire time course of the experiment independent of the stimulus given (data not shown). These results suggest that Dex cooperates with TNF-α to increase the TLR2 mRNA level in A549 cells, where it acts as an anti-inflammatory agent opposing the well-known action of TNF-α on the IL-8 gene.
We next evaluated whether Dex binding to its receptor is necessary for the cooperative regulation of both IL-8 and TLR2 mRNA expression. For this purpose, we employed RU486, a specific GR antagonist, in combination with TNF-α and/or Dex (28). Addition of RU486 alone to the cells did not alter IL-8 mRNA levels (Fig. 2B, upper graph). However, cotreatment of RU486 and TNF-α plus Dex for 8 h blocked the Dex-induced repression in IL-8 mRNA (Fig. 2B, upper graph). The TNFα-induced increase in the IL-8 mRNA level was only slightly reduced by the glucocorticoid antagonist RU486 (Fig. 2B). These data are in accord with the counteracting effect of the anti-inflammatory molecule Dex on the TNF-α induction of proinflammatory molecules, such as IL-8 and suggest that these biological effects require the binding of glucocorticoid to its receptor. Using the same RNA samples from A549 cells, the TLR2 mRNA levels were also measured after an 8-h TNF-α and/or Dex treatment in the presence or absence of RU486. The addition of RU486 itself to the cells did not affect the TLR2 mRNA levels, whereas RU486 blocked the increase in TLR2 mRNA levels induced by Dex and significantly inhibited the cooperative activation of Dex on TNFα-induced TLR2 mRNA (Fig. 2B, lower graph). No changes were observed in the cyclophilin B mRNA levels with any of the cell treatments (data not shown). Together, these data indicate that the differential regulation of IL-8 and TLR2 expression by TNF-α and Dex are both occurring in a GR-dependent manner.
Considering our findings on the expression of TLR2 mRNA by TNF-α and Dex, we next sought to determine if these changes in mRNA were also reflected by alterations in TLR2 protein. To undertake these investigations, we treated A549 cells with 10 ng of TNF-α per ml and/or 100 nM Dex for a total of 6 h, pulsing the cells with [35S]Cys-[35S]Met during the last hour of treatment. Endogenous TLR2 protein was then immunoprecipitated from cell lysates using an anti-TLR2 antibody raised against the cytoplasmic tail of the receptor (19). Treatment of cells with either TNF-α or Dex induced an increase in the amount of TLR2 immunoprecipitated protein (Fig. 2C), whereas cotreatment of the cells with TNF-α and Dex induced a further increase in the amount of immunoreactive TLR2 protein (Fig. 2C). To ascertain the identity of this immunoprecipitated band, we used the complete TLR2 cDNA sequence that contains an amino-terminal Flag epitope in the pCMV1 vector that was transiently expressed in A549 cells. Both the endogenous and overexpressed receptor, immunoprecipitated with an anti-Flag or anti-TLR2 antibody, showed similar electrophoretic mobilities (Fig. 2C, right gel). The densitometric analysis of the immunoprecipitated bands after TNF-α and/or Dex treatment revealed a four- and fivefold increase, respectively, in the amount of TLR2 protein. However, when both pro- and anti-inflammatory molecules were given together to the cells, a 13-fold increase in the amount of TLR2 immunoprecipitated protein was observed (Fig. 2C, graph). These findings indicate that the cooperative interaction between TNF-α and Dex is also reflected by an increase in the amount of TLR2 protein.
TNF-α and Dex cooperatively activate TLR2 promoter activity in A549 cells through a mechanism that involves GR.
As shown above, the TLR2 endogenous gene cooperatively responds to TNF-α and Dex, two regulatory molecules that are intuitively antagonistic. The evaluation of the TLR2 protein and analysis of its promoter activity by TNF-α in adipose cells and macrophages has previously been studied, but its regulation in other cell types such as those derived from the lungs is largely unknown (19, 24). Thus, to assess whether the TNF-α and/or Dex-induced TLR2 expression occurs by regulation of TLR2 gene transcription, we used the full-length TLR2 promoter construct (pGL3-1486) cloned upstream of a luciferase reporter gene in the pGL3-basic vector. Using this construct in addition to a series of deletion mutants, we evaluated the effect of TNF-α and/or Dex on the TLR2 promoter activity to identify putative responsive elements within this gene. A diagram of the known molecular organization of the TLR2 promoter with their potential transcription factor-binding sites is shown in Fig. 3A (24). The deletion promoter constructs used for this study are also noted. Cells transiently transfected with the intact promoter TLR2 promoter deletion constructs were stimulated with 10 ng of TNF-α per ml and/or 100 nM Dex for 16 h, a time when luciferase levels reach a maximum (data not shown). The luciferase activity was then determined, and the activities were normalized for each control condition. Treatment of A549 cells with TNF-α increased the luciferase reporter activity of all TLR2 promoter deletion constructs, although the response in pGL3-201 was attenuated in comparison to that in the intact promoter and the other deletion mutants (Fig. 3A). Interestingly, treatment of transfected cells with Dex alone had little or no effect on any of the TLR2 promoter constructs or mutants, suggesting that the GR alone does not efficiently regulate our transfected gene. In contrast, addition of TNF-α and Dex to cells expressing the TLR2 promoter constructs induced a cooperative increase of the luciferase activity in all the 5′ TLR2 promoter deletion constructs, except for the pGL3-201-expressing cells, which did not respond to the combined treatment with TNF-α and Dex (Fig. 3A). These data clearly show that the combination of TNF-α and Dex can increase the TLR2 expression by enhancing the TLR2 gene transcription whereas Dex alone is largely inefficient in its effect on the transfected reporter genes under our experimental conditions. Additionally, the deletion mutant pGL3-297 could fully recapitulate the cooperative effects of TNF-α and Dex observed using the intact promoter. Together, the data shown in this figure indicate that the 297 bp 5′ upstream region of the TLR2 gene is sufficient for the cooperative induction by TNF-α and Dex (diagram in Fig. 3B).
