Abstract
Background
The single nucleotide polymorphism 5p12-rs10941679has been found to be associated with risk of breast cancer, particularly estrogen receptor (ER)-positive disease. We aimed to further explore this association overall, and by tumor histopathology, in the Breast Cancer Association Consortium.
Methods
Data were combined from 37 studies, including 40,972 invasive cases, 1,398 cases of ductal carcinoma in situ (DCIS) and 46,334 controls, all of white European ancestry, as well as 3,007 invasive cases and 2,337 controls of Asian ancestry. Associations overall and by tumor invasiveness and histopathology were assessed using logistic regression.
Results
For white Europeans, the per-allele odds ratio (OR) associated with 5p12-rs10941679 was 1.11 (95% confidence interval [CI] =1.08–1.14, P=7×10−18) for invasive breast cancer and 1.10 (95%CI=1.01–1.21, P=0.03) for DCIS. For Asian women, the estimated OR for invasive disease was similar (OR=1.07, 95%CI=0.99–1.15, P=0.09). Further analyses suggested that the association in white Europeans was largely limited to progesterone receptor (PR)-positive disease (per-allele OR=1.16, 95%CI=1.12–1.20, P=1×10−18 versus OR=1.03, 95%CI=0.99–1.07, P=0.2 for PR-negative disease; P-heterogeneity=2×10−7); heterogeneity by estrogen receptor status was not observed (P=0.2) once PR status was accounted for. The association was also stronger for lower-grade tumors (per-allele OR [95%CI]=1.20 [1.14–1.25], 1.13 [1.09–1.16] and 1.04 [0.99–1.08] for grade 1, 2 and 3/4, respectively; P–trend=5×10−7).
Conclusion
5p12 is a breast cancer susceptibility locus for PR-positive, lower gradebreast cancer.
Impact
Multi-centre fine-mapping studies of this region are needed as a first step to identifying the causal variant or variants.
Keywords: Breast cancer, SNP, susceptibility, disease subtypes
Introduction
Genome-wide association studies (GWAS) have identified several single-nucleotide polymorphisms (SNPs) associated with breast cancer risk. Key to these findings, and to the more precise estimation of the associated relative risks, has been their replication in large independent case-control series. SNPs in or close to the genes LSP1, MAP3K1, FGFR2, TOX3, MRPS30, COX 11, MRPS30, and SLC4A7, and in chromosomal regions 8p24 and 2q35 have all been replicated in the Breast Cancer Association Consortium (BCAC) (1–4)
An Icelandic GWAS and replication study of 5,028 breast cancer cases and 32,090 controls revealed multiple signals in the 5p12 chromosomal region (5). The most strongly associated SNP was rs10941679, the minor G allele being associated with an estimated per-allele odds ratio (OR) of 1.19 (95% confidence interval [CI]=1.11–1.26, P=2.9×10−11). The authors reported that the increased risk was particularly marked for estrogen receptor (ER)-positive disease (N=2,726 cases, per-allele OR=1.27, 95%CI=1.19–1.35, P=2.5×10−12; P-heterogeneity=0.004) (5). In addition, our previous breast cancer GWAS reported evidence of an association with another SNP on 5p12, rs981782 (P=9×10−6), based on data from 23,408 cases and 24,636 controls from 21 BCAC studies (2). The Icelandic-led study evaluated both these SNPs in multivariable models and observed that only 5p12-rs10941679 was independently associated with breast cancer risk (5). This SNP was not among those genotyped in the first phase of our previous GWAS (2).
The aims of the present study were to estimate the relative risk of breast cancer associated with 5p12-rs10941679 in a much larger case-control series comprising studies participating in the BCAC, to evaluate its independence of any association with 5p12-rs981782, and to assess associations by disease subtypes defined by histopathology.
Materials and Methods
Thirty-seven studies from Europe, North America, Australia, and Asia participated in genotyping for the present study via the BCAC, contributing a total of 41,243 invasive breast cancer cases, 1,406 cases of ductal carcinoma in-situ (DCIS) and 46,621 controls of white European origin and 3,082 invasive cases and 2,402 controls of Asian origin. All 37 studies genotyped 5p12-rs10941679. Seven studies also genotyped 5p12-rs981782 in a total of 8,247 invasive cases and 10,363 controls, all of European origin. Fourteen of the 37 participating studies had already genotyped 5p12-rs981782 in white Europeans as part of the previously published study (2). Descriptions of studies are provided in Supplemental Table 1 and final sample sizes are presented in Supplemental Table 2.
