Abstract
The 12p13 ETV6 (TEL) gene is frequently targeted by chromosomal translocations in human malignancies, resulting in the formation of oncogenic ETV6 gene fusions. Many of the known partner genes encode protein tyrosine kinases (PTKs), generating fusion proteins that function as chimeric PTKs. ETV6-NTRK3 (EN), comprised of the ETV6 SAM domain fused to the NTRK3 PTK, is unique among ETV6 chimeric oncoproteins, as it is expressed in cancers of multiple lineages. We initially hypothesized that, similar to other ETV6-PTK chimeras, SAM-mediated dimerization of EN leads to constitutive activation of the PTK and downstream signaling cascades. However, when the EN SAM domain was replaced with an inducible FK506 binding protein (FKBP) dimerization system, resulting FKBP-NTRK3 chimeras failed to transform NIH 3T3 cells even though PTK activation was preserved. It was recently shown that the ETV6 SAM domain has two potential interacting surfaces, raising the possibility that this domain can mediate protein polymerization. We therefore mutated each EN SAM binding interface in a manner shown previously to abolish self-association of wild-type ETV6. Each mutation completely blocked the ability of EN to polymerize, to activate its PTK, and to transform NIH 3T3 cells. Furthermore, EN itself formed large polymeric structures within cells while mutant EN proteins were present only as monomers. Finally, we observed a dominant negative effect on the transformation of isolated SAM domains coexpressed in EN-transformed cells. Taken together, our results suggest that higher-order polymerization may be a critical requirement for the transformation activity of EN and possibly other ETV6-PTK fusion proteins.
Many chromosomal translocations described in human malignancies lead to the expression of chimeric oncogenes resulting from the in-frame fusion of coding sequences from two different partner genes (30). Dimerization or oligomerization through the interaction of domains contributed by one of the partner genes has been shown to be an important activating mechanism for many chimeric oncoproteins. For BCR-ABL and other chimeric protein tyrosine kinases (PTKs), oligomerization leads to ligand-independent auto- or cross-phosphorylation of the kinase domain and constitutive PTK activation (2). Moreover, oligomerization appears to have additional roles in facilitating oncogenesis, such as by stabilizing the binding of other proteins to the oncoprotein complex. For example, evidence suggests that binding of the SMRT corepressor to the STAT5-RARα fusion protein is facilitated by its oligomerization (23). In general, however, structural aspects and stoichiometric relationships within oligomeric complexes of these chimeric oncoproteins remain very poorly understood.
The ETS family transcription factor ETV6 (or TEL), required for developmental processes such as hematopoiesis and yolk sac angiogenesis (50), consists of an N-terminal (SAM) domain and a C-terminal DNA-binding ETS domain (36). The ETV6 gene, found on chromosome 12p13, is disrupted by translocations in numerous human leukemias as well as in solid tumors, generating many different ETV6 fusion genes (3). Many of these ETV6 chimeras encode fusion oncoproteins in which the ETV6 SAM domain is fused either to a PTK, including PDGFRβ (8), Abl (9, 13), ARG (4, 12), Jak2 (28), FGFR3 (51), or NTRK3 (19, 49), or to transcription factors such as AML1 (7) or ARNT (34). These chimeric proteins appear to utilize the ETV6 SAM domain for self-association, which, for chimeric tyrosine kinases, induces constitutive PTK activation. The SAM domain, also known as the Pointed or PNT domain, is a ca. 75-amino-acid module that mediates a wide variety of homo- and heterotypic protein-protein interactions (17, 22, 29, 31, 35). This domain is found in a large number of proteins, including a subset of ETS transcription factors (14, 38), Eph family receptor tyrosine kinases (42), diacylglycerol kinases (26, 33), serine threonine kinases (48), Polycomb group (PcG) proteins (20), yeast mating type signaling proteins (31, 48), the p73 tumor suppressor (5), and the RNA-binding protein Smaug (10). Crystal structures of the EphB2 and EphA4 receptor SAM domains revealed that the domain has two distinct binding interfaces, each providing a possible site for intermonomeric association (41, 44). The EphB2 crystal structure also suggests that its SAM domain could form extended polymeric structures (43). Subsequently, Kim et al. and Tran et al. demonstrated that the isolated ETV6 SAM domain, which forms an insoluble homopolymer when expressed in bacterial cells, also self-associates in a head-to-tail fashion to crystallize as an extended helical polymer (16, 47). Furthermore, mutations of single amino acids within either hydrophobic polymerization interface (alanine 93 and valine 112 in ETV6 cDNA numbering) to charged residues renders the SAM domain soluble and monomeric. These studies provide evidence that higher-order polymer formation might be essential for the normal function of ETV6. Similar behavior was also reported for the SAM domain of the PcG protein polyhomeotic (15).
The above findings for the wild-type (WT) ETV6 SAM domain raise the possibility that ETV6 SAM-containing chimeric oncoproteins also form higher-order polymeric structures and that this may be important for transformation. This possibility was suggested by earlier findings demonstrating that ETV6-PDGFRβ forms higher-order structures in hematopoietic cells (37). To test this further, we analyzed complexes of the ETV6-NTRK3 (EN) fusion protein associated with the t(12;15)(p13;q25) translocation. This oncoprotein, originally described in the pediatric mesenchymal malignancy congenital fibrosarcoma, contains the ETV6 SAM domain fused to the PTK domain of the neurotrophin 3 receptor NTRK3 (19). EN expression was subsequently described for congenital mesoblastic nephroma (18), acute myeloid leukemia (6), and recently, the secretory variant of ductal breast carcinoma (46). EN is therefore unique among known chimeric oncoproteins, as it is expressed in malignancies derived from multiple cell lineages. We previously reported that EN self-association through its SAM domain is required for PTK activation (49). This in turn leads to activation of WT NTRK3 downstream pathways such as the Ras-Erk1/2 mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)-AKT cascades that are essential for EN transformation of NIH 3T3 cells (45).
In the present study we wished to further explore the role of oligomerization in EN transformation. We therefore made chimeric proteins in which the EN SAM domain was replaced with the inducible dimerization domain of the FK506 binding protein (FKBP) polypeptide. Upon induction, FKBP-NTRK3 expression in NIH 3T3 cells resulted in PTK activation and transient activation of Ras-Erk1/2 and PI3K-AKT pathways but failed to transform these cells. This suggested that activation through dimerization is not sufficient for EN transformation or that the ETV6 SAM domain may contribute additional functions. We now show that both binding interfaces of the SAM domain must be intact for EN transformation and that mutating key amino acids within either of these interfaces abolishes the ability of EN to form large polymeric structures. Taken together, our results suggest that polymerization through the ETV6 SAM domain is important for EN transformation and that targeting the regions mediating this property may represent a novel therapeutic strategy for inhibiting fusion proteins containing the ETV6 SAM domain.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells were obtained from the American Type Culture Collection and maintained at low confluence in 10% calf serum (CS; Gibco BRL) and Dulbecco's modified Eagle medium (DMEM; Gibco BRL). The BOSC23 packaging cell line was obtained from Rob Kay (Terry Fox Laboratory, Vancouver, Canada) and grown in 10% fetal CS (Gibco BRL) and DMEM. 293T cells were obtained from the American Type Culture Collection and grown in 10% fetal CS-DMEM.
Inducible dimerizer system.
The FKBP inducible dimerization system was a generous gift from Ariad Pharmaceuticals, Inc. (Cambridge, Mass.) FKBP1E-NTRK3 and FKBP2E-NTRK3 fusion constructs were created by amplifying the NTRK3 region of ETV6-NTRK3 (with the primer set NTRK3-1601-XbaI [5′-GCTCTAGAGATGTGCAGCACATTAAGAGG-3′] and NTRK3-2490rev-SpeI [5′-GGACTAGTGCCAAGAATGTCCAGG-3′]), cutting the resultant product with XbaI/SpeI (NEB), and fusing it in frame with pC4M-FV1E or pC4M-FV2E (renamed FKBP1E and FKBP2E, respectively). The fusion constructs were verified by sequence analysis. An EcoRI/ApoI or partial EcoRI/ApoI digest facilitated cloning of the fusion constructs into the MSCVpuro retroviral vector (Clontech). Control FKBP1E and FKBP2E retroviral constructs were created by performing EcoRI/ApoI or EcoRI/ApoI partial digests. Stable cell lines expressing either FKBP1E, FKBP2E, FKBP1E-NTRK3, or FKBP2E-NTRK3 alone were generated, and protein expression was determined either via immunoprecipitation (IP) followed by Western blot analysis with anti-hemagglutinin (HA) antibodies (BabCO) (for FKBP1E constructs) or via anti-HA Western blot analysis (for FKBP2E constructs). Experiments were performed with a range of AP20187 dimerizer concentrations (from 100 to 1,000 nM). Soft agar experiments were performed as described below with the addition of 100 nM AP20187 in both the lower and upper layers. The top layer was replenished daily with media containing 100 nM AP20187. Wells were stained with 0.005% crystal violet in 75% methanol for 1 h and then rinsed with water for destaining.
Site-directed mutagenesis.
Point mutations within the SAM domain of EN were generated by using the QuikChange site-directed mutagenesis kit (Stratagene). Three different SAM domain mutations were made by using the following primers: A93D (5′-GAATGGCAAAGATCTCCTGCTGCTG-3′), V112E (5′-TTCAGGTGATGAGCTCTATGAACTCC-3′), and V112R (5′-TTCAGGTGATCGGCTCTATGAACTCC-3′). Mutagenesis was performed with EN pBS KS+ as a template. Each construct was sequenced, cut with EcoRI, and cloned into MSCVpuro.
Retroviral expression system.
A cDNA encoding EN was inserted into the retroviral vector MSCVpuro or MSCVneo at the EcoRI site. cDNAs encoding isolated WT, A93D mutant, and V112E mutant SAM domains were cut with BglII/BamHI out of pET-22b(+) (Novagen) and cloned into BglII-cut MSCVpuro. The MSCVneo green fluorescent protein (GFP) vector was created by cutting out the GFP from pEGFP-N3 (EcoRI/HpaI; Clontech) and inserting it into the MSCVneo vector.
Generation of retrovirally transduced cell lines.