FIG. 3.
Glucocorticoids cooperate in the TNF-α-induced increase in TLR2 promoter activity. Functional analysis of 5′ deletion constructs of the TLR2 promoter is shown. (A) Scheme of the 5′ TLR2 promoter deletion luciferase constructs (top). Binding sites for different transcription factors are indicated. The number of each construct corresponds to its 5′ end. The luciferase activity of each of the 5′ deletion constructs of the TLR2 promoter transfected into A549 cells is shown in the graph. Cells were treated for 16 h with 10 ng of TNF-α per ml or 100 nM Dex or a combination and then harvested for luciferase activity determination. Bars represent the ratio between relative luciferase units (RLU) to protein content and normalized to the control (fold induction). All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. (B) Study of the role of GR in the activation of the TLR2 promoter. A diagram of the pGL3-297 TLR2 promoter construct used to study GR participation in the enhancement induced by 10 ng of TNF-α per ml plus 100 nM Dex is shown (top). The luciferase activity of the pGL3-297 TLR2 promoter construct transfected into A549 cells is shown in the graph. The transfected cells were left untreated or treated for 16 h with 10 ng of TNF-α per ml, 100 nM Dex, 1 μM RU486, or a combination of them, as indicated. Bars represent the ratio between relative luciferase units (RLU) and protein content and normalized to the control (fold induction). All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test) to each control condition.
It is interesting that neither this construct nor the intact promoter showed any response to Dex alone, although computer analysis of this DNA fragment indicated that it does contain a sequence resembling a GRE. Thus, to elucidate the role of GR in the cooperative activation of the TLR2 promoter, we first evaluated if RU486 can block the effect of Dex and TNF-α. The effect of the GR antagonist RU486 administered simultaneously with TNF-α and/or Dex was studied after 16 h of treatment of cells expressing the pGL3-297 TLR2 promoter construct. RU486 inhibited the induction of TLR2 promoter activity, as judged by the luciferase reporter activity, when coadministered with TNF-α plus Dex but did not alter the effect of TNF-α alone (Fig. 3B). This finding strongly suggests that the cooperative action of Dex and TNF-α on TLR2 promoter activity requires Dex binding to GR, although this interaction alone appears to be insufficient to activate this reporter gene.
The downstream NF-κB consensus site is essential for the cooperative effect between TNF-α and Dex.
We next evaluated the role of the NF-κB-binding sites located in the 5′ upstream region of the TLR2 gene, on the cooperative response of TNF-α and Dex on the TLR2 promoter activity. Selective point mutations in the NFκB-binding site were created at −160 in both the pGL3-1486 (Fig. 4A) and pGL3-297 (Fig. 4B) TLR2 promoter constructs. In addition, pGL3-1486 contains the NF-κB site at −1115 while it is deleted in the pGL3-297 (NFm) construct. Using these TLR2 promoter mutants (pGL3-1486/NFm and pGL3-297/NFm), we assessed the contribution of the NF-κB-binding site on the synergistic effect of TNF-α and/or Dex on the TLR2 gene activation. Deletion of the NF-κB consensus-binding site at position −1115 had no impact on the TNF-α effect or the cooperative effect with Dex (data not shown). In contrast, mutation of the NF-κB site at position −160 eliminated both responses. These data indicate that the 3′ NF-κB-binding site is necessary for the increase in the transcriptional response to TNF-α alone or in combination with Dex whereas the 5′ site does not appear to contribute to this response.
FIG. 4.
Role of the NF-κB transcription factor in TLR2 promoter activity in A549 cells. (A) The TLR2 promoter construct pGL3-1486 was used as a wild type to produce the NF-κB deletion construct (pGL3-1486/NFm). The X indicates that the NF-κB transcription factor site has been deleted. Bars represent the fold induction by TNF-α, Dex, or both after 16 h. (B) The shortened TLR2 promoter construct, pGL3-297wt TLR2, was used as a template to construct pGL3-297/NFm, in which the 3′ NF-κB consensus site was deleted, as indicated. The bar graph represents the luciferase activity of the wild type (wt), mutated constructs, and pure pGL3-basic vector (empty vector) normalized to untreated cells. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. (C) A549 cells were transiently cotransfected with the pGL3-297wt TLR2 reporter construct and the IκBα superrepressor expression plasmid that was ramped from 0 to 5 μg. The fold induction of luciferase activities by TNF-α, Dex, or both is shown. The luciferase activity of the pGL3-297 TLR2 construct in the presence of different concentrations of the IκBα superrepressor expression plasmid is shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test) to each control condition.