All studies provided information on disease status and self-reported race/ethnicity for all subjects and age at diagnosis for cases. All but five studies (BIGGS, FBCS, HUBCS, RBCS and UCIBCS) also provided age at data collection for controls. “Selected cases” were a subset of 7,128 invasive cases selected for inclusion or oversampled by nine studies because they had bilateral breast cancer, a family history of breast cancer and/or other characteristics that suggested they were at increased genetic risk (all cases from BBCS, FBCS, GC-HBOC, kConFab/AOCS, MBCSG, NC-BCFR, OFBCR and RBCS and 211 cases from CNIO-BCS, see Supplemental Table 1). ER and progesterone receptor (PR) status were provided for a subset of cases, respectively, as were human epidermal growth factor receptor 2 (HER2) status and other histological features including axillary node status and tumor grade, size, and morphology (see Supplemental Table 3). This histopathology information was generally abstracted from medical reports.
Subjects who reported having ethnicity other than white European were excluded, with the exception of those from the three Asian studies, for which only subjects of Asian origin were included. Only subjects from studies that genotyped at least 30 cases of DCIS were included in the analyses of risk of DCIS. All subjects gave written informed consent, where applicable, and each study was approved by the relevant local institutional review boards.
Most studies carried out genotyping using Taqman nuclease assay (Taqman®), with reagents designed by Applied Biosystems as Assays-by-Design™ and genotyping performed using the ABI PRISM 7900HT, 7700 or 7500 Sequence Detection Systems according to manufacturer’s instructions. Three studies used Sequenom’s MassARRAY system and iPLEX technology (Sequenom, San Diego, CA, USA), with oligonucleotides design carried out according to the guidelines of Sequenom and performed using MassARRAY Assay Design software (version 3.1). The method used by each study is identified in Supplemental Table 2. All studies complied with BCAC genotyping quality control (QC) standards by including at least 2% of samples in duplicate and a common set of 93 CEPH DNAs used by the HapMap Consortium (HAPMAPPT01, Coriell Institute for Medical Research, Cambden, NJ).
Statistical Methods
Departure from Hardy-Weinberg equilibrium (HWE) was tested for in controls from each centre using Pearson/s χ2 test (1df). The association of each SNP with breast cancer risk was assessed by estimating genotype-specific and per-allele ORs using logistic regression, adjusted for study. The exclusion of studies for which the age of controls was not known and the additional adjustment for age (in 5-year categories and as a continuous covariate) made no substantial difference to the results. Between-study heterogeneity in ORs was assessed using a likelihood ratio test (LRT) comparing the model with interaction terms for the per-allele log-OR by study to the model with no interaction terms. Differences in ORs by ethnicity (white European, Asian) and age (<40, 40–49, 50–59, 60–69, ≥70 years) were evaluated using a similar LRT, the latter modeled as a linear trend by fitting the median age for each of the defined categories.
ORs specific to disease subtypes defined by ER, PR and HER2 status (positive, negative), by combinations of these markers, and by axillary node status (none, ≥1 affected), tumor grade (1, 2, ≥3), tumor size (≤10, 11–20, >20mm) and tumor morphology (ductal, lobular), were estimated for white Europeans using polytomous logistic regression with control status as the reference outcome. Heterogeneity in the OR by subtypes was tested for by applying polytomous logistic regression to cases-only, treating the number of minor alleles as the outcome and restricting, for each explanatory variable, the beta coefficient for the comparison of 2 to 0 minor alleles to be double that for the comparison of 1 to 0 minor alleles. This is equivalent to modeling a log-additive per-allele OR and allows multiple tumor markers to be modeled simultaneously. Linear trends were tested for grade by fitting values 1, 2, and 3, and for size by fitting the median value, for the defined categories, respectively. Enrichment of the risk allele in “selected cases” was assessed using the likelihood ratio test comparing polytomous logistic regression models with and without the per-allele OR constrained to be equal for selected and unselected cases, relative to controls (1df). We minimised bias in the estimation of OR by repeating all analyses after excluding “selected cases”. All statistical tests were two-sided. The term “genome-wide” statistically significant is taken to imply p<10−7; otherwise “statistically significant” implies p<0.05. All analyses were carried out using Stata: Release 10 (6).
Meta-analyses were carried out using metan command based on log-transformed OR estimates and their 95%CI from different reports.
Results
Minimum genotype concordance of 98% for duplicate samples and 95% for the CEPH samples was observed in all studies, as were minimum genotype calls of 97% for both SNPs in cases and controls. Evidence of departure from HWE was observed for 5p12-rs10941679 in controls from two studies (FBCS [P = 0.02] and HABCS [P=0.005], Supplemental Table 2); for both studies, cluster plots were double-checked visually and determined to be of high quality, and all their genotype data were therefore included in the final analysis.
