Abstract
Comprehensive characterization of the antigen-specific B cells induced during infections or following vaccination would facilitate the discovery of novel antibodies and inform how interventions shape protective humoral responses. The analysis of human B cells and their antibodies has been performed using flow cytometry to evaluate memory B cells and expanded plasmablasts, while microtechnologies have also provided a useful tool to examine plasmablasts/plasma cells after vaccination. Here we present an integrated analytical platform using arrays of subnanoliter wells (nanowells) for constructing detailed profiles for human B cells comprising the immunophenotypes of the cells, the distribution of isotypes of secreted antibodies, the specificity and relative affinity for defined antigens, and for a subset of cells, the genes encoding the heavy and light chains. The approach combines on-chip image cytometry, microengraving, and single-cell RT-PCR. Using clinical samples from HIV-infected subjects, we demonstrate that the method can identify antigen-specific neutralizing antibodies, is compatible with both plasmablasts/plasma cells and activated memory B cells, and is well-suited for characterizing the limited numbers of B cells isolated from tissue biopsies (e.g., colonic biopsies). The technology should facilitate detailed analyses of human humoral responses for evaluating vaccines and their ability to raise protective antibody responses across multiple anatomical compartments.
Keywords: Microengraving, humoral responses, immune profiling, plasma and memory B cells analysis, nanowells
1. Introduction
Characterizing the nature and breadth of the antibody responses generated in humans is important for understanding how vaccines elicit prophylactic protection and for developing new insights to designing effective vaccines against diseases such as HIV, hepatitis C, tuberculosis, and malaria. Despite strong correlates of protection associating humoral responses with common vaccines, it is still unclear how to elicit such responses by rational design. Strategies for reverse engineering of immunogens for vaccines depend on efficient means for identifying and characterizing neutralizing antibodies from infected patients [1, 2]. Furthermore, the enumeration of novel antibodies with useful properties (e.g., broad and potent neutralizing activity) directly from humans may also provide new candidates for therapies and diagnostics. In this paper, we present a process for evaluating antigen-specific B cells from humans using a dense array of subnanoliter wells (nanowells) and an integrated set of analytical operations.
The most developed approaches to identify and recover individual antigen-specific B cells from primary samples use fluorescence-activated cell sorting (FACS). Sorting of individual memory B cells labeled with fluorescent antigen bound to surface-expressed immunoglobulins (Igs) has enabled the discovery of novel antibodies, including new broadly neutralizing antibodies from HIV-infected subjects [3, 4]. Following acute infection or vaccination, indiscriminate sorting of plasma cells or plasmablasts has also enabled the recovery of native human antibodies to pathogens like influenza [5–7]. For chronic diseases like HIV, however, this approach is relatively inefficient since the antigen-specific cells of interest represent only a minor fraction of all activated B cells [8]. The ELISPOT-like immunospot array assay on a chip (ISAAC) has also facilitated the recovery of antigen-specific antibody-secreting cells (ASCs) from recently vaccinated subjects [9].
We have previously demonstrated the use of arrays of nanowells to recover mouse hybridomas producing antigen-specific antibodies [10, 11], and to characterize the diversity of isotypes and affinities among the antigen-specific antibodies produced by primary mouse B cells following immunization [12]. Here we present a new integrated analytical process that extends these approaches to efficiently and comprehensively evaluate human B cells. The process combines image-based cytometry, microengraving, and automated micromanipulation to yield multidimensional data on the immunophenotypes of the B cells, the distribution of isotypes of secreted antibodies and the relative affinity of the secreted antibodies for specific antigens for thousands of cells in parallel. In addition, for antigen-specific antibodies, the method allows the recovery of the paired genes encoding the variable regions of the heavy and light chains. We show that the approach can be applied to characterize ASCs and activated memory B cells from the same individual, as well as B cells isolated from mucosal tissue biopsies. The flexibility and compatibility of the technique with small samples makes this approach a useful complement to existing methods for evaluating humoral responses in humans, and should provide a rapid and cost-effective technology for monitoring responses to vaccines and pathogens across different compartments.