To further validate the role of NF-κB in the cooperative effect of TNF-α and Dex on TLR2 gene activation, we employed the use of the IκBα superrepressor of NF-κB signaling (30). This mutant IκB cannot be degraded by the proteosome and thus sequesters NFκB in the cytoplasm (10). A549 cells transiently coexpressing the pGL3-297 TLR2 promoter construct and the IκBα superrepressor cDNA in a range from 0 to 5 μg significantly inhibited the effect of TNF-α and/or Dex on the TLR2 promoter activity at all concentrations of transfected superrepressor (Fig. 4C). Together, these data indicate that the TLR2 gene activation by TNF-α alone or in combination with Dex requires both activation and translocation of NF-κB to the nucleus.
Role of the STAT consensus site in the cooperative effect of TNF-α and Dex on TLR2 gene activation.
We next evaluated the importance of the STAT consensus sequence on the cooperative effect of TNF-α and Dex on the regulation of the TLR2 promoter. Previously, an interactive mechanism had been reported for STAT and GR that promotes STAT phosphorylation and synergistic activation of the β-casein gene by prolactin and glucocorticoids (38). In that model system, a functional GRE in the β-casein gene was shown to be important but not essential for the synergistic effect of prolactin and glucocorticoids (38). Although the TLR2 promoter does not contain a canonical GRE consensus site, it does contain a degenerate half GRE in this region of the promoter. This element alone, however, is insufficient to induce TLR2 mRNA in response to glucocorticoids alone when reporter genes are transfected into A549 cells. To fully understand the basis for this regulation, we first determined if the STAT consensus site is necessary for the cooperative effect of TNF-α and Dex on TLR2 gene activation by using the pGL3-297 TLR2 promoter construct deletion mutant, which lacks the STAT consensus sequence (pGL3-Δ285-269). Deletion of the STAT site markedly reduced the magnitude of but did not abolish the TNF-α and Dex effect and slightly decreased the effect of TNF-α alone (Fig. 5A), suggesting that a STAT does play a role in the cooperation between these pro- and anti-inflammatory signaling molecules. As an alternative approach to this question, we investigated whether dimerization of STAT5 is critical for the cooperative action of TNF-α and Dex on TLR2 gene activation. A549 cells were cotransfected with the pGL3-297 TLR2 promoter construct and a STAT5b cDNA containing a point mutation at tyrosine 699 (STAT5b-Y699F), which is known to act as a dominant negative STAT due to its inability to dimerize with itself or with endogenous STATs (16). Coexpression of the dominant negative STAT5b significantly reduced the cooperative effect between TNF-α and Dex on the regulation of the TLR2 gene (Fig. 5B). Preliminary studies using immunocytochemistry approaches revealed that A549 cells contain STAT3, STAT5, and STAT6 but not STAT1 (data not shown), so that these molecules are the likely targets for the action of the dominant negative STAT.
FIG. 5.
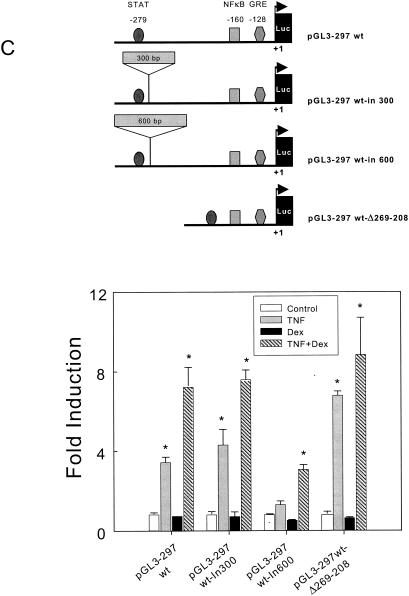
Analysis of the role of STAT transcription factor in the activation of the TLR2 promoter in A549 cells. (A) The TLR2 promoter construct pGL3-297 was used as the wild type to produce the deletion construct pGL3-297wt-Δ269-208, in which a specific deletion was conducted to remove the STAT-binding site, as indicated in Materials and Methods. The fold induction of luciferase by TNF-α, Dex, or both is shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. Luciferase activities of the wild type (wt), mutated constructs, and pGL3-basic vector are shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. (B) The pGL3-297wt TLR2 promoter construct was cotransfected with the STAT5b (Y699F) dominant negative expression plasmid, ramped from 0 to 5 μg. The fold induction of luciferase activities by TNF-α, Dex, or both is shown. Luciferase activities of all constructs and the pGL3-basic vector are shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. (C) Interaction between NF-κB and STAT transcriptional factor binding sites during TLR2 gene transcription. A diagram of the pGL3-297 TLR2 promoter construct used as the wild type and the insertion and deletion constructs is shown (top). The shortened luciferase reporter construct pGL3-297 TLR2 was used as a template to construct pGL3-297wt-in300 and pGL3-297wt-in600 (containing a 300- and a 600-bp fragment, respectively, inserted between the NF-κB and STAT sites). The pGL3-297Δ285-269 was obtained after deletion of the DNA fragment between the NF-κB and STAT sites. The fold induction of luciferase activities by TNF-α, Dex, or both is shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test) to each control condition.