Thirty-four studies successfully genotyped 5p12-rs10941679 in a total of 40,972 invasive breast cancer cases (7,037 of which were selected for increased genetic risk), 1,398 DCIS cases, and 46,334 controls of white European origin. Genotypes were also obtained for this SNP for 3,007 cases and 2,337 controls of Asian origin recruited by three studies. A total of 8,213 invasive cases and 10,340 controls were successfully genotyped for 5p12-rs981782 by seven studies. Previously published data for this SNP were included from 14 studies to obtain genotypes for both SNPs in 5p12 for 23,548 invasive cases and 28,142 controls, all of white European origin. Genotype counts by study are provided in Supplemental Table 4.
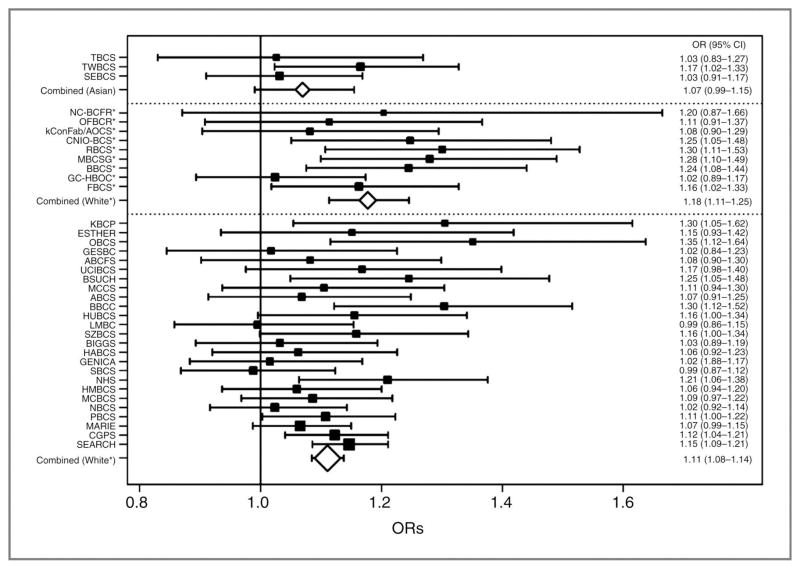
The minor G allele of 5p12-rs10941679 was less frequent in white European women (26%) than in Asian women (49%). Estimated ORs, for invasive breast cancer and DCIS, and by ethnicity, are presented in Table 1. The genotype-specific OR estimates were consistent with a log-additive model for white Europeans. The per-G-allele OR estimate was 1.12 (95%CI=1.10–1.15, P=6×10−24), and there was no evidence of heterogeneity in the OR among studies overall (P=0.1) or among studies of Asians (P=0.3), studies of white Europeans that included “selected cases” (P=0.3) or studies of white Europeans without “selected cases” (P=0.2) (Figure 1). This association was maintained at genome-wide statistical significance when SNP 5p12-rs981782 was included as a covariate in the logistic regression model (OR=1.11, 95%CI=1.09–1.15, P=6×10−13). For 5p12-rs10941679 the estimated per-G-allele OR for DCIS was similar to that for invasive disease (OR=1.10, 95%CI=1.01–1.21, P=0.03). The per-G-allele OR for invasive breast cancer did not appear to be different for Asian women (P-heterogeneity=0.3), but the estimate was lower and not statistically significant (1.07; 95%CI=0.99–1.15, P=0.09). All further analyses were based on white European women only.
Table 1.
SNP 5p12-rs10941679 and risk of breast cancer
| Group/invasiveness/genotype | Controls, N (%) | Cases, N (%) | OR (95%CI) | P |
|---|---|---|---|---|
| White European women | ||||
| Invasive disease (34 studies) | ||||
| AA | 25,622 (55) | 21,241 (52) | 1.00 | |
| AG | 17,668 (38) | 16,671 (41) | 1.14 (1.11–1.17) | 9×10−19 |
| GG | 3,044 (6.6) | 3,060 (7.5) | 1.22 (1.15–1.29) | 2×10−12 |
| per G-allele | 1.12 (1.10–1.15) | 6×10−24 | ||
| Adjusted for 5p12-rs981782* | ||||
| AA | 15,648 (56) | 12,243 (52) | 1.00 | |
| AG | 10,684 (38) | 9,598 (41) | 1.14 (1.10–1.18) | 2×10−11 |
| GG | 1,810 (6.4) | 1,707 (7.3) | 1.19 (1.11–1.28) | 3×10−6 |
| per G-allele | 1.11 (1.09–1.15) | 6×10−13 | ||
|
| ||||
| White European women | ||||
| DCIS (15 studies) | ||||
| AA | 14,187 (55) | 726 (52) | 1.00 | |
| AG | 9,733 (38) | 578 (41) | 1.17 (1.04–1.31) | 0.009 |
| GG | 1,662 (6.5) | 94 (6.7) | 1.10 (0.88–1.38) | 0.4 |
| per G-allele | 1.10 (1.01–1.21) | 0.03 | ||
|
| ||||
| Asian women | ||||
| Invasive disease (3 studies) | ||||
| AA | 612 (26) | 765 (25) | 1.00 | |
| AG | 1,155 (49) | 1,460 (49) | 1.03 (0.90–1.18) | 0.6 |
| GG | 570 (24) | 782 (26) | 1.14 (0.98–1.33) | 0.09 |
| per G-allele | 1.07 (0.99–1.15) | 0.09 | ||
OR, odds ratio; CI, confidence interval; DCIS, ductal carcinoma in situ
SNP 5p12-981782 included under a codominant model (2df), based on data on both variants available from 19 studies, including previously published data on 5p12-rs198782 (2).