Retroviral vector plasmid DNA was transfected into the BOSC23 ecotropic retroviral packaging cell line by using calcium phosphate precipitation as described by Pear et al. (27). Supplemental Gag/Pol (pGP1) and Env plasmids were used during the transfection procedure to increase viral titers. Retrovirus-containing supernatants were collected from the BOSC23 cells 48 h after transfection and used to infect NIH 3T3 cells. Infected cells were selected for by using the appropriate antibiotic (puromycin [2 μg/ml; Sigma] for 48 h or Geneticin [900 μg/ml; Gibco BRL] for 7 to 9 days). Protein expression was determined by Western blotting. Cells coexpressing two different constructs were made by transfecting and selecting for the control (MSCV) or EN constructs first and then coexpressing and selecting for the second construct (i.e., isolated SAM domains). Expression of both proteins was confirmed by Western blot analysis for the EN protein and Tricine gel electrophoresis for the isolated SAM domains.
Co-IP experiments.
cDNAs encoding WT or mutant SAM domain EN constructs were C-terminally tagged with either an HA or V5/His tag by using the mammalian expression vectors pMH (Roche) and pcDNA3.1/V5-His-TOPO (Invitrogen), respectively. Different combinations of HA- and V5-tagged constructs were transiently cotransfected in 293T cells with the FuGENE6 transfection reagent (Roche). Anti-HA IPs were performed on lysates collected 36 h after transfection followed by anti-V5 Western blotting (see below).
Lysate preparation, IP, and immunoblotting (IB).
Cells were rinsed twice with phosphate-buffered saline (PBS) and lysed with 500 μl of phosphorylation solubilization buffer (50 mM HEPES, 100 mM NaF, 10 mM Na4P2O7, 2 mM Na3VO4, 2 mM EDTA, 2 mM NaMoO4 · 2H2O, 0.5% Nonidet P-40) or radioimmunoprecipitation assay buffer (20 mM Tris [pH 7.4], 120 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 10% glycerol, 5 mM EDTA, 50 mM NaF, 0.5 mM Na3VO4) containing protease inhibitors (leupeptin [10 μg/ml], aprotinin [10 μg/ml], and phenylmethylsulfonyl fluoride [250 μM]). The cells were solubilized for 30 min at 4°C on a shaking platform. Lysates were cleared by centrifugation at 12,000 × g for 10 min at 4°C. Protein quantification of the lysates was performed with a DC protein assay kit from Bio-Rad. IPs were performed on 500 μg of total cell lysate with the appropriate amount of antibody (see “Antibodies” below) and 20 μl of protein A-Sepharose (Amersham Pharmacia Biotech) or protein G beads (Sigma). Lysate-antibody-bead mixtures were nutated for 4 h at 4°C. Beads were spun out at 2,000 × g for 1 min and then washed three times in wash buffer (50 mM HEPES, 100 mM NaF, 10 mM Na4P2O7, 2 mM Na3VO4, 2 mM EDTA, 2 mM NaMoO4 · 2H2O, 0.1% Nonidet P-40). The final wash was removed from the beads, 50 μl of Laemmli buffer was added, and the samples were boiled for 5 min before loading. For Western blot analysis, 30 μg of total cell lysate was mixed with Laemmli buffer and electrophoresed overnight on SDS-10 to 12% polyacrylamide gels according to standard methods. Electrophoresed proteins were transferred to Immobilon-P membranes (Millipore) prior to immunoblot analysis with the indicated antibodies. Proteins were visualized by enhanced chemiluminescence (Amersham) according to the manufacturer's protocols.
Antibodies.
The following antibodies were utilized in the Western blotting experiments described in this paper: phospho-Mek1/2 (Ser217/221) (immunoblot [IB], 1:1,000 dilution; Cell Signaling), phospho-Akt (ser473) (IB, 1:1,000 dilution; Cell Signaling), Akt (IB, 1:1,000 dilution; Cell Signaling), RC20 (IB, 1:2,500 dilution; Transduction Labs), TrkC (NTRK3, C14) (Western blotting, 1:1,000 dilution; IP, 5 μl/IP; Santa Cruz Biotechnology), cyclin D1/2 (IB, 1:2,000 dilution; Upstate Biotechnology), glutathione S-transferase-Tel (ETV6)-helix-loop-helix (IB, 1:5,000 dilution; IP, 2 μl/IP; a generous gift from Peter Marynen, Center for Human Genetics, Institute for Biotechnology, Flanders Interuniversity, Leuven, Belgium), HA antibody (HA.11) (IB, 1:2,000 dilution; IP, 5 μl of 1/50 dilution/IP; BabCO), V5 (IB, 1:5,000 dilution; IP, 1.5 μl/IP; Invitrogen), and GFP (IB, 1:1,000 dilution; Clontech).
GFP-tagged constructs and localization experiments.
WT and mutant SAM domain EN constructs were C-terminally tagged with GFP by using PCR and the following GFP tagging primers: 5′GFP primer, 5′-ACGAATTCCTGATGTCTGAGACTCCTGCTGCTC-3′; 3′GFP primer, 5′-CGTGAATTCAGGCCAAGAAGTTCCAGGTAG-3′. PCR products were cut with EcoRI and inserted into the MSCVneo GFP vector (see “Retroviral expression system” above). NIH 3T3 cell lines stably expressing the GFP constructs seeded onto glass coverslips were rinsed once with PBS, fixed with 4% paraformaldehyde for 20 min at room temperature, rinsed three times with PBS, and mounted onto slides with Vectashield mounting media containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc.). All image capturing was done with a Zeiss Axioplan2 epifluorescence microscope with selective filters, a cooled charge-coupled device camera (Sensys Photometrics), and Quips-Vysis software.
Monolayer morphology and soft agar assays.
The morphology of NIH 3T3 cells expressing WT and mutant SAM domain EN constructs was documented by seeding equivalent numbers of cells in tissue culture dishes. Twenty-four hours after plating, the cells were photographed with a Zeiss Telaval 3 inverted microscope equipped with an FM2 Nikon camera. Soft agar assays were performed as described previously (25). Cells were seeded in triplicate at a concentration of approximately 8 × 104 cells per 35-mm-diameter dish. Bottom layers were made up of 0.4% agar in 9% CS-DMEM. Cells were resuspended in a top layer of 0.2% agar in 9% CS-DMEM. Cells were fed every other day by placing two drops of media on the top layer. After 2 weeks at 37°C, the number of single cells and colonies per high-power view were counted. Results were formulated as percentages of colonies formed per total number of cells plated.
Tumor growth in nude mice.
Two million NIH 3T3 cells expressing either (i) MSCVneo alone, (ii) MSCVneo WT EN alone, (iii) MSCVneo- plus MSCVpuro-isolated V112R SAM domain, or (iv) EN MSCVneo- plus MSCVpuro-isolated V112R SAM domain were injected subcutaneously into the flanks of nude mice (Charles River Laboratories). Tumor injection sites were monitored three times weekly, and when the tumors were large enough to assay, caliper measurements were taken. Tumors were excised and weighed after 20 days, and an average tumor size was calculated. Tumor growth (in milligrams) over time was estimated by using the following equation: tumor length × tumor width2 × 0.5236 mg/mm3.
Characterization of complexes formed in mammalian lysates by FPLC.
Lysates prepared from NIH 3T3 cells expressing various constructs (described above) were fractionated by fast-performance liquid chromatography (FPLC) by using a Superose 6 HR 10/30 gel filtration column (Amersham) and the following buffer: 1.5 mM MgCl2, 150 mM NaCl, 50 mM HEPES (pH 7.3). The flow rate was 0.5 ml/min, and 0.5-ml fractions were collected. Fractions 13 to 35 were analyzed by Western blot analysis as described above with the following antibodies: anti-ETV6 helix-loop-helix, anti-V5, and anti-GFP.
Generation and analysis of His6-tagged fusion proteins by gel filtration chromatography.
N-terminally His6-tagged proteins of WT EN, A93D EN, V112E EN, and V112R EN were created by PCR amplification of each construct, followed by EcoRI digestion and ligation into pET-22b(+) (Novagen). Tagged fusion construct plasmids were used to transform Escherichia coli BL21(λDE3) cells, resulting in fusion protein expression representing greater than 50% of total cellular proteins. Cells were gently lysed, followed by protein purification with Ni2+ affinity chromatography with a buffer of 50 mM HEPES (pH 7.5), 500 mM NaCl, and 5% glycerol with 5, 30, and 250 mM imidazole for binding, wash, and elution, respectively. The isolated protein was further purified by FPLC with a 100-cm S-200 Sepharose column (Amersham Pharmacia Biotech) with 20 mM potassium phosphate (pH 7.5) and 150 mM NaCl, and the elution profile was monitored by densitometric analysis of a Coomassie-stained SDS-10% polyacrylamide gel electrophoresis gel. Elution volumes were compared to previously run sizing standards. The eluted WT EN was further characterized by loading 50 μl of a 20 μM protein solution mixed with an equal amount of size standards (composed of blue dextran [2,000 kDa], ferritin, aldolase, catalase, bovine serum albumin, and orange G) onto a 30- by 1-cm BioGel A-1.5 column (Bio-Rad) and separated by using the same buffer as described above at a flow rate of 0.5 ml/min, with elution monitored by densitometric analysis of a Coomassie-stained SDS-10% polyacrylamide gel electrophoresis gel.
Creation of isolated SAM mutants for electron microscopy (EM).
The isolated murine ETV6 SAM domain was amplified from a 10.5-day-old mouse cDNA library with NdeI and HindIII restriction sites introduced by PCR to allow for directional cloning into pET22b (+) (Novagen). The A93D mutation was created by using the QuikChange protocol (Stratagene). The protein was purified by FPLC with an SP-Sepharose column and a buffer of 20 mM potassium phosphate (pH 7.0). Elution of the protein by a salt gradient occurred at an NaCl concentration of 50 mM. Following purification, the isolated SAM mutant was dialyzed into a buffer containing 20 mM potassium phosphate (pH 7.5) and 50 mM NaCl. Insoluble A93D SAM filaments were generated by a change in pH to 6.0, which led to protonation of the introduced Asp93 residue and restoration of the hydrophobic surface region required for SAM domain oligomerization (16).
Visualization of expressed proteins by EM.