Based on the data presented in the previous figures, it is clear that both NF-κB and STAT transcription factors are important in the transcriptional regulation of the TLR2 gene by the combination of TNF-α and Dex. To elucidate the mechanisms involved in this regulation, we created and utilized mutants where the spacing between the transcription factor-binding sites has been altered by introducing 300 and 600 bp of DNA. A detailed diagram of the mutations and design description is shown in Fig. 5C and detailed in Materials and Methods. When spacing between the NF-κB and STAT consensus binding sites was increased by 300 bp (pGL3-297wt-in300), no difference in the transcriptional activity of the TLR2 promoter mutant was observed when TNF-α alone or in combination with Dex was used to activate TLR2 transcription (Fig. 5C). In contrast, when the spacing between the two sites was increased by 600 bp (pGL3-297wt-in600), a significant reduction in the enhanced transcriptional activity of this TLR2 construct was observed when TNF-α alone or in combination with Dex was given to the cells (Fig. 5C). TNF-α alone still increased, albeit slightly, the transcriptional activity of this TLR2 mutant promoter construct, and when it was used in combination with Dex, a cooperative effect on the transcriptional activity was observed. Finally, when the spacing between the NF-κB and the STAT transcription factor-binding sites was reduced (pGL3-297wt-Δ269-208), the cells had an enhanced response to TNF-α and the cooperative effect of Dex was retained. Together, these results indicate that the spatial distribution of the transcription factor-binding sites for NF-κB and STAT is important for a composite transcriptional regulation of the TLR2 gene by TNF-α and Dex.
The AF-1 site of the GR and an intact GRE in the TLR2 promoter are required for the TNF-α- and Dex-induced cooperative effect on TLR2 transcriptional activation.
Transcriptional synergy of some genes, such as the casein gene, has been reported to involve interaction between STAT and GR transcription factors that results in STAT phosphorylation (38). The AF-1 domain in the N terminus of the GR is a critical region for the transcriptional synergy that takes place between prolactin and glucocorticoids in mammary cells (11, 31-33). To explore the role of the N-terminal domain of GR in the cooperative effect of TNF-α and Dex on TLR2 gene activation, we transiently cotransfected A549 cells with the pGL3-297 TLR2 promoter construct and either a wild-type version of GR (wtGR) or a deletion mutant with a mutation of the AF-1 region of the GR cDNA (GRΔ77-262). When the pGL3-297 TLR2 promoter construct was cotransfected with wtGR, an increased transcriptional activity was observed after treatment of the cells with Dex alone or in combination with TNF-α (Fig. 6A). The enhanced glucocorticoid response seen under these conditions probably reflects the overexpressing GR. In contrast, the mutant GR missing the AF-1 domain inhibited the expression of the pGL3-297 TLR2 promoter by TNF-α and Dex (Fig. 6A). This result indicates that the transcriptional cooperation of TLR2 gene induced by TNF-α and Dex requires an intact AF-1 domain of GR, which is also essential for interaction with STATs. Consistent with these data, we mutated the GRE-like element present in the construct by using both pGL3-1486wt and pGL3-297wt TLR2 promoter constructs to generate the GRE mutants (Fig. 6B, top panel) and showed that while both of these mutant constructs retained responsiveness to TNF-α, they both failed to show the cooperative effect between Dex and TNF-α. These results indicate that the GRE-like sequence present in the 3′ end of the TRL2 promoter is also required for transcription induced by TNF-α and Dex together, although it is not sufficient for Dex regulation alone.
FIG. 6.
Study of the role of the GR and the TLR2 GRE-like element in the cooperative effect between TNF-α and Dex on TLR2 gene activation. (A) The pGL3-297 TLR2 reporter construct was cotransfected with the full-length GR cDNA (wtGR) expression plasmid or with the AF-1 deletion mutant of the glucocorticoid receptor (GRΔ77-262) expression plasmid. The fold induction of luciferase activities by TNF-α, Dex, or both is shown. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. (B) The TLR2 promoter constructs pGL3-1486 and pGL3-297 were used as wild type to produce the mutant GRE construct (pGL3- 1486/GREm and pGL3-297/GREm). The X indicates that the GRE site has been mutated. The graph represents the luciferase activity induced by treatment by TNF-α, Dex, or both, after 16 h, of the wild-type and mutated constructs normalized to untreated cells. All samples were analyzed in duplicate, and the values are the mean ± SE of three experiments. *, P < 0.05 for pair comparison analysis (Tukey-Kramer test) to each control condition.