Figure 1. Association of 5p12-rs10941679 with risk of invasive breast cancer, by study.
Per-allele odds ratios (OR) and 95% confidence intervals (CI), with studies grouped into those that recruited Asian women and white European women. The latter group is further divided into those including “selected cases” likely to be at higher genetic risk (marked by an asterisk) and those with unselected cases. The area of the box/diamond is inversely proportional to the standard error of the log OR estimate.
We observed weak evidence that the G allele of 5p12-rs10941679 was enriched in “selected cases” (P=0.06). The estimated per-allele OR was 1.18 (95%CI=1.11–1.24, P=1×10−8) when comparing cases selected for increased genetic risk to controls from the same studies and 1.11 (95%CI=1.08–1.14, P=7×10−18) when they were excluded. There was evidence of an increase in the per-allele OR associated with 5p12-rs10941679 with age (P=0.01), with estimates of 1.10 (95%CI=1.01–1.19), 1.08 (95%CI=1.02–1.15), 1.14 (95%CI=1.09–1.19), 1.12 (1.07–1.17) and 1.16 (95%CI=1.06–1.26) for white European women aged <40, 40–49, 50–59, 60–69 and ≥70, respectively. This trend was also observed when age was modeled in years (P=0.02), with an estimated interaction OR of 1.02 (95%CI=1.00–1.04) per G allele, per 10-year increase in age. The same trend was observed after excluding “selected cases” (interaction OR=1.02, 95%CI=1.00–1.05, P=0.03).
Results from analyses by breast cancer subtypes defined by histopathological features are presented in Tables 2 and 3. The number of cases from each study for which this information was available is presented in Supplemental Table 3. We observed strong evidence that the per-allele OR associated with 5p12-rs10941679 differed by ER status, PR status and ER/PR combined (all P<10−5, Table 2). When ER and PR status were modeled together in the case-only analysis only the association with PR status was maintained (P=6×10−4, compared to P=0.2 for ER status). The OR estimates by combined ER/PR status were consistent with the heterogeneity being present by PR rather than ER status. Furthermore, heterogeneity in the OR by PR status was observed when only cases with ER-positive disease were considered (P=0.005), but not by ER status when only cases with PR-positive disease were considered (P=0.8). After excluding “selected cases”, the per-G-allele OR estimates were 1.03 (95%CI=0.99–1.07, P=0.2) for PR-negative disease and 1.16 (95%CI=1.12–1.20, P=1×10−18) for PR-positive disease. We observed no further heterogeneity by HER2 status (P=0.3). The trend of increasing per-allele OR with age remained apparent when only the risk of PR-positive disease was considered (interaction OR=1.03, 95%CI=1.00–1.06, P=0.05).
Table 2.