Drops of purified His-tagged EN and SAM domain mutant proteins were placed on the surface of Formvar-coated 200-mesh nickel grids. The grids were then rinsed with 2 hanging drops of protein resuspension buffer [25 mM KPO4 (pH 8), 300 mM NaCl, and 1 mM 1,1-bis(4-chlorophenyl)-2,2,2-trichloroethane] and negatively stained with 10 hanging drops of 1% aqueous uranyl acetate. The excess of the uranyl acetate was removed by blotting the edges of the grids with filter paper and air drying for 5 min. The grids were examined, and images were digitally collected with a Hitachi H7600 transmission electron microscope.
RESULTS
Inducible dimerization of FKBP-NTRK3 fails to transform NIH 3T3 cells.
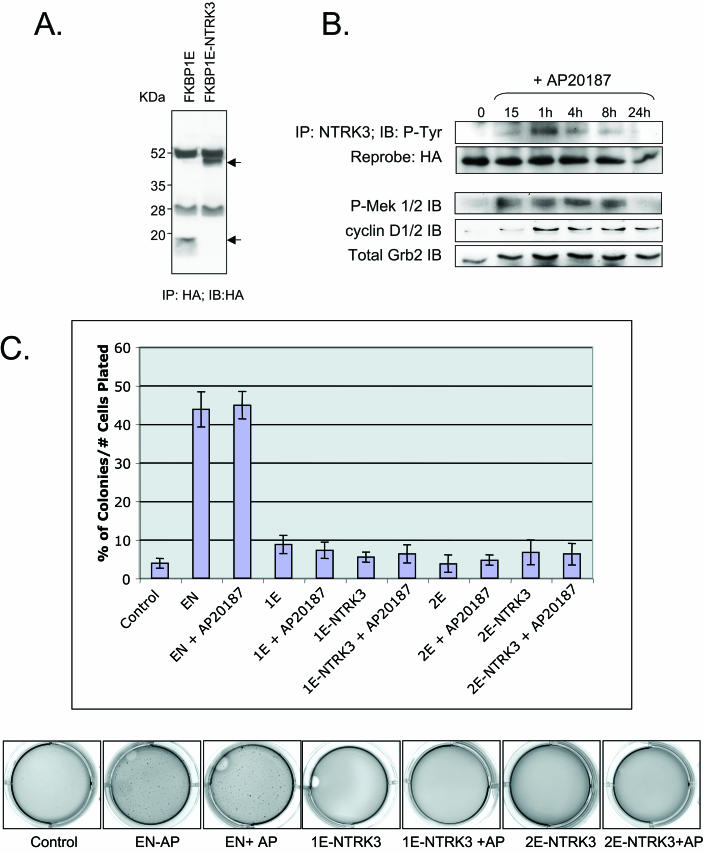
It was previously demonstrated that EN self-associates through its SAM domain and that this function is essential for EN PTK constitutive activation and transformation activity (49). A simple explanation for this observation is that ETV6 contributes only a dimerization motif to EN, thus facilitating PTK activation and stimulation of downstream NTRK3 signaling pathways. To test this hypothesis, we utilized the FKBP inducible dimerization system (Ariad Inc.). In this system, either one or two ligand binding domains of the FK506 binding protein (FKBP1E or FKBP2E) are fused to a protein of interest such that self-association is induced in the presence of a bivalent analog of FK506 (AP20187) (40). We created chimeras in which the entire ETV6 portion of EN, including the SAM domain, was replaced with one or two FKBP domains and expressed these fusion proteins or the FKBP motifs alone in NIH 3T3 cells (shown for FKBP1E-NTRK3 in Fig. 1A). Structurally, FKBP1E-NTRK3 is only capable of dimerizing, whereas in principle, FKBP2E-NTRK3 can tetramerize or form higher-order structures. However, previous studies showed that FKBP1E-ABL and FKBP2E-ABL both exist as dimers of similar size when expressed in mammalian cells (39), suggesting that either system is appropriate for studying dimerization. The addition of 100 nM AP20187 induced tyrosine phosphorylation of FKBP1E-NTRK3 (Fig. 1B) or FKBP2E-NTRK3 (data not shown) within 15 min of treatment. The dimerizer also induced MEK1/2 activation by 15 min and elevated cyclin D1 expression by 1 h (Fig. 1B). However, in contrast to EN, in which tyrosine phosphorylation is constitutive, as are MEK1/2 activation and cyclin D1 elevation (45), both FKBP-NTRK3 chimera were only transiently tyrosine phosphorylated, as were MEK1/2 phosphorylation and cyclin D1 elevation. Moreover, cells expressing either chimera failed to undergo morphological transformation even after continuous addition of 100 nM dimerizer for up to 5 days (data not shown). We then assessed FKBP-NTRK3 chimera expressing cells for their ability to form colonies in soft agar. Macroscopic colonies failed to form even when 100 nM AP20187 was added daily for up to 2 weeks, whereas this agent had no effects on EN transformation activity (Fig. 1C). Higher concentrations of AP20187 up to 1 μM were also tested, but these caused marked cytotoxicity in all cell types when added for extended periods of time (data not shown). Taken together, these results suggest that replacement of the ETV6 SAM domain with a surrogate dimerization module is insufficient for EN transformation.
FIG. 1.
Swapping the SAM domain of EN with an inducible FKBP1E dimerization module is insufficient to restore transformation activity. (A) Protein expression in FKBP1E cell lines. Anti-HA IP followed by anti-HA Western blotting was performed on lysates from FKBP1E control and FKBP1E-NTRK3-expressing NIH 3T3 cells. Arrows indicate HA-tagged proteins. (B) Autophosphorylation of FKBP1E-NTRK3 upon addition of dimerizer. AP20187 stimulation (100 nM) of FKBP1E-NTRK3-expressing NIH 3T3 cells induces FKBP1E-NTRK3 phosphorylation as shown by anti-HA IP and antiphosphotyrosine (RC20) IB. FKBP1E-NTRK3 cells exhibit transient MEK phosphorylation and low levels of cyclin D1 expression upon dimerizer addition. Equal protein levels were determined by using a Grb2 antibody. (C) Results of soft agar assays containing AP20187 dimerizer. Graph depicts the percentage of colonies formed per number of cells plated for cell lines tested with or without dimerizer. Soft agar wells were stained with crystal violet and are depicted below the graph. The addition of the dimerizer had no effect on EN-mediated transformation and did not induce transformation in 1E-NTRK3- or 2E-NTRK3-expressing cells.
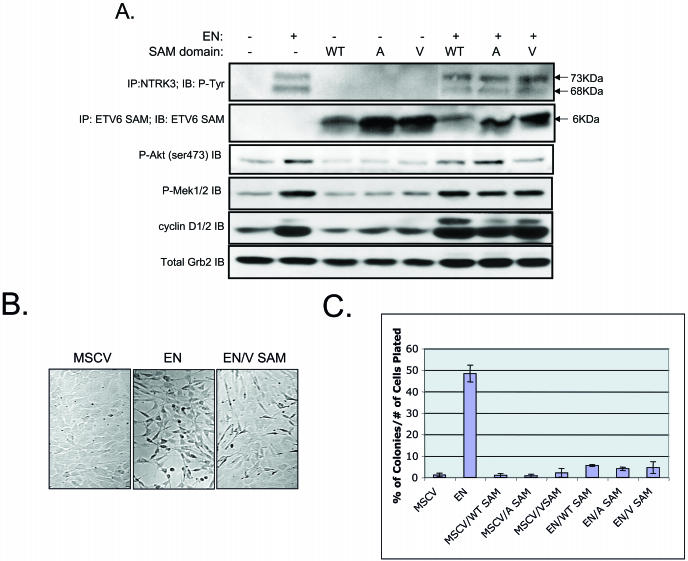
Mutations within the EN SAM domain block EN tyrosine phosphorylation, induction of cyclin D1 expression, and cell transformation.
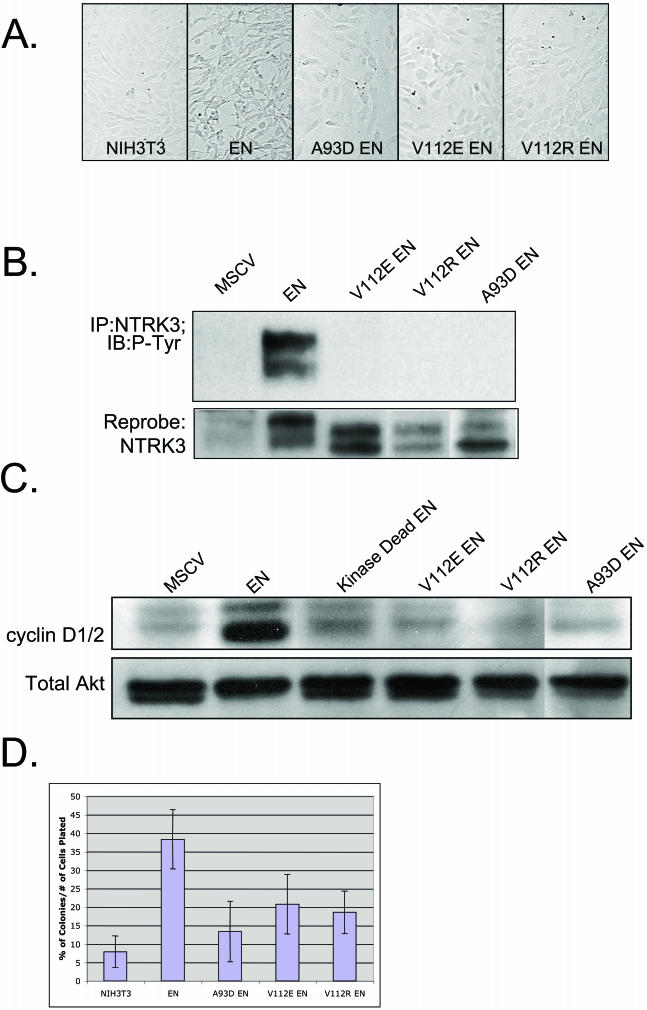
Kim et al. (16) reported that mutation of residues within either self-association interface of the ETV6 SAM domain, namely alanine 93 to aspartate (A93D) or valine 112 to glutamate or arginine (V112E or R), could abolish the ability of WT ETV6 to polymerize. We hypothesized that these mutations would similarly affect EN self-association and that this would, in turn, inhibit the transforming activity of the fusion protein. Using site-directed mutagenesis, we introduced each of these amino acid substitutions within the SAM domain of EN to generate A93D-EN, V112E-EN, and V112R-EN. Each construct was confirmed to be free of any additional PCR errors by DNA sequencing. All three mutant constructs failed to induce morphological transformation of NIH 3T3 cells when expressed at levels similar to those of nonmutated EN (Fig. 2A). The proteins isolated from these cells remained in a non-tyrosine-phosphorylated state (Fig. 2B), suggesting that mutations in the SAM domain abolished the ability of EN to cross- or autophosphorylate itself. Moreover, cells expressing A93D-EN, V112E-EN, or V112R-EN expressed markedly lower levels of cyclin D1 compared to those expressing nonmutated EN (Fig. 2C). Loss of transformation activity was confirmed by soft agar colony assays, as NIH 3T3 cells expressing any one of the three EN SAM domain mutants showed a marked reduction in the ability to form colonies compared to cells expressing WT EN and a small increase relative to the control cells (Fig. 2D). Therefore both SAM binding interfaces must be intact for full EN-transforming activity.