Dex regulates MKP-1 expression in A549 cells.
The anti-inflammatory actions of glucocorticoids have recently been suggested to be mediated via the regulation of MKP-1 in several model systems (15, 18, 29). Thus, it was important for us to determine if this regulation was occurring under the conditions of our experiments. We therefore examined the effect of Dex treatment on MKP-1 mRNA expression by using real-time PCR analysis. Dex treatment for 8 h, with increasing concentrations from 1 to 1,000 ng/ml, led to an increase in the level of MKP-1 mRNA (Fig. 7A). A549 cells were treated with 100 nM Dex and harvested at the indicated time points (0, 1, 2, 4, 6, 8, and 24 h). MKP-1 mRNA induction by Dex was time dependent, as shown in Fig. 7B. The Dex-induced effect on MKP-1 mRNA is probably being mediated via the classical GR, since RU486, a specific GR antagonist, was effective in repressing MKP-1 mRNA levels (Fig. 7C). The constitutively expressed transcript, cyclophilin B, was also evaluated in parallel for each sample, and MKP-1 mRNA expression was normalized to cyclophilin B mRNA expression, since no significant changes were observed for this gene with Dex treatment. Asterisks denote a significant increase between untreated and Dex-treated cultures as determined by Tukey-Kramer pair comparison analysis (P < 0.05).
Considering our results for the expression of MKP-1 mRNA by Dex, we wanted to determine if these changes in mRNA were also reflected by alterations in the levels of MKP-1 protein. To undertake these investigations, we treated A549 cells with 100 nM Dex and/or 1 μM RU486 for a total of 8 h. Endogenous MKP-1 protein was then immunoprecipitated from cells lysates by using an anti-MKP-1 antibody. Treatment of cells with Dex induced an increase in the amount of immunoprecipitated MKP-1 (Fig. 7D), whereas cotreatment of the cells with Dex and RU486 reversed the immunoreactive MKP-1 protein increase induced by Dex (Fig. 7D). The densitometric analysis of the immunoprecipitated bands after Dex treatment revealed a fourfold increase in the amount of MKP-1 protein (Fig. 7D, graph). These findings indicate that Dex is also affecting MKP-1 protein under conditions where it is increasing TLR2 in the same cells.
NF-κB, STAT, and GR occupancy in the endogenous and transfected TLR2 promoters.
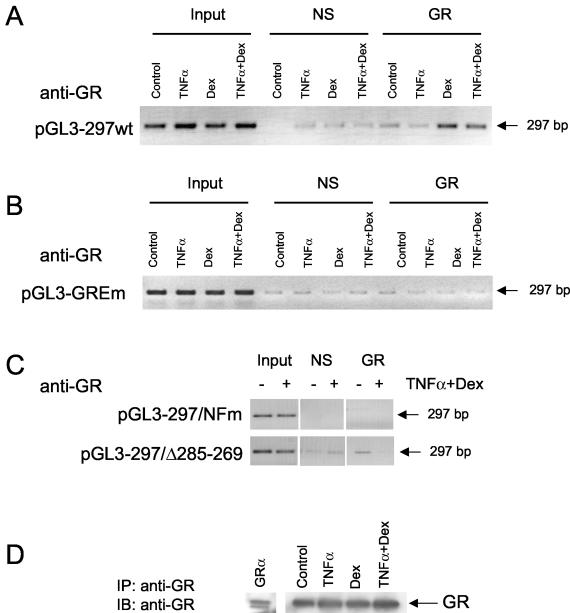
We next examined the ability of endogenous GR, p65, and STAT5 to interact with both the endogenous TLR promoter and wild-type or mutant versions of pGL3-297wt TLR2 transfected promoter construct in A549 cells. The goal of these ChIP assay experiments is to determine if the NF-κB and the STAT transcription factor-binding sites in this region of the promoter sequester the GR to the GRE-like element in the TLR2 promoter. The ChIP assay was first conducted to determine GR, p65, and STAT5 occupancy of the endogenous TLR2 promoter. Minimal occupancy of the TLR2 promoter by GR was detected in the absence of hormone or when TNF-α was added alone to the untransfected cells (fig. 8, top gel). However, treatment with Dex alone or TNF-α plus Dex resulted in a substantial increase of GR recruitment to the TLR2 promoter (top gel). When the endogenous TLR2 promoter occupancy by p65 or STAT was examined, minimal amplification was detected in the absence of glucocorticoid or when Dex was added (middle and bottom gels). However, treatment with TNF-α alone or TNF-α plus Dex resulted in a substantial increase of p65 or STAT recruitment to the TLR2 promoter (middle and bottom gels). These data for the endogenous TLR2 promoter suggest that all three transcription factors can be found at the promoter following treatment with TNF-α and Dex in combination but not with either Dex or TNF alone.
FIG. 8.