SNP 5p12-rs10941679 and risk of invasive breast cancer for white Europeans, by ER, PR and HER2 status
| N Participants | Log-additive Model | Case-only P-heterogeneity | |||
|---|---|---|---|---|---|
|
| |||||
| Cases | Controls | OR (95%CI) | P | ||
| ER Status (29 studies) | |||||
| ER+ | 20,044 | 41,436 | 1.15 (1.12–1.19) | 7×10−24 | |
| ER− | 6,201 | 41,436 | 1.04 (0.99–1.08) | 0.1 | 4×10−6 |
|
| |||||
| PR Status (28 studies) | |||||
| PR+ | 14,557 | 40,577 | 1.16 (1.12–1.19) | 8×10−20 | |
| PR− | 7,744 | 40,577 | 1.04 (1.00–1.08) | 0.08 | 5×10−7 |
|
| |||||
| ER/PR Status (28 studies) | |||||
| ER+/PR+ | 13,517 | 40,577 | 1.16 (1.12–1.20) | 3×10−19 | |
| ER+/PR− | 3,104 | 40,577 | 1.06 (1.00–1.13) | 0.04 | |
| ER−/PR+ | 960 | 40,577 | 1.13 (1.02–1.26) | 0.02 | |
| ER−/PR− | 4,558 | 40,577 | 1.02 (0.97–1.07) | 0.5 | 6×10−6 |
|
| |||||
| ER, PR and HER2 Status (16 studies) | |||||
| (ER+ or PR+)&HER2− | 8,078 | 25,726 | 1.15 (1.10–1.20) | 2×10−11 | |
| (ER+ or PR+)&HER2+ | 1,188 | 25,726 | 1.11 (1.02–1.22) | 0.02 | |
| ER−/PR−/HER2− | 1,641 | 25,726 | 1.04 (0.95–1.12) | 0.4 | |
| ER−/PR−/HER2+ | 750 | 25,726 | 0.93 (0.83–1.05) | 0.3 | |
OR, odds ratio; CI, confidence interval; ER, estrogen receptor; PR progesterone receptor; HER2, human epidermal growth factor receptor 2
Table 3.
SNP 5p12-rs10941679 and risk of invasive breast cancer for white Europeans, by pathology characteristics
| N Participants | Log-additive Model | Case-only P-heterogeneity* | |||
|---|---|---|---|---|---|
|
| |||||
| Cases | Controls | OR (95%CI) | P | ||
| Axillary node status (27 studies) | |||||
| Negative | 16,566 | 39,148 | 1.12 (1.09–1.15) | 3×10−13 | |
| Positive | 9,762 | 39,148 | 1.13 (1.09–1.17) | 1×10−10 | 0.9 |
|
| |||||
| Grade (26 studies) | |||||
| 1 | 5,032 | 38,601 | 1.20 (1.14–1.25) | 1×10−13 | |
| 2 | 12,204 | 38,601 | 1.13 (1.09–1.17) | 1×10−12 | |
| 3/4 | 7,802 | 38,601 | 1.04 (1.00–1.08) | 0.05 | 5×10−7 |
|
| |||||
| Size (19 studies) | |||||
| ≤10mm | 3,707 | 28,102 | 1.16 (1.09–1.22) | 3×10−7 | |
| 11–20mm | 7,964 | 28,102 | 1.11 (1.07–1.16) | 1×10−7 | |
| >20mm | 6,823 | 28,102 | 1.11 (1.07–1.16) | 1×10−6 | 0.4 |
|
| |||||
| Morphology (26 studies) | |||||
| Ductal | 20,283 | 38,447 | 1.12 (1.09–1.15) | 2×10−15 | |
| Lobular | 3,829 | 38,447 | 1.07 (1.02–1.13) | 0.01 | 0.1 |
OR, odds ratio; CI, confidence interval
For size and grade, a linear trend was tested by fitting the median values for each category as a continuous variable.
There was also clear evidence that 5p12-rs10941679 was more strongly associated with the risk of lower-grade breast cancer (P=5×10−7, Table 3). When grade and PR status were modeled together in a case-only analysis, evidence of heterogeneity was observed for both tumor characteristics (P<0.002). After excluding “selected cases”, the per-G-allele OR estimates were 1.20 (95%CI=1.14–1.25), OR=1.13 (95%CI=1.09–1.16) and OR=1.04 (95%CI=0.99–1.08) for grade 1, 2 and 3/4 disease, respectively). Further restriction to PR-positive cases gave consistent results [OR=1.22 [95%CI=1.14–1.30], OR=1.15 [95%CI=1.10–1.20] and OR=1.07 [95%CI=1.00–1.15], respectively). We also assessed the association of 5p12-rs10941679 with risk of different grades of DCIS and, while based on limited data (N=210, 102 and 124 cases with grade 1, 2 and 3 DCIS, respectively), the per-allele OR estimates were consistent with the trend observed for invasive disease (OR=1.33 [95%CI=0.88–2.02], OR=1.08 [95%CI=0.78–1.49] and OR=0.98 [95%CI=0.72–1.33] for grade 1, 2 and 3 DCIS, respectively). There was no evidence of heterogeneity in per-G-allele OR by nodal status, tumor size or tumor type.