FIG.2.
Characteristics of EN SAM variants with point mutations that disrupt the self-association of the SAM domain. (A) Morphology of NIH 3T3 cells expressing WT EN and EN SAM domain mutants (A93D-EN, V112E-EN, and V112R-EN). WT EN transforms NIH 3T3 cells, whereas the EN SAM domain mutants do not. (B) NTRK3 IP of NIH 3T3 cells expressing WT EN and EN SAM domain mutants probed with antiphosphotyrosine (RC20) and anti-NTRK3 antibodies. Mutant EN SAM proteins are not phosphorylated. Doublets are observed due to an alternative start site within the ETV6 gene. (C) Levels of cyclin D1, detected by Western blotting, in cells expressing WT EN, kinase-dead EN, A93D-EN, V112E-EN, and V112R-EN. WT EN cells express high levels of cyclin D1, but mutant EN cells do not. Equal protein levels were determined with a total AKT antibody. (D) Results of soft agar assay. Cells expressing mutant EN SAM domains form fewer colonies in soft agar than those expressing WT EN.
Altered cellular localization of EN SAM domain mutants.
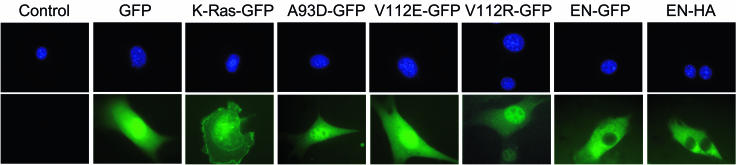
One explanation for the above findings is that mutations within the SAM domain of EN affect subcellular localization of this chimeric protein. To localize the different fusion proteins, WT EN, A93D-EN, V112E-EN, and V112R-EN were each C-terminally tagged with GFP, and cells expressing these constructs were fixed and visualized by fluorescent microscopy (Fig. 3). GFP alone and a GFP-tagged prenylated K-Ras construct were used as controls. The nonmutated EN-GFP protein was almost exclusively cytoplasmic, whereas the GFP-tagged SAM domain mutants each localized to the nucleus as well as to the cytoplasm. Expression of GFP-tagged proteins was confirmed by anti-NTRK3 IP followed by anti-GFP and anti-NTRK3 Western blotting (data not shown). Nonmutated EN was also tagged with HA and transiently expressed in NIH 3T3 cells. Cells were fixed, and HA-specific and fluorescein isothiocyanate-conjugated antibodies were used to determine the localization of the tagged protein. As with GFP-tagged EN, HA-tagged EN proteins were observed to be primarily cytoplasmic. These studies indicate that the presence of two functional SAM domain self-association interfaces is associated with retention of EN in the cytoplasm.
FIG. 3.
Differential localization of GFP-tagged WT EN and EN SAM domain mutants. NIH 3T3 cells were engineered to stably express all GFP constructs, except for the EN-HA construct, which was expressed by using a transient protocol. Cells were fixed and counterstained with DAPI. DAPI images (top) and corresponding GFP images (bottom) are shown. Cells expressing EN-HA were fixed, and the protein was detected by using anti-HA and fluorescein isothiocyanate-conjugated antibodies. Both EN-GFP and EN-HA are localized primarily to the cytoplasm, whereas all three EN SAM domain mutants (A93D-GFP, V112E-GFP, and V112R-GFP) are found in the cytoplasm and nucleus. GFP alone and K-Ras-GFP were used as controls.
A93D and V112R EN SAM domain mutants bind to each other but fail to self-associate.
Alanine 93 and valine 112 are located on opposite oligomerization interfaces of the ETV6 SAM domain. Thus, the A93D and V112R mutants of this domain are incapable of self-associating but can form a tight heterodimer by virtue of their complementary WT interfaces (16, 47). To test whether EN would be similarly affected by such mutations, EN, A93D-EN, and V112R-EN proteins were differentially tagged with either HA or V5 histidine. Various combinations, consisting of one HA-tagged construct along with one V5-tagged construct, were transiently cotransfected into 293T cells. IPs were performed on total cell lysates from these cells with anti-HA antibodies, followed by Western blotting with anti-V5 antibodies. As shown in Fig. 4, V5-tagged EN could be immunoprecipitated with HA-tagged EN, further confirming earlier observations that EN can self-associate (49). We also observed that EN can associate with either A93D-EN or V112R-EN (Fig. 4). In contrast, A93D-EN can bind to either V112R-EN or V112E-EN, but none of these proteins are capable of self-associating. These studies indicate that EN variants with an inactivating mutation in their complementary SAM domain binding interfaces can still interact in a heterotypic fashion, but they can no longer self-associate. Based on the reported properties of the isolated WT and mutant ETV6 SAM domains (16, 47), as well as further results shown below, these data are entirely consistent with the conclusion that EN self-associates to produce an open-ended head-to-tail polymer (see below), whereas variants with mutations of A93 and V112 are restricted to forming heterodimers.
FIG. 4.
Association of EN requires the presence of complementary WT SAM domain binding interfaces. NIH 3T3 cells expressing various combinations of V5- or HA-tagged proteins were anti-HA immunoprecipitated, followed by anti-V5 IB. WT EN is able to bind to itself and to each EN SAM domain mutant. Although A93D-EN and V112R-EN SAM are unable to self-associate, A93D-EN can form complexes with V112R-EN and V112E-EN.
Dimerization is insufficient for EN-mediated transformation.
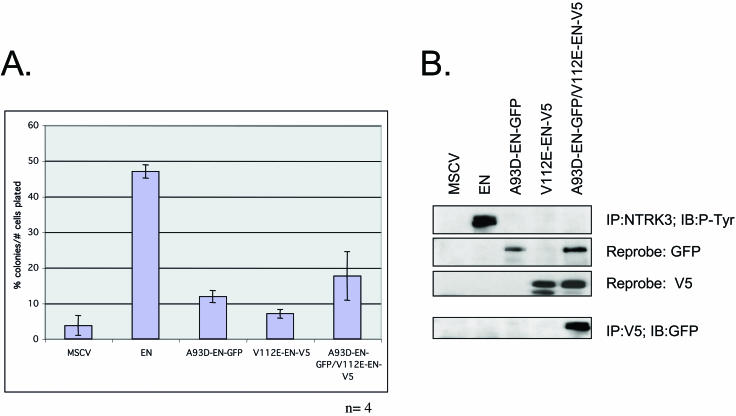
If the A93D and V112R EN mutants fail to self-associate but are capable of heterodimerizing (Fig. 4), then coexpression of these mutants would be expected to transform cells if dimerization is sufficient for this activity of EN. To test this hypothesis, we stably coexpressed A93D-EN-GFP and V112R-EN-V5 in NIH 3T3 cells and assessed the ability of these constructs to promote colony growth in soft agar (Fig. 5A). Western blotting with anti-NTRK3 antibodies confirmed equivalent expression levels (data not shown). Compared to cells expressing EN itself, fibroblasts coexpressing these constructs were markedly reduced in their colony-forming ability. The expression of each of the proteins was confirmed by Western blot analysis (Fig. 5B). Interestingly, even though V112E-EN and A93D-EN associated with each other as shown by co-IP (Fig. 5B, bottom), there was a complete absence of tyrosine phosphorylation of the fusion proteins observed in the doubly transfected cells (Fig. 5B, top). Also of note was the finding that while transformation activity in the presence of the two mutants was markedly reduced compared with WT EN, it was still slightly higher than in vector alone cells (see Discussion). Taken together, these results corroborate those from the FKBP-NTRK3 studies described above (Fig. 1), and both sets of results indicate that EN dimerization is insufficient for full EN transformation.
FIG. 5.
SAM domain-mediated dimerization is insufficient for EN transformation. (A) Soft agar data from cells expressing MSCV control, WT EN, A93D-EN-GFP alone, or V112E-EN-V5 alone or NIH 3T3 cells expressing both A93D-EN-GFP and V112E-EN-V5. (B) NTRK3 IP of all cell lines followed by anti-phosphotyrosine Western blotting (top panel). Only WT EN protein is phosphorylated. Protein expression of all constructs was assayed by IB with anti-GFP and -V5 antibodies (middle panels). The association of A93D-EN-GFP with V112-EN-V5 was confirmed by performing anti-V5 IP followed by anti-GFP Western blots (bottom panel). Equal protein levels were determined with an anti-Grb2 antibody.
Coexpression of isolated WT or mutant SAM domains in EN-expressing cells blocks transformation.