Study of transcription factor occupancy of the TLR2 promoter after TNF-α and Dex treatment. A549 cells were lysed and analyzed for the ChIP assay. After a 1-h treatment with TNF-α and/or Dex, cells were sonicated and immunoprecipitated with specific antibodies. Thereafter, the immunoprecipitated TLR2 promoter was identified by PCR. Endogenous GR, p65, and STAT5 binding to the endogenous TLR2 promoter was explored after treatment with TNF-α and/or Dex. The PCR amplification product from immunoprecipitated endogenous TLR2 promoter from A549 cells using the specific GR antibody Ab57 (anti-GR), the antibody against p65 (anti-p65), or the anti-STAT5 antibody (anti-STAT5) is shown.
We next sought to extend these observations by evaluating the occupancy by a ChIP assay of the wild-type and mutant TLR2 promoters used in our transfection assays. When we examined the overexpressed TLR2 promoter mutants, we detected minimal occupancy of the TLR2 promoter by GR in the absence of hormone or when TNF-α was added alone to the cells (Fig. 9A). However, treatment with Dex alone or with TNF-α plus Dex resulted in a substantial increase in recruitment of GR to the TLR2 promoter (Fig. 9A). Mutation of the GRE-like element within this construct destroyed the binding of Dex, alone or in combination with TNF-α, to this fragment (Fig. 9B). Similarly, no binding of GR to either pGL3-297/NFm or pGL3-297wtΔ285-269 TLR2 promoter amplified by PCR was observed (Fig. 9C). A representative immunoblot from immunoprecipitated endogenous GR revealed no differences between the treatments (Fig. 9D). A transiently transfected hGRα construct in Cos cells was used as the control (Fig. 9D, lane GRα). These results highlight the fact that endogenous GR is recruited to the exogenously expressed TLR2 promoter in response to a brief exposure to TNF-α and/or Dex. Moreover, the NF-κB and STAT transcription factor and the GRE-like element are all critical for this interaction.
FIG. 9.
GR association with overexpressed TLR2 promoter mutants after TNF-α and Dex treatment. Transfected A549 cells were lysed and analyzed for the ChIP assay. After a 1-h treatment with TNF-α and/or Dex, cells were sonicated and immunoprecipitated with the specific anti-GR antibody, Ab57. Thereafter, the immunoprecipitated TLR2 promoter mutants was identified by PCR. (A) PCR amplification product from immunoprecipitated A549 cells transiently expressing the pGL3-297wt TLR2 promoter constructs after TNF-α and/or Dex treatment. (B) PCR Amplification products from immunoprecipitated A549 cells transiently expressing the pGL3-297GREm TLR2 promoter constructs after TNF-α and/or Dex treatment. (C) PCR amplicon from immunoprecipitated A549 cells transiently expressing the pGL3-297/NFm or the pGL3-297Δ285-269 TLR2 promoter mutants. (D) The immunoprecipitated (IP) GR content was similar for all the treatment conditions. A transiently transfected hGRα construct was used as the control (first lane). IB, western blotting.
DISCUSSION
The airway epithelium is one of the first lines of defense against invasion by airborne pathogens, such as endotoxins. These toxins are recognized by PRR, represented in mammals by the TLRs. The activation of the TLRs triggers proinflammatory reactions that induce the production of proinflammatory cytokines such as TNF-α, and ILs, such as IL-8. These cytokines produced at the site of inflammation in turn activate macrophages and lymphocytes, which also activate the hypothalamic-pituitary axis to induce the synthesis and release of anti-inflammatory glucocorticoids. This feedback system provides fine homeostatic control over inflammation and prohibits an overresponse to inflammatory signals. The cellular mechanisms that regulate these processes are multifaceted and highly conserved among cells. Similarly, pathogenic endotoxins are also known to increase the expression of GRs, which sensitizes the cells to glucocorticoids, thereby counteracting the effect of proinflammatory molecules such as TNF-α and the induction of ILs. The interaction between glucocorticoids and cytokines is often cell type specific and depends on the physiologic context of the cell. The antagonizing effect of glucocorticoids on TNF-α-induced proinflammatory reactions involves the interaction between the GR and proinflammatory transcription factors bound to the promoter region of different cytokine and proinflammatory genes. In contrast, cooperative regulation of the TLR2 gene at both the mRNA and protein levels in response to TNF-α and Dex, as shown here, represents a novel signaling mechanism that involves the recruitment of NF-κB and STAT transcription factors as well as the GR.
A significant increase in TLR2 expression was observed after TNF-α treatment of A549 cells, consistent with observation made with activated lymphocytes and macrophages, where the TLR2 is important for the recognition pattern of gram-positive bacteria (19). In this regard, different cell types up-regulate TLR2 at the protein and transcriptional levels in response to bacterial products including TNF-α and other cytokines (7, 19, 23, 24). Glucocorticoids have been widely used as anti-inflammatory agents that down-regulate NF-κB-dependent gene transcription. However, we show here that NF-κB is required for TNF-α-regulated expression of the TLR2 gene and, further, that glucocorticoids enhance this effect in the same cells where it represses the production of IL-8 mRNA and induces MKP-1. This result is consistent with recent work performed with macrophages, where glucocorticoids were shown to induce TLR2, mRNA, and protein (29), although such mechanisms do not appear to prevail in hepatocytes (21).