Further to our previous report of a possible association with breast cancer risk of variant rs981782, also in 5p12, we genotyped this SNP in an additional 8,213 invasive cases and 10,340 controls from studies participating in the BCAC and found no evidence of association (per-G-allele OR=0.97, 95%CI=0.93–1.02, P=0.2, Table 4). When we added in data for participants genotyped as part of the previous study (2), to give a combined total of 23,548 invasive cases and 28,142 controls of white European origin, the per-G-allele OR estimate was 0.95 (95%CI=0.92–0.97, P=2×10−5). The MAF for controls genotyped using Taqman, iPlex and the GWAS SNP-array (2) were similar (0.47, 0.45 and 0.50, respectively). This possible association was attenuated, but still nominally statistically significant, when 5p12-rs10941679 was also included in the logistic regression model (OR=0.97, 95%CI=0.94–0.99, P=0.007). The per-allele OR estimate for each SNP adjusted for the other was unchanged after excluding “selected cases”. The estimated linkage disequilibrium coefficient (r2) between the two SNPs was 0.04, both across all white European subjects genotyped (N=52,506) and for controls only (N=28,142).
Table 4.
SNP 5p12-rs981782 and risk of invasive breast cancer in white European women
| Group/genotype | Controls, N (%) | Cases, N (%) | OR (95%CI) | P |
|---|---|---|---|---|
| Subjects genotyped for the present study (7 studies) | ||||
| AA | 2,845 (28) | 2,310 (28) | 1.00 | |
| AG | 5,162 (50) | 4,048 (49) | 0.95 (0.89–1.02) | 0.2 |
| GG | 2,333 (23) | 1,855 (23) | 0.94 (0.87–1.03) | 0.2 |
| per G-allele | 0.97 (0.93–1.01) | 0.2 | ||
|
| ||||
| Combined analysis* (19 studies) | ||||
| AA | 7,815 (28) | 6,890 (29) | 1.00 | |
| AG | 14,056 (50) | 11,626 (49) | 0.93 (0.90–0.97) | 0.001 |
| GG | 6,271 (22) | 5,032 (21) | 0.90 (0.86–0.95) | 3×10−5 |
| per G-allele | 0.95 (0.92–0.97) | 2×10−5 | ||
|
| ||||
| Adjusted for rs10941679* (19 studies) | ||||
| AA | 7,815 (28) | 6,890 (29) | 1.00 | |
| AG | 14,056 (50) | 11,626 (49) | 0.95 (0.91–0.99) | 0.02 |
| GG | 6,271 (22) | 5,032 (21) | 0.93 (0.89–0.98) | 0.009 |
| per G-allele | 0.97 (0.94–0.99) | 0.007 | ||
OR, odds ratio; CI, confidence interval
based on white European subjects genotyped for 5p12-rs10941679 and 5p12-rs981782, including previously published data on the latter (2)
Discussion
This analysis of 40,972 invasive cases and 46,334 controls from 37 studies definitively confirms the 5p12-rs10941679 as a breast cancer susceptibility locus in white European women. After excluding cases selected for increased genetic susceptibility, we estimated an OR for white European women of 1.11 per G allele (95%CI=1.08–1.14). This association was independent of a possible association with 5p12-rs981782 and appeared to be stronger for older women.
The estimated OR for Asian women (3,007 cases and 2,337 controls) was consistent with that for European women, but the confidence limits were too wide to demonstrate a definite association (OR=1.07, 95%CI=0.99–1.15). The Shanghai Breast Cancer Study also estimated a per-G-allele OR of 1.07 (95%CI=0.99–1.15) using data from 2,950 cases and 2,986 controls (7). Assessed together, these two large studies suggest that 5p12-rs10941679 is associated with increased breast cancer risk in Asian women (meta-analysis OR=1.07 (95%CI=1.02–1.13, P=0.01), although even larger studies will be required to precisely estimate the corresponding OR.
The initial paper on 5p12-rs10941679, based on 6,145 cases and 33,016 controls, reported a larger per-G-allele OR for breast cancer in general of 1.19 (95%CI=1.11–1.28) (5). This larger initial estimate probably reflects “winner’s curse” whereby the estimated OR in the study discovering the association tends to be overestimated (8). Stacey et al. (5) also reported that the increased risk associated with 5p12-rs10941679 was limited to ER-positive disease and consistent results have been observed in other studies (7, 9). We replicated this finding, but our larger sample size enabled us to carry out a more detailed analysis that revealed a stronger association with PR-positive disease (per-G-allele OR=1.16, 95%CI=1.12–1.20). The heterogeneity in the OR by ER status appeared to be driven by the correlation between ER and PR status. Furthermore, we have established that the association is also stronger for lower grade tumors in general, and lower grade PR-positive tumors specifically. Stacey et al. (5), also noted a possibly larger OR for lower grade tumors, but not after ER-status was taken into account. This multifaceted heterogeneity has also been observed for 10q26-rs2981582, which appears to be associated with increased risk of lower grade ER-positive tumors (10).