If EN polymerizes through its SAM domain, then it would be predicted that isolated SAM domains may be able to interrupt this association and thus inhibit transformation. To test this hypothesis, and to determine whether there are different activities associated with WT versus mutant SAM domain polypeptides, control (MSCV) and EN-expressing cells were cotransfected with constructs encoding either an isolated WT ETV6 SAM domain (WT-SAM), an isolated ETV6 SAM domain with the A93D mutation (A-SAM), or an isolated ETV6 SAM domain with the V112R mutation (V-SAM). EN expression was confirmed by IP with anti-NTRK3 antibodies followed by IB with RC20 anti-phosphotyrosine antibodies (Fig. 6A). Isolated SAM domain expression was confirmed by anti-ETV6 IP followed by IB with the same antibody (Fig. 6A). The levels of EN and each coexpressed isolated SAM domain protein appeared to be roughly equivalent. NIH 3T3 cells engineered to express the SAM domains alone (WT or mutant) were morphologically identical to untransformed control cells (data not shown). EN cells expressing either the isolated WT-SAM, A-SAM, or V-SAM domains demonstrated a more flattened appearance than and decreased refractility compared to EN cells alone, as shown by a representative experiment for V-SAM (Fig. 6B). Interestingly, cells coexpressing EN and SAM domain peptides still show similar levels of EN phosphorylation (Fig. 6A). Moreover, they maintain similar levels of activated AKT and MEK1/2 as well as elevation of cyclin D1. These cells were then plated in soft agar to determine their abilities to form anchorage-independent colonies (Fig. 6C). EN cells coexpressing either WT or mutant SAM domain constructs all showed the same result, with a dramatic reduction in soft agar colony formation compared to EN cells alone. These results indicate that coexpression of the isolated SAM domains, whether WT or mutant, has a strong dominant negative effect on EN-transforming activity.
FIG. 6.
Coexpression of isolated WT and mutant SAM domains blocks EN-mediated transformation. (A) Western blot analysis of EN and isolated SAM domain-expressing NIH 3T3 cells. NIH 3T3 cells were engineered to express MSCVneo alone, WT EN alone, MSCVneo plus isolated SAM domains (either WT-SAM, A-SAM, or V-SAM), or EN plus isolated SAM domains. Cells engineered to express WT EN exhibited a phosphorylated doublet of 68 and 73 kDa. The doublet is observed due to the presence of an alternative start site in the ETV6 portion of EN. Cells engineered to express isolated SAM domains produced a 6-kDa protein identified by anti-ETV6 SAM IP followed by Tricine gel electrophoresis and IB with an anti-ETV6 SAM antibody. Coexpression of isolated SAM domains in EN-expressing cells had no effect on downstream EN-mediated signaling as assessed by anti-phospho (ser 473) AKT, anti-phospho MEK1/2, or anti-cyclin D1/2 Western blot analysis. +, present; −, absent. (B) EN cells engineered to coexpress the isolated SAM domains (data only shown for EN and V SAM) exhibited a flatter shape than cells expressing EN alone. (C) Coexpression of isolated SAM domain mutants in transformed EN cells blocks colony formation in soft agar.
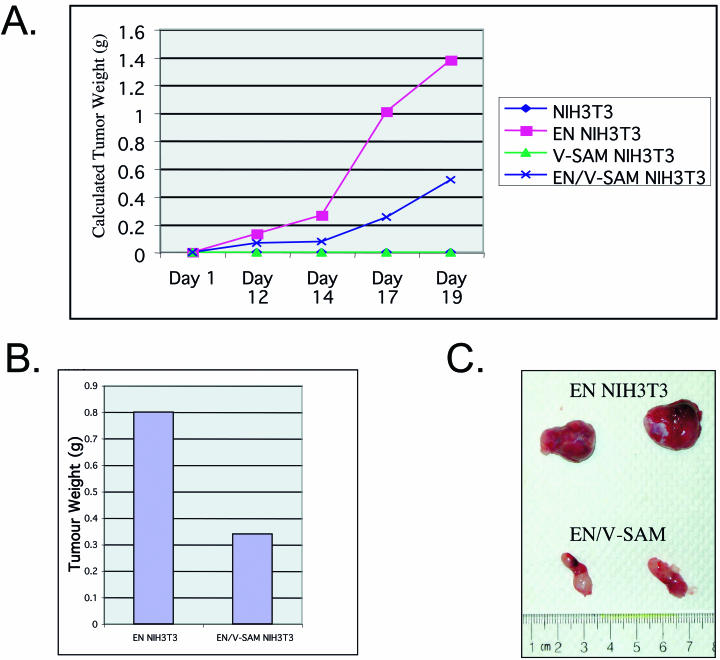
Coexpression of the isolated V-SAM domain in EN-expressing cells slows tumor growth in nude mice.
To assess the effects of coexpressing isolated SAM domains on EN transformation in vivo, we used a nude mouse tumor model. Cells were injected into the flanks of nude mice, and the injection sites were monitored for tumor growth over a period of 21 days. Control NIH 3T3 cells (vector plus or minus V-SAM) were injected into the left flanks of mice, and test cells (EN plus or minus V-SAM) were injected into the right flanks of the same mice. Caliper measurements were taken three times weekly to calculate the tumor growth over time. Control cells produced no visible tumors in mice, whereas EN-expressing cells produced large tumors by day 21. In comparison, EN plus V-SAM cells produced tumors at approximately half the rate of EN cells (Fig. 7A). By day 21, the average tumor masses were 0.8 and 0.35 g for EN and EN plus V-SAM cells, respectively (Fig. 7B). Figure 7C illustrates representative tumors excised from the mice. These results confirm an in vivo dominant negative effect on EN transformation by coexpression of isolated SAM domains.
FIG. 7.
EN-induced tumor formation in nude mice is attenuated by the presence of the isolated ETV6 SAM domain. (A) Coexpression of the isolated V-SAM domain mutant in NIH 3T3 cells transformed by EN blocks tumor formation in nude mice. Two million cells were injected subcutaneously into the flanks of male nude mice. Caliper measurements of the tumors were taken, and an estimated tumor weight was calculated over time. Control or isolated V-SAM-expressing cells did not form tumors in the mice. WT EN rapidly formed tumors over a 20-day period. WT EN/V-SAM-expressing cells also formed tumors but at a slower rate. (B) Average tumor weight after excision. Control cells did not form tumors. V-SAM-expressing WT EN cells produced tumors that were on average ca. half the size of those produced by WT EN-expressing cells. (C) Representative tumor sizes produced from WT EN-expressing cells (top set of tumors) and doubly transfected WT EN/V-SAM domain-expressing cells (bottom set of tumors).
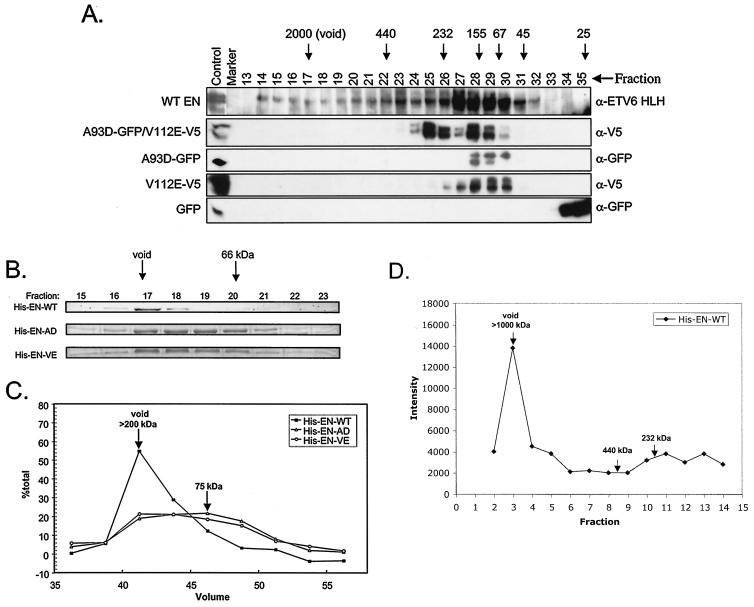
EN produces large multimeric complexes in vitro.
The above results indicate that both binding surfaces of the EN SAM domain are necessary for EN transformation. One explanation for this finding is that EN SAM domains mediate higher-order complex formation that is essential for transformation activity. To assess this possibility, lysates from NIH 3T3 cells expressing EN, EN-GFP, EN-A93D-GFP, or EN-V112E-V5 or coexpressing EN-A93D-GFP and EN-V112E-V5 were fractionated by FPLC with a Superose 6 HR 10/30 gel filtration column. Fractions were analyzed by Western blot analysis with anti-ETV6, anti-GFP, or anti-V5 antibodies. As shown in Fig. 8A, both EN-A93D-GFP and EN-V112E-V5 eluted in a peak near fraction 28, representing monomeric complexes. Coexpression of EN-A93D-GFP and EN-V112E-V5 resulted in an additional earlier peak centered at fraction 25, most likely representing dimeric complexes. In marked contrast, EN complexes began to elute as early as fraction 14 in the void volume and were present in a broad peak well past fraction 28 (Fig. 8A). An identical elution profile was observed for EN-GFP (data not shown). These findings strongly suggest that at least a portion of EN forms higher-order complexes and that this property is abolished by SAM domain mutations. To probe complex sizes in more detail, N-terminal His-tagged EN with WT or mutant SAM domains were bacterially expressed and then purified by Ni2+ affinity chromatography. Passage of these proteins through an S-200 Sepharose sizing column revealed that the A93D and V112E EN His-tagged mutants eluted as broad peaks centered at a volume approximating that of the bovine serum albumin standard (66 kDa) (Fig. 8B and C). This indicates that each exists predominantly in a monomeric form, given that the molecular mass of EN is ∼72 kDa. Consistent with the Superose gel filtration data, His-tagged EN eluted in the column void volume, indicating a significant increase in its apparent molecular size, as might arise from SAM domain-mediated polymerization (Fig. 8B and C). To further characterize these EN complexes, the S-200 purified protein was analyzed by additional chromatography on a Bio-Rad A-1.5 gel filtration column to provide a wider range of size determination. His-tagged EN again eluted in the void volume, in this case, calibrated with blue dextran (2,000 kDa). This indicates that EN complexes were at least larger than the upper limit of the Bio-Rad A-1.5 separation media (i.e., >1,000 kDa) (Fig. 8D). These results are consistent with EN forming large polymeric structures potentially incorporating more than 12 copies of EN protein per complex.
FIG. 8.