Because of our findings that Dex cooperatively enhances TNF-α-induced TLR2 up-regulation at the level of mRNA and protein in A549 cells via the activation of GR, the potential of these molecules to regulate the transcription of the TLR2 gene was investigated. A cooperative enhancement of the TNF-α-induced increase in TLR2 promoter activity was observed when Dex was administered with the proinflammatory molecules. Similarly, a synergistic role of Dex in the increase of TLR2 expression mediated by H. influenzae endotoxins has been suggested; although the mechanisms appear to involve posttranscriptional regulation via MAPK (29) or MKP-1 (3). These data are consistent with our findings and other recent reports showing that glucocorticoids increase MKP-1 expression, thus providing the potential to inhibit MAPK p38 activity and be anti-inflammatory (18). However, these results do not exclude other potential mechanisms for the regulation of TLR2 by TNF-α and glucocorticoids. Here we demonstrate that TNF-α and Dex do activate unique intracellular mechanisms that promote a transcription of the TLR2 gene, although other mechanisms of control exist since Dex alone does not appear to regulate transfected reporter genes in these cells. Our data indicate that TNF-α and Dex induce NF-κB and STAT binding to their respective consensus sequences in the TLR2 promoter. The lack of direct regulation of this promoter by glucocorticoids alone suggests that the single GRE found in this region is not a bona fide GRE, although it is necessary for the cooperation between TNF-α and Dex with regard to the expression of the TLR2 gene regulation. In addition, the presence of the GRE-like element, which alone is insufficient to cause glucocorticoid regulation of TLR2 gene, indicates that novel mechanisms exist for regulation by these three transcription factors. The cooperation between NF-κB and the STATs is reminiscent of TNF-α and gamma interferon synergy that occurs on the transcriptional activation of the MUC1 gene in human breast cells (17). GRs also appear to be contributing to the effect of TNF-α and Dex on the TLR2 gene, since removal of the AF-1 region of human GR induced a dose-dependent inhibition of the synergistic response of TNF-α and Dex. Previous studies have shown that the AF-1 region in the N terminal of GR is necessary for the transcriptional synergy between GR and STAT5b (38). Another example of such synergistic action is the induction of the β-casein gene by glucocorticoids and prolactin (PRL) (31, 33). After PRL binding to the PRL receptor (PRL-R) and dimerization of the PRL-R, the Jak/STAT signal transduction cascade becomes active. After this activation of the Jak/STAT signaling cascade, STAT5 translocates into the nucleus to bind DNA consensus sites, where it interacts with GR (31). In conclusion, our finding that endogenous GR is recruited to the endogenous TLR2 promoter in response to TNF-α and Dex and that this interaction involves the STAT and NF-κB transcription elements as well as a GRE-like element supports the hypothesis that these three transcription factors are involved in the cooperative effects of TNF-α and Dex on TLR2 gene activation.
REFERENCES
- 1.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 2.An, H., Y. Yu, M. Zhang, H. Xu, R. Qi, X. Yan, S. Liu, W. Wang, Z. Guo, J. Guo, Z. Qin, and X. Cao. 2002. Involvement of ERK, p38 and NF-kappaB signal transduction in regulation of TLR2, TLR4 and TLR9 gene expression induced by lipopolysaccharide in mouse dendritic cells. Immunology 106:38-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 4.Chrousos, G. P. 1995. The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. N. Engl. J. Med. 332:1351-1362. [DOI] [PubMed] [Google Scholar]
- 5.Chuang, T., and R. J. Ulevitch. 2001. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim. Biophys. Acta 1518:157-161. [DOI] [PubMed] [Google Scholar]
- 6.Cidlowski, J. A., D. L. Bellingham, F. E. Powell-Oliver, D. B. Lubahn, and M. Sar. 1990. Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Mol. Endocrinol. 4:1427-1437. [DOI] [PubMed] [Google Scholar]
- 7.Faure, E., L. Thomas, H. Xu, A. Medvedev, O. Equils, and M. Arditi. 2001. Bacterial lipopolysaccharide and IFN-gamma induce Toll-like receptor 2 and Toll-like receptor 4 expression in human endothelial cells: role of NF-kappa B activation. J. Immunol. 166:2018-2024. [DOI] [PubMed] [Google Scholar]
- 8.Flo, T. H., O. Halaas, S. Torp, L. Ryan, E. Lien, B. Dybdahl, A. Sundan, and T. Espevik. 2001. Differential expression of Toll-like receptor 2 in human cells. J. Leukoc. Biol. 69:474-481. [PubMed] [Google Scholar]
- 9.Galon, J., D. Franchimont, N. Hiroi, G. Frey, A. Boettner, M. Ehrhart-Bornstein, J. J. O'Shea, G. P. Chrousos, and S. R. Bornstein. 2002. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J. 16:61-71. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109 Suppl:S81-S96. [DOI] [PubMed] [Google Scholar]