In our previous GWAS, we observed that the minor G allele of 5p12-rs981782 in the same region was associated with reduced risk of breast cancer. The estimated per-G-allele OR for white European women was 0.94 (95%CI=0.91–0.97, P=9×10−6), based on data from 20,649 cases of invasive breast cancer and 22,578 controls from 19 studies participating in the replication stages (2 and 3) (2). We did not replicate this association in the present study of 8,247 cases and 10,363 controls, but our result (OR=0.97, 95%CI=0.93–1.02, P=0.2) was not inconsistent with the existence of a modest protective effect. Reeves et al. (11) studied 5p12-rs981782 in 10,306 cases and 10,309 controls from the Million Women Study and estimated a per-G-allele OR of 0.94 (95%CI=0.91–0.98, P=0.003). Stacey et al. (5) also evaluated this SNP in 5,028 cases and 32,090 controls and reported the corresponding OR to be 0.96 (95%CI=0.92–1.01, P=0.1). The Cancer Genetic Markers of Susceptibility (CGEMS) project studied SNP 5p12-rs4866929, which is in high linkage disequilibrium with 5p12-rs981782 (r2=0.96), in a total of 5,692 cases and 5,576 controls and obtained an OR estimate of 0.97 (95%CI=0.91–1.03, P=0.4) (3, 9). When combined together in a meta-analysis, these results for 5p12-981782 (excluding those from CGEMS) give an estimated OR of 0.95, 95%CI=0.93–0.97, P=5×10−8), suggesting that this SNP is associated with a modest reduction in breast cancer risk.
It is less clear whether the association between 5p12-rs981782 and breast cancer risk is independent of the effect marked by 5p12-rs10941679. In contrast to the findings of Stacey et al. (5), in the present study (23,548 cases and 28,142 controls) the evidence of association for 5p12-rs981782 remained after adjusting for 5p12-rs10941679 (P=0.007), although the effect was slightly attenuated. Furthermore, the correlation between these two SNPs was too weak (r2=0.04, N=52,506) for the association seen with 5p12-r981782 to be due to confounding by 5p12-rs10941679. However, it remains possible that these two associations are driven by a variant correlated with both SNPs. Stacey et al. (5) found that a third SNP nearby, 5p12-rs4415084 was associated with breast cancer risk, independently of 5p12-rs10941679 (r2=0.51 between the two SNPs). Multi-centre fine-mapping studies of this region are needed to obtain the samples sizes required to identify the causal variant or variants behind these multiple small signals.
As discussed by Stacey et al. (5), 5p12-rs10941679 resides between two genes: MRPS30, which encodes the smaller 28S subunit of the mitochondrial ribosome and has pro-apoptotic properties (12), and FGF10, a fibroblast growth factor which is amplified in around 10% of breast cancers (13), although there is a recombination hotspot separating this SNP from FGF10. Therefore, as for the vast majority of low-penetrance breast cancer susceptibility loci identified by GWAS, an implicated gene or functional mechanism remains to be elucidated.
A potential limitation of our study was that information on tumor histopathology was not available for all cases and, where it was, these data were predominantly abstracted from medical records, rather than being obtained through a standardised pathology review. Thus some misclassification may have been present and this may have attenuated some associations with tumor phenotype. However the strong association with PR and ER status suggests that this effect was minimal, at least for the features examined here.
In conclusion, 5p12-rs10941679 is a confirmed marker of breast cancer susceptibility. In white European women it is associated with increased risk of lower-grade, PR-positive tumors. These findings suggest that commonly collected tumor characteristics other than ER status can be used to identify subtypes of breast cancer with distinct etiology, and that large collaborative studies are required to identify the genetic contributions specific to each of these.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. In particular, we thank: Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez, Anna González-Neira, Michael Bremer, Sue Higham, Helen Cramp, Dan Connley, The Wellcome Trust Case Control Consortium (see the WTCCC website for a full list of contributing investigators), Maggie Angelakos, Judi Maskiell, Gillian Dite, Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel, Kathleen Corthouts, Ursula Eilber, Tanya Koehler, Marco Pierotti, Bernard Peissel, Daniela Zaffaroni, Bernardo Bonanni, Loris Bernard, the personnel of the Cancer Genetics Testing laboratory at the IFOM-IEO campus, Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner, Hartwig Ziegler, Sonja Wolf, Volker Hermann, Teresa Selander, Nayana Weerasooriya, Eija Myöhänen, Helena Kemiläinen, Bernd Frank, Eileen Williams, Elaine Ryder-Mills, Kara Sargus, Tracy Slanger, Elke Mutschelknauss, Ramona Salazar, S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit, Irene Masunaka, Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones, Richard van Hien, Sten Cornelissen, Linde Braaf, Flora van Leeuwen (NKI-AVL), Bas Bueno-de-Mesquita (RIVM; for the release of control samples), the SEARCH and EPIC teams, Heather Thorne, Eveline Niedermayr, the AOCS Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb), the ACS Management Group (A Green, P Parsons, N Hayward, P Webb, D Whiteman), The GENICA network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany; [CJ, Hiltrud Brauch], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Bonn, Germany [Hand-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [UH] and Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany [Thomas Bruening, Beate Pesch, Sylvia Rabstein, Anne Spickenheuer, VH], Meeri Otsukka, Kari Mononen, Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk and the Erasmus MC Family Cancer Clinic.