Comparison of relative sizes of WT and mutant EN proteins by gel filtration. (A) EN protein obtained from mammalian cell lysates are found in high-molecular-weight fractions as assessed by FPLC. FPLC was performed on lysates made from NIH 3T3 cells engineered to express the following proteins: WT EN, EN-GFP, A93D-GFP/V112EV5, A93D-GFP, V12E-V5, and GFP alone. Positive control, marker, and fraction numbers are listed above each lane. Molecular weight markers are shown with arrows. Western blotting was performed on the fractions, and antibodies used to detect the proteins are listed to the right (α, anti). Both WT EN and GFP-tagged EN proteins (data not shown) are found in higher-molecular-weight complexes. A93D-GFP/V112E-V5 proteins were found in higher-molecular-weight complexes than A93D-GFP or V112E-V5 alone. GFP protein was found in fractions corresponding to very small-molecular-mas complexes of approximately 30 kDa. Calculated polypeptide masses of the various constructs are as follows: WT EN and V5-tagged V112E EN, 77.2 kDa; GFP-tagged A93D EN, 106.2 kDa; and FP alone, 30 kDa. (B) WT EN proteins produced in bacteria form large complexes when assayed with protein sizing columns. His-EN-WT, His-EN-A93D, and His-EN-V112E protein fractions were collected from an S-200 Sepharose protein fractionation column. A Coomassie blue-stained gel of fractions from this column reveals that His-EN-WT forms a high-molecular-weight complex that elutes mainly in the void fraction. In contrast, His-EN-A93D and His-EN-V112E eluted as broad peaks centered at a volume approximating that of the bovine serum albumin standard (66 kDa). (C) Graphic representation of the amount of isolated protein in each fraction calculated as a percentage of the total protein. (D) His-EN-WT protein also elutes in the void volume from a BioGel A-1.5 column, suggesting that it forms large complexes of over 1,000 kDa in mass.
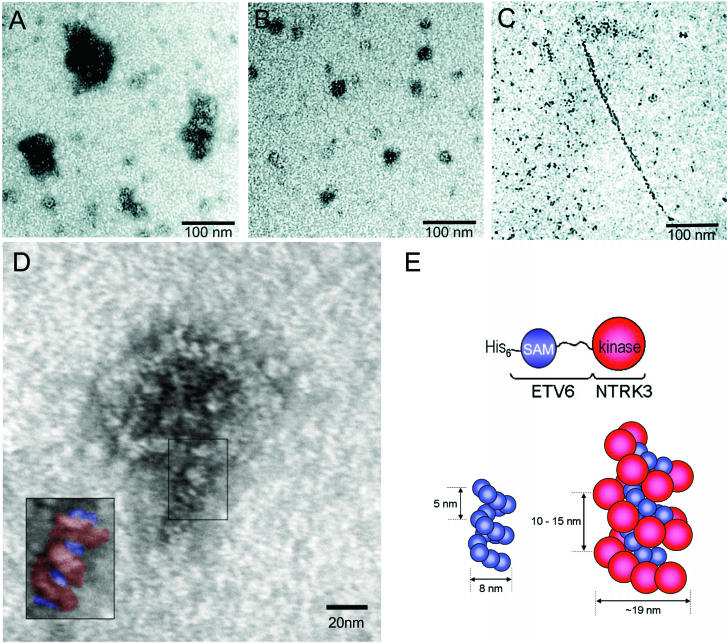
EM of EN complexes.
To directly visualize EN complexes, we performed EM on S-200 Sepharose-purified His-EN-WT. In keeping with the large size observed by gel filtration, His-EN-WT produced globular complexes with overall dimensions on the order of 50 to 100 nm (Fig. 9A). Because previous experiments indicated that coexpression of EN with the isolated ETV6 SAM domains results in reduced EN transformation (Fig. 6 and 7), we hypothesized that EN polymerization could be interrupted by the competitive binding of mutant ETV6 SAM domains to the EN SAM domains. To visualize this possibility, a 10-fold molar excess of isolated A93D SAM domain was coincubated with S-200 Sepharose-purified His-EN-WT for 48 h prior to microscopic analysis. Consistent with the hypothesis, resulting EM images indicate that the size of the EN complex is reduced on the order of 2.5-fold in size in the presence of an excess of A93D SAM domains (Fig. 9B). For comparison, EM was also performed on WT isolated ETV6 SAM domains. These peptides formed long, thin filaments with a diameter of ∼10 nm when polymerization was induced by a reduction in sample pH (Fig. 9C). This filamentous structure is in keeping with the previously published crystal structure and EM studies of the ETV6 SAM domain (16). Taken together, these EM data are consistent with EN forming higher-order polymeric structures in vitro that can be interrupted by the presence of ETV6 SAM domains with mutations within their binding interfaces. However, the globular EN polymers differ from the long filaments formed by the ETV6 SAM domain, possibly due to steric interactions involving the NTRK3 kinase domains.
FIG. 9.
EM of the ETV6 and EN polymers. Proteins were spotted onto grids and visualized by transmission EM after negative staining with uranyl acetate. (A) His-EN-WT formed large globular structures. (B) The His-EN-WT complexes were reduced in size upon addition of 10 M equivalents of the isolated A93D SAM domains. (C) For comparison, the isolated A93D SAM domains can form a polymeric filament when the pH is reduced to 6.0. (D) Higher-magnification scan of WT EN protein. The boxed insert highlights the repeating helical pattern. (E) Schematic model of the WT EN polymer. ETV6 SAM domains are diagrammed in blue, and NTRK3 kinase domains are diagrammed in red. Representations of both WT ETV6 and WT EN polymers, consistent with the EM data, are shown for size comparison.
DISCUSSION
All translocation-associated chimeric PTKs studied to date contain protein-protein interaction domains contributed by one of the partner genes. Accordingly, it is generally assumed that oligomerization mediated by these domains leads to constitutive kinase activation and subsequent transformation via stimulation of downstream signaling pathways. It was previously shown that EN, a chimeric oncoprotein expressed in malignancies of multiple cell lineages, self-associates through its ETV6 SAM domain and that this association is essential for its transformation activity (49). Oncogenesis associated with other ETV6 fusion proteins such as ETV6-PDGFRβ, ETV6-AML1, and ETV6-ABL also requires SAM domain-mediated self-association (7-9). It is therefore of considerable interest to better understand this self-association process, as its inhibition offers therapeutic potential in blocking transformation by ETV6 fusion oncoproteins.
Recent biochemical and structural studies revealed that the ETV6 SAM domain has two binding interfaces located on opposite surfaces of the native protein that lead to the formation of open-end head-to-tail polymeric structures in solution (16, 47). We therefore asked whether this SAM domain also mediates polymerization of ETV6-PTK fusion proteins such as EN and whether this is important for transformation. In this report we show that both SAM domain binding interfaces are essential for full EN transformation activity. EN mutants containing substitutions of charged residues (A93D and V112E or V112R) within either SAM domain hydrophobic binding interface fail to self-associate, do not become tyrosine phosphorylated, and lack transforming ability in NIH 3T3 cells. Moreover, sizing chromatography and EM suggest that, while EN chimera polymerize to form large multimeric complexes both in vitro and in vivo, EN variants with mutations within their SAM domains appear to exist in solution predominantly as monomeric species.
Several other findings in our study support the notion that higher-order polymer formation is important for EN transformation. First, chemically induced dimerization of FKBP-NTRK3 chimeras in which the ETV6 SAM domain was replaced by one or two FKBP binding domains did not result in NIH 3T3 cell transformation, even though abundant chimeric protein was expressed and became tyrosine phosphorylated after the addition of dimerizer. While it is possible that domain swapping might have introduced conformational changes affecting transformation even while maintaining an active NTRK3 PTK domain, other studies indicate that FKBP-ABL chimeras form dimers and retain transformation activity (39). A more likely explanation is based on our observation that chemically induced FKBP-NTRK3 complexes only stimulate transient autophosphorylation of the NTRK3 PTK. We also observed only transient MEK1 activation, cyclin D1 up-regulation (Fig. 1B), and AKT activation (data not shown) in these cells after the addition of AP20187. This suggests that chemically induced FKBP-NTRK3 dimers lack the ability to constitutively activate the Ras-MAPK and PI3K-AKT cascades, in contrast to EN (45). Second, while the stable coexpression of EN V-SAM and EN A-SAM mutants in NIH 3T3 cells resulted in heterodimerization of these proteins, cells remained nontransformed. Moreover, neither mutant became tyrosine phosphorylated under these conditions (Fig. 5) nor did they induce cyclin D1 expression (data not shown). Taken together, these findings strongly indicate that dimerization is insufficient for EN transformation. It is possible that higher-order polymer formation somehow facilitates or sustains activation of key transformation pathways such as Ras-MAPK and PI3K-AKT. One plausible mechanism would be by providing a scaffold for interaction with other molecules required to link EN to these cascades. For example, it was previously shown that the insulin-like growth factor 1 receptor substrate IRS-1 associates with EN and acts as an adaptor protein to couple EN to both Ras-MAPK and PI3K-AKT pathways in an insulin-like growth factor 1 receptor-dependent manner (20a,25). Further studies are necessary to determine whether nonpolymerizing EN mutants are deficient in binding IRS-1 or other EN-associated signaling molecules.
There are several paradoxical findings in this study. First, heterodimers of A93D-EN-GFP and V112E-EN-V5 did not become tyrosine phosphorylated (Fig. 5). One explanation is that EN polymerization somehow facilitates sustained PTK activation, possibly by providing an ordered grid of juxtaposed EN molecules for efficient auto- or cross-phosphorylation (see below). In this respect, the artificial FKBP1E-NTRK3 system may orient the kinase domains such that phosphorylation can occur within the context of an isolated homodimer, yet transformation does not result due to the lack of such polymerization-dependent ordering. Alternatively, polymerization may function to localize EN molecules subcellularly in close proximity to key signaling complexes required for sustained PTK activation or excluding an as yet unknown EN tyrosine phosphatase. Consistent with this was our finding that GFP-tagged EN is almost exclusively cytosolic while the GFP-tagged EN SAM domain mutant also localizes to the nucleus. It is therefore possible that polymerization is necessary for retention of EN in the cytoplasm. A second paradoxical finding is the observation that neither EN phosphorylation nor activation of downstream pathways was appreciably affected by coexpression of isolated SAM domains, suggesting additional complexity in how polymerization might influence EN transformation activity. Under these conditions there may be a heterogeneous population of EN polymers, SAM polymers, and SAM-EN dimers and oligomers in the cells. Consistent with this, Superose gel filtration demonstrated elution of EN in a range of sizes from large to dimeric or monomeric complexes, suggesting that a dynamic equilibrium exists between large polymers and smaller oligomers. Therefore, different threshold degrees of polymerization may be necessary for phosphorylation versus transformation, such that SAM domain polypeptides act to shift that equilibrium below that necessary for transformation but not phosphorylation. Another possibility is that in the presence of excess SAM domains, different tyrosine residues become phosphorylated in EN than in the absence of these polypeptides, which could be missed with standard anti-phosphotyrosine antibodies.