- 11.Groner, B. 2002. Transcription factor regulation in mammary epithelial cells. Domest. Anim. Endocrinol. 23:25-32. [DOI] [PubMed] [Google Scholar]
- 12.Heck, S., M. Kullmann, A. Gast, H. Ponta, H. J. Rahmsdorf, P. Herrlich, and A. C. Cato. 1994. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 13:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 15.Imasato, A., C. Desbois-Mouthon, J. Han, H. Kai, A. C. Cato, S. Akira, and J. D. Li. 2002. Inhibition of p38 MAPK by glucocorticoids via induction of MAPK phosphatase-1 enhances nontypeable Haemophilus influenzae-induced expression of toll-like receptor 2. J. Biol. Chem. 277:47444-47450. [DOI] [PubMed] [Google Scholar]
- 16.Kabotyanski, E. B., and J. M. Rosen. 2003. Signal transduction pathways regulated by prolactin and Src result in different conformations of activated Stat5b. J. Biol. Chem. 278:17218-17827. [DOI] [PubMed] [Google Scholar]
- 17.Lagow, E. L., and D. D. Carson. 2002. Synergistic stimulation of MUC1 expression in normal breast epithelia and breast cancer cells by interferon-gamma and tumor necrosis factor-alpha. J. Cell. Biochem. 86:759-772. [DOI] [PubMed] [Google Scholar]
- 18.Lasa, M., S. M. Abraham, C. Boucheron, J. Saklatvala, and A. R. Clark. 2002. Dexamethasone causes sustained expression of mitogen-activated protein kinase (MAPK) phosphatase 1 and phosphatase-mediated inhibition of MAPK p38. Mol. Cell. Biol. 22:7802-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, Y., H. Lee, A. H. Berg, M. P. Lisanti, L. Shapiro, and P. E. Scherer. 2000. The lipopolysaccharide-activated toll-like receptor (TLR)-4 induces synthesis of the closely related receptor TLR-2 in adipocytes. J. Biol. Chem. 275:24255-24263. [DOI] [PubMed] [Google Scholar]
- 20.Matsuguchi, T., T. Musikacharoen, T. Ogawa, and Y. Yoshikai. 2000. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J. Immunol. 165:5767-5772. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura, T., A. Ito, T. Takii, H. Hayashi, and K. Onozaki. 2000. Endotoxin and cytokine regulation of toll-like receptor (TLR) 2 and TLR4 gene expression in murine liver and hepatocytes. J. Interferon Cytokine Res. 20:915-921. [DOI] [PubMed] [Google Scholar]
- 22.McKay, L. I., and J. A. Cidlowski. 1998. Cross-talk between nuclear factor-kappa B and the steroid hormone receptors: mechanisms of mutual antagonism. Mol. Endocrinol. 12:45-56. [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov, R., and C. A. Janeway, Jr. 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91:295-298. [DOI] [PubMed] [Google Scholar]
- 24.Musikacharoen, T., T. Matsuguchi, T. Kikuchi, and Y. Yoshikai. 2001. NF-kappa B and STAT5 play important roles in the regulation of mouse Toll-like receptor 2 gene expression. J. Immunol. 166:4516-4524. [DOI] [PubMed] [Google Scholar]
- 25.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheinman, R. I., P. C. Cogswell, A. K. Lofquist, and A. S. Baldwin, Jr. 1995. Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science 270:283-286. [DOI] [PubMed] [Google Scholar]
- 29.Shuto, T., A. Imasato, H. Jono, A. Sakai, H. Xu, T. Watanabe, D. D. Rixter, H. Kai, A. Andalibi, F. Linthicum, Y. L. Guan, J. Han, A. C. Cato, D. J. Lim, S. Akira, and J. D. Li. 2002. Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced Toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J. Biol. Chem. 277:17263-17270. [DOI] [PubMed] [Google Scholar]
- 30.Stein, B., A. S. Baldwin, Jr., D. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 12:3879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stocklin, E., M. Wissler, F. Gouilleux, and B. Groner. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726-728. [DOI] [PubMed] [Google Scholar]
- 32.Stoecklin, E., M. Wissler, R. Moriggl, and B. Groner. 1997. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 17:6708-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoecklin, E., M. Wissler, D. Schaetzle, E. Pfitzner, and B. Groner. 1999. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. J. Steroid Biochem. Mol. Biol. 69:195-204. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, O., T. Kawai, H. Sanjo, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. Takeda, and S. Akira. 1999. TLR6: A novel member of an expanding toll-like receptor family. Gene 231:59-65. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Q., R. Dziarski, C. J. Kirschning, M. Muzio, and D. Gupta. 2001. Micrococci and peptidoglycan activate TLR2→MyD88→IRAK→TRAF→NIK→IKK→NF-kappaB signal transduction pathway that induces transcription of interleukin-8. Infect. Immun. 69:2270-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilder, R. L. 1995. Neuroendocrine-immune system interactions and autoimmunity. Annu. Rev. Immunol. 13:307-338. [DOI] [PubMed] [Google Scholar]
- 38.Wyszomierski, S. L., J. Yeh, and J. M. Rosen. 1999. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13:330-343. [DOI] [PubMed] [Google Scholar]