Grant support
Part of this work was supported by the European Community’s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The BCAC is funded by CR-UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). D.F.E. is a Principal Research Fellow of CR-UK. The ABCS was supported by the Dutch Cancer Society [grants NKI 2001-2423; 2007-3839] and the Dutch National Genomics Initiative. The ABCFS, NC-BCFR and OFBCR work was supported by the United States National Cancer Institute, National Institutes of Health (NIH) under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Northern California Cancer Center (U01 CA69417), University of Melbourne (U01 CA69638). Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], the Cancer Council of Tasmania and Cancer Foundation of Western Australia and the NHMRC [199600]. G.C.T. and P.W. are supported by the NHMRC. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital. The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI081583 and PI081120). The ESTHER study was supportd by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The FBCS is supported by funds from Cancer Research UK (C8620/A8372 and C8620/A8857), a US Military Acquisition (ACQ) Activity, Era of Hope Award (W81XWH-05-1-0204) and the Institute of Cancer Research (UK). C.T. is funded by a Medical Research Council (UK) Clinical Research Fellowship. The FBCS acknowledges NHS funding to the Royal Marsden / Institute of Cancer Research NIHR Specialist Cancer Biomedical Research Centre. The GC-HBOC was supported by Deutsche Krebshilfe [107054], the Dietmar-Hopp Foundation, the Helmholtz society and the German Cancer Research Centre (DKFZ). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The GESBC was supported by the Deutsche Krebshilfe e. V. [70492] and genotyping in part by the state of Baden-Württemberg through the Medical Faculty of the University of Ulm [P.685]. The HABCS study was supported by an intramural grant from Hannover Medical School. The HMBCS was supported by short-term fellowships from the German Academic Exchange Program [to N.B], and the Friends of Hannover Medical School [to N.B.]. The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/017). The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, The Academy of Finland and by the strategic funding of the University of Eastern Finland. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I], the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG was supported by grants from Ministero della Salute ( Extraordinary National Cancer Program 2006 “Alleanza contro il Cancro”, and “Progetto Tumori Femminili” to PR), Ministero dell’Universita’ e Ricerca (RBLAO3-BETH to PR), Fondazione Italiana per la Ricerca sul Cancro (Special Project “Hereditary tumors”), Associazione Italiana per la Ricerca sul Cancro (4017 to PP) and by funds from Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5×1000” ). The MCBCS was supported by the NIH grants [CA122340, CA128978] and a Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201]. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. SEE ABCFS. The NBCS was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to ALBD, FUGE-NFR 181600/V11 to VNK and a Swizz Bridge Award to ALBD. The NHS was funded by NIH grant CA87969. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the Academy of Finland, the University of Oulu, and the Oulu University Hospital. The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). The SBCS was supported by Yorkshire Cancer Research and the Breast Cancer Campaign. SEARCH is funded by programme grants from Cancer Research UK [C490/A10124, C8197/A10123, C1287/A10118]. The SEBCS was supported by the Korea Health 21 R&D Project [AO30001], Ministry of Health and Welfare, Republic of Korea. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004; Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University, supported by the Polish Foundation of Science. The TBCS was funded by The National Cancer Institute Thailand. The TWBCS is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre.
Footnotes
Conflicts of interest: None to declare
References
- 1.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23. 2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 5.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 6.StataCorp. Stata Statistical Software: Release 10.0. College Station, Texas (USA): Stata Corporation LP; 2007. [Google Scholar]
- 7.Long J, Shu XO, Cai Q, Gao YT, Zheng Y, Li G, et al. Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers Prev. 2010;19:2357–65. doi: 10.1158/1055-9965.EPI-10-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraft P. Curses--winner’s and otherwise--in genetic epidemiology. Epidemiology. 2008;19:649–51. doi: 10.1097/EDE.0b013e318181b865. discussion 657–8. [DOI] [PubMed] [Google Scholar]
- 9.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24. 1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: Findings from the Breast Cancer Association Consortium. Human Molecular Genetics. doi: 10.1093/hmg/ddr228. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K, et al. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–34. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- 12.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–74. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 13.Theodorou V, Boer M, Weigelt B, Jonkers J, van der Valk M, Hilkens J. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene. 2004;23:6047–55. doi: 10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.