Evidence that higher-order complex formation of an ETV6 fusion protein may be essential for transformation has previously been reported (37), in which an ETV6-PDGFRβ mutant lacking the PTK domain blocked large complex formation and transformation. Replacement of the ETV6 SAM domain with the homologous N-terminal sequence of other ETS proteins led to the loss of transformation activity of the ETV6-PDGFRβ fusion protein (14, 37), again highlighting the importance of self-association of the ETV6 SAM domain in this process. Moreover, a recent study by Beissert et al. found that swapping the BCR coiled-coil domain of BCR-ABL with the SAM domain of ETV6 resulted in a shift from lower-molecular-mass complexes (∼2 to 4 monomers) to higher-molecular-mass complexes that eluted in the void volume of sizing columns (i.e., ∼2,000 kDa) (1). This corroborates our data suggesting higher-order polymer formation of EN proteins. Limited oligomerization is well documented as a requirement for transformation by chimeric oncoproteins (7-9, 11, 21, 24, 49). For example, a tetrameric structure for the BCR-ABL chimeric protein mediated by the BCR coiled-coil region was previously demonstrated biochemically (24) and then confirmed by crystallographic analysis (52). Although these ABL chimeras failed to transform Rat-1 fibroblasts, there was an inverse relationship between the size of the complexes and the ability of the kinase inhibitor STI571 to inhibit chimeric PTK autophosphorylation. On the other hand, it was also reported that replacement of BCR with the FKBP polypeptide resulted in chemically inducible dimers of FKBP-c-ABL that could transform BaF3 cells and fibroblasts (39), suggesting that dimerization is sufficient for this process. Moreover, Zhao et al. showed that the FKBP-JAK2 chimera in which the ETV6 SAM domain was replaced by FKBP2E (as well as the transmembrane and extracellular portions of low-affinity nerve growth factor receptor) was transforming (52). This is somewhat surprising in view of our findings, although these experiments were performed with a different system than ours, namely a hematopoietic system screening for interleukin-3 dependence in BaF3 cells. Therefore, polymerization may only be required for transformation by some SAM-containing ETV6 chimeras and in certain cellular contexts.
Polymerization also appears to influence the function of WT ETV6. It is postulated that higher-order polymer formation of ETV6 at the level of DNA affects chromatin organization and may underlie, at least in part, the transcriptional repressor function of ETV6 (16, 47). A similar chromatin-associated structure was found for SAM-containing members of a family of proteins that are unrelated to ETV6, the PcG of repressor proteins (15). This suggests that polymerization may be a more general phenomenon in SAM domain-containing proteins. This may also help to explain why the A93D-EN and V112E-EN/V112R-EN mutants retain some residual transforming activity in NIH 3T3 cells (Fig. 2). WT ETV6 is thought to function as a tumor suppressor, and haploinsufficiency for the ETV6 gene is described for childhood acute leukemias (32). Both EN SAM mutants are expected to retain the ability to bind WT ETV6, and therefore, partial inactivation of WT ETV6 by these overexpressed mutants may underlie their residual transformation activity. Alternatively, Kim et al. (16) showed that similar mutants of WT ETV6 self-associate under conditions of lower pH, such that there could be slight polymerization of the EN mutants in vivo.
When we performed EM of S-200 Sepharose-purified EN proteins isolated from bacteria, we observed 50- to 100-nm globular complexes that were reduced in size by approximately 2.5-fold in the presence of excess WT SAM domain peptides. In contrast, WT ETV6 SAM domains alone formed long polymeric filaments with a thickness of ∼10 nm, in keeping with the previously published crystal structure of the ETV6 SAM domain (16). If such polymerization is retained in EN complexes, then we can make preliminarily predictions as to the structure of EN globular complexes. If formation of the ∼10-nm-thin filament, as observed for isolated SAM domains, is preserved in EN polymers and if it is assumed that the diameter of the NTRK3 PTK domain is ∼5 nm (based on dimensions of other tyrosine kinase domains in the Protein Data Bank [www.rcsb.org/pdb]), then the PTK domain would be expected to coat the outside of the SAM domain filament due to steric constraints. We therefore constructed a potential composite model, based on the crystal structure of the ETV6 SAM domain polymer (16, 47) along with the estimated PTK domain size (Fig. 9E). In this model, the NTRK3 PTK domain is postulated to sterically alter packing within the polymer such that formation of elongated filaments is prevented and a globular shape of the complexes is favored. This putative large helical patterning is at least one possible explanation for the ∼20-nm-wide coiled and bent structure observed in higher magnification images of EN complexes (Fig. 9D). To further speculate on this model, it is possible that the helical shape facilitates optimal positioning of PTK domains for cross-phosphorylation. In the EN polymer, perhaps the PTK phosphorylation partners are not nearest neighbors in a linear sense but are located one helical turn apart for proper positioning as substrates for each other. This may provide an additional explanation as to why the A93D and V112E heterodimer is not tyrosine phosphorylated, as the PTK domains in the dimer are not properly juxtaposed for cross-phosphorylation. However, we do not rule out other interpretations, as additional mutational or structural analyses are required to further probe the nature of EN polymerization complexes.
We found that expression of either WT or mutated SAM domains in EN-transformed NIH 3T3 cells dramatically reduced EN-mediated morphological transformation and soft agar colony formation. Moreover, these cells were markedly attenuated in their ability to form tumors in nude mice. This indicates that isolated SAM domains can have a dominant negative effect on EN transformation. In keeping with the proposed role of polymerization in EN transformation, isolated SAM domains were able to reduce the size of high-molecular-weight EN complexes, as discussed above. Taken together, our results suggest that polymerization is a critical step in EN-mediated transformation and that targeting the dimerization interfaces of the ETV6 SAM domain may be a potential strategy for blocking the transforming activity of ETV6 SAM domain-containing fusion proteins. The two binding interfaces of the ETV6 SAM domain have recently been analyzed structurally by Tran et al. (47). Although no obvious pockets were identified within these interfaces that could act as binding sites for small-molecule inhibitors of polymerization, additional studies may reveal other potential targets for molecular therapeutic intervention. Alternatively, the use of SAM domain peptides may be a more effective strategy for blocking polymerization, although size considerations may necessitate new strategies for cellular uptake. However, the data presented in this paper demonstrate that isolated SAM domains can indeed be effectively utilized to competitively inhibit the polymerization and transforming activity of an oncogenic ETV6-PTK fusion protein.
Acknowledgments
We thank Martin Gleave and Mary Bowden for assistance with the mouse studies, Wayne Vogl for technical assistance, and Philip Webster for help with protein expression.
This work was supported by funds from the Canadian Institutes for Health Research (CIHR) (to P.H.B.S.), the CIHR/Candlelighter's Fund (to C.E.T.), the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to L.P.M.), the Natural Sciences and Engineering Research Council of Canada (to C.D.M.), the NCIC and Michael Smith Foundation (to A.M.S.), and the Johal Program in Pediatric Oncology Basic and Translational Research at the BC Research Institute for Children's and Women's Health. L.P.M. is a CIHR Scientist.
REFERENCES
- 1.Beissert, T., E. Puccetti, A. Bianchini, S. Guller, S. Boehrer, D. Hoelzer, O. G. Ottmann, C. Nervi, and M. Ruthardt. 2003. Targeting of the N-terminal coiled coil oligomerization interface of BCR interferes with the transformation potential of BCR/ABL and increases sensitivity to STI571. Blood 102:2985-2993. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 3.Bohlander, S. K. 2000. Fusion genes in leukemia: an emerging network. Cytogenet. Cell Genet. 91:52-56. [DOI] [PubMed] [Google Scholar]
- 4.Cazzaniga, G., S. Tosi, A. Aloisi, G. Giudici, M. Daniotti, P. Pioltelli, L. Kearney, and A. Biondi. 1999. The tyrosine kinase abl-related gene ARG is fused to ETV6 in an AML-M4Eo patient with a t(1;12)(q25;p13): molecular cloning of both reciprocal transcripts. Blood 94:4370-4373. [PubMed] [Google Scholar]
- 5.Chi, S. W., A. Ayed, and C. H. Arrowsmith. 1999. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 18:4438-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eguchi, M., M. Eguchi-Ishimae, A. Tojo, K. Morishita, K. Suzuki, Y. Sato, S. Kudoh, K. Tanaka, M. Setoyama, F. Nagamura, S. Asano, and N. Kamada. 1999. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood 93:1355-1363. [PubMed] [Google Scholar]
- 7.Golub, T. R., G. F. Barker, S. K. Bohlander, S. W. Hiebert, D. C. Ward, P. Bray-Ward, E. Morgan, S. C. Raimondi, J. D. Rowley, and D. G. Gilliland. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 92:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golub, T. R., G. F. Barker, M. Lovett, and D. G. Gilliland. 1994. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77:307-316. [DOI] [PubMed] [Google Scholar]
- 9.Golub, T. R., A. Goga, G. F. Barker, D. E. Afar, J. McLaughlin, S. K. Bohlander, J. D. Rowley, O. N. Witte, and D. G. Gilliland. 1996. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol. Cell. Biol. 16:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, J. B., C. D. Gardner, R. P. Wharton, and A. K. Aggarwal. 2003. RNA recognition via the SAM domain of Smaug. Mol. Cell 11:1537-1548. [DOI] [PubMed] [Google Scholar]
- 11.Grignani, F., V. Gelmetti, M. Fanelli, D. Rogaia, S. De Matteis, F. F. Ferrara, D. Bonci, C. Nervi, and P. G. Pelicci. 1999. Formation of PML/RAR alpha high molecular weight nuclear complexes through the PML coiled-coil region is essential for the PML/RAR alpha-mediated retinoic acid response. Oncogene 18:6313-6321. [DOI] [PubMed] [Google Scholar]
- 12.Iijima, Y., T. Ito, T. Oikawa, M. Eguchi, M. Eguchi-Ishimae, N. Kamada, K. Kishi, S. Asano, Y. Sakaki, and Y. Sato. 2000. A new ETV6/TEL partner gene, ARG (ABL-related gene or ABL2), identified in an AML-M3 cell line with a t(1;12)(q25;p13) translocation. Blood 95:2126-2131. [PubMed] [Google Scholar]
- 13.Janssen, J. W., S. A. Ridge, P. Papadopoulos, F. Cotter, W. D. Ludwig, C. Fonatsch, H. Rieder, W. Ostertag, C. R. Bartram, and L. M. Wiedemann. 1995. The fusion of TEL and ABL in human acute lymphoblastic leukaemia is a rare event. Br. J. Haematol. 90:222-224. [DOI] [PubMed] [Google Scholar]
- 14.Jousset, C., C. Carron, A. Boureux, C. T. Quang, C. Oury, I. Dusanter-Fourt, M. Charon, J. Levin, O. Bernard, and J. Ghysdael. 1997. A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFR beta oncoprotein. EMBO J. 16:69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, C. A., M. Gingery, R. M. Pilpa, and J. U. Bowie. 2002. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9:453-457. [DOI] [PubMed] [Google Scholar]
- 16.Kim, C. A., M. L. Phillips, W. Kim, M. Gingery, H. H. Tran, M. A. Robinson, S. Faham, and J. U. Bowie. 2001. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 20:4173-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klambt, C. 1993. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117:163-176. [DOI] [PubMed] [Google Scholar]
- 18.Knezevich, S. R., M. J. Garnett, T. J. Pysher, J. B. Beckwith, P. E. Grundy, and P. H. Sorensen. 1998. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 58:5046-5048. [PubMed] [Google Scholar]
- 19.Knezevich, S. R., D. E. McFadden, W. Tao, J. F. Lim, and P. H. Sorensen. 1998. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 18:184-187. [DOI] [PubMed] [Google Scholar]
- 20.Kyba, M., and H. W. Brock. 1998. The Drosophila Polycomb group protein Psc contacts ph and Pc through specific conserved domains. Mol. Cell. Biol. 18:2712-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Lannon, C. L., M. J. Martin, C. E. Tognon, W. Jin, S. J. Kim, and P. H. Sorensen. 2004. A highly conserved NTRK3 C-terminal sequence in the ETV6-NTRK3 oncoprotein binds the PTB domain of IRS-1: an essential interaction for transformation. J. Biol. Chem. 279:6225-6234. [DOI] [PubMed] [Google Scholar]
- 21.Lin, R. J., and R. M. Evans. 2000. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol. Cell 5:821-830. [DOI] [PubMed] [Google Scholar]
- 22.Mackereth, C. D., M. Scharpf, L. N. Gentile, and L. P. McIntosh. 2002. Chemical shift and secondary structure conservation of the PNT/SAM domains from the ets family of transcription factors. J. Biomol. NMR 24:71-72. [DOI] [PubMed] [Google Scholar]
- 23.Maurer, A. B., C. Wichmann, A. Gross, H. Kunkel, T. Heinzel, M. Ruthardt, B. Groner, and M. Grez. 2002. The Stat5-RARalpha fusion protein represses transcription and differentiation through interaction with a corepressor complex. Blood 99:2647-2652. [DOI] [PubMed] [Google Scholar]
- 24.McWhirter, J. R., D. L. Galasso, and J. Y. Wang. 1993. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol. Cell. Biol. 13:7587-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison, K. B., C. E. Tognon, M. J. Garnett, C. Deal, and P. H. B. Sorensen. 2002. ETV6-NTRK3 transformation requires insulin-like growth factor 1 receptor signaling and is associated with constitutive IRS-1 tyrosine phosphorylation. Oncogene 21:5684-5695. [DOI] [PubMed] [Google Scholar]
- 26.Nagaya, H., I. Wada, Y. J. Jia, and H. Kanoh. 2002. Diacylglycerol kinase delta suppresses ER-to-Golgi traffic via its SAM and PH domains. Mol. Biol. Cell 13:302-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters, P., S. D. Raynaud, J. Cools, I. Wlodarska, J. Grosgeorge, P. Philip, F. Monpoux, L. Van Rompaey, M. Baens, H. Van den Berghe, and P. Marynen. 1997. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15:12) in a myeloid leukemia. Blood 90:2535-2540. [PubMed] [Google Scholar]
- 29.Ponting, C. P. 1995. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4:1928-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabbitts, T. H. 1994. Chromosomal translocations in human cancer. Nature 372:143-149. [DOI] [PubMed] [Google Scholar]
- 31.Ramachander, R., C. A. Kim, M. L. Phillips, C. D. Mackereth, C. D. Thanos, L. P. McIntosh, and J. U. Bowie. 2002. Oligomerization-dependent association of the SAM domains from Schizosaccharomyces pombe Byr2 and Ste4. J. Biol. Chem. 277:39585-39593. [DOI] [PubMed] [Google Scholar]
- 32.Raynaud, S., H. Cave, M. Baens, C. Bastard, V. Cacheux, J. Grosgeorge, C. Guidal-Giroux, C. Guo, E. Vilmer, P. Marynen, and B. Grandchamp. 1996. The 12;21 translocation involving TEL and deletion of the other TEL allele: two frequently associated alterations found in childhood acute lymphoblastic leukemia. Blood 87:2891-2899. [PubMed] [Google Scholar]
- 33.Sakane, F., M. Kai, I. Wada, S. Imai, and H. Kanoh. 1996. The C-terminal part of diacylglycerol kinase alpha lacking zinc fingers serves as a catalytic domain. Biochem. J. 318(Pt 2):583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomon-Nguyen, F., V. Della-Valle, M. Mauchauffe, M. Busson-Le Coniat, J. Ghysdael, R. Berger, and O. A. Bernard. 2000. The t(1;12)(q21;p13) translocation of human acute myeloblastic leukemia results in a TEL-ARNT fusion. Proc. Natl. Acad. Sci. USA 97:6757-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz, J., C. P. Ponting, K. Hofmann, and P. Bork. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6:249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharrocks, A. D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827-837. [DOI] [PubMed] [Google Scholar]
- 37.Sjoblom, T., A. Boureux, L. Ronnstrand, C. H. Heldin, J. Ghysdael, and A. Ostman. 1999. Characterization of the chronic myelomonocytic leukemia associated TEL-PDGF beta R fusion protein. Oncogene 18:7055-7062. [DOI] [PubMed] [Google Scholar]
- 38.Slupsky, C. M., L. N. Gentile, L. W. Donaldson, C. D. Mackereth, J. J. Seidel, B. J. Graves, and L. P. McIntosh. 1998. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc. Natl. Acad. Sci. USA 95:12129-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, K. M., and R. A. Van Etten. 2001. Activation of c-Abl kinase activity and transformation by a chemical inducer of dimerization. J. Biol. Chem. 276:24372-24379. [DOI] [PubMed] [Google Scholar]
- 40.Spencer, D. M., T. J. Wandless, S. L. Schreiber, and G. R. Crabtree. 1993. Controlling signal transduction with synthetic ligands. Science 262:1019-1024. [DOI] [PubMed] [Google Scholar]
- 41.Stapleton, D., I. Balan, T. Pawson, and F. Sicheri. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44-49. [DOI] [PubMed] [Google Scholar]
- 42.Tessier-Lavigne, M. 1995. Eph receptor tyrosine kinases, axon repulsion, and the development of topographic maps. Cell 82:345-348. [DOI] [PubMed] [Google Scholar]
- 43.Thanos, C. D., S. Faham, K. E. Goodwill, D. Cascio, M. Phillips, and J. U. Bowie. 1999. Monomeric structure of the human EphB2 sterile alpha motif domain. J. Biol. Chem. 274:37301-37306. [DOI] [PubMed] [Google Scholar]
- 44.Thanos, C. D., K. E. Goodwill, and J. U. Bowie. 1999. Oligomeric structure of the human EphB2 receptor SAM domain. Science 283:833-836. [DOI] [PubMed] [Google Scholar]
- 45.Tognon, C., M. Garnett, E. Kenward, R. Kay, K. Morrison, and P. H. Sorensen. 2001. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 61:8909-8916. [PubMed] [Google Scholar]
- 46.Tognon, C., S. R. Knezevich, D. Huntsman, C. D. Roskelley, N. Melnyk, J. A. Mathers, L. Becker, F. Carneiro, N. MacPherson, D. Horsman, C. Poremba, and P. H. Sorensen. 2002. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2:367-376. [DOI] [PubMed] [Google Scholar]
- 47.Tran, H. H., C. A. Kim, S. Faham, M. C. Siddall, and J. U. Bowie. 2002. Native interface of the SAM domain polymer of TEL. BMC Struct. Biol. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu, H., M. Barr, D. L. Dong, and M. Wigler. 1997. Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol. 17:5876-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wai, D. H., S. R. Knezevich, T. Lucas, B. Jansen, R. J. Kay, and P. H. B. Sorensen. 2000. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene 19:906-915. [DOI] [PubMed] [Google Scholar]
- 50.Wang, L. C., W. Swat, Y. Fujiwara, L. Davidson, J. Visvader, F. Kuo, F. W. Alt, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1998. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 12:2392-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yagasaki, F., D. Wakao, Y. Yokoyama, Y. Uchida, I. Murohashi, H. Kayano, M. Taniwaki, A. Matsuda, and M. Bessho. 2001. Fusion of ETV6 to fibroblast growth factor receptor 3 in peripheral T-cell lymphoma with a t(4;12)(p16;p13) chromosomal translocation. Cancer Res. 61:8371-8374. [PubMed] [Google Scholar]
- 52.Zhao, X., S. Ghaffari, H. Lodish, V. N. Malashkevich, and P. S. Kim. 2002. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat. Struct. Biol. 9:117-120. [DOI] [PubMed] [Google Scholar]