2. Materials and methods
A detailed description of methods used is included as supplementary information in Appendix.
2.1. Ethics statement
Patient samples were obtained following approval by the institutional review boards at Massachusetts General Hospital (MGH), Boston; Massachusetts Institute of Technology (MIT), Cambridge; and Brigham and Women’s Hospital (BWH), Boston. Written informed consent was obtained from study participants prior to enrollment in the study.
2.2. Peripheral blood and mucosal tissue samples
Blood and intestinal biopsies were collected from HIV-infected individuals from a cohort of controllers (HIV viral load <2,000 RNA copies/ml in the absence of ART) from MGH [13]. PBMCs from blood samples were isolated using Histopaque®-1077 (Sigma-Aldrich). Cells were then either analyzed fresh or from frozen aliquots of 107 cells. Intestinal biopsies were obtained by upper and lower endoscopy. Tissue was disaggregated using both mechanical disruption and enzymatic digestion, based on published protocols [14]. Recovered mucosal cells were washed in media and used fresh. PBMCs from healthy donors were obtained from BWH or purchased from SeraCare Life Sciences Inc. (Milford, MA). Before screening populations of ASCs, PBMCs were thawed and rested for 1 h in complete media (RPMI 1640 media supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin; 37°C, 5% CO2).
2.3 Activation of memory B cells
A mixture of stimulatory molecules based on a previous report for polyclonal activation of B cells [15] was used to induce antibody secretion by resting memory B cells. ~3 × 106 PBMCs were seeded in 5 mL round-bottom tubes (BD Falcon) and incubated for 3–7 days in 500 μL of complete media, containing CD40L (2.5 μg/mL, Peprotech), IL-21 (50 ng/mL, eBioscience), anti-human APO1 (2.5 μg/mL, eBioscience) and EBV (5 μg, Advanced Biotechnology Inc.).
2.4. Antibody-producing cell lines
Chinese hamster ovary (CHO) cell lines producing b12 (anti-gp120) and 2F5 (anti-gp41) antibodies (courtesy of D. Burton, Scripps Institute) were cultured in ProCHO-5 media (Lonza) with 3% FBS, 1× HT supplement (Gibco), 1× GS supplement (Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin and 50 μM L-methionine sulfoxime (Sigma-Aldrich). A human B cell hybridoma cell line producing the 4D20 (anti-hemagglutinin) antibody [16] (courtesy of J. Crowe, Vanderbilt University) was adapted to grow in RPMI 1640 media with 15% (v/v) FBS, 2 mM L-glutamine and 1 mM sodium pyruvate. Cultures were passaged every 3–5 days and used in experiments when cultures were 70–80% confluent.
2.5. Fabrication of arrays of nanowells
Arrays with ~80,000–250,000 wells (50 × 50 × 50 μm3 or 30 × 30 × 30 μm3 in dimension, respectively) were manufactured as previously described using poly (dimethylsiloxane) (Sylgard 184) [11, 17]. After curing in custom-made injection molds, arrays were removed, covered and stored at ambient conditions until use. Arrays were treated with oxygen plasma (PDC-001, Harrick Plasma) immediately prior to use.
2.6. Image-based cytometry
PBMCs were stained for viability and surface-expressed proteins with CellTrace™ Calcein Violet AM and a panel of mouse anti-human antibodies – anti-CD19 (clone SJ25-C1, Qdot® 605 conjugate), anti-CD20 (clone 2H7, Alexa Fluor® 488 conjugate), anti-CD38 (clone HB7, PerCP-eFluor® 710 conjugate), and anti-CD138 (clone B-B4, APC conjugate), using manufacturers’ recommended concentrations. Cells were labeled for 30 min at room temperature in complete media. After staining, cells were deposited onto an array of nanowells to obtain a density of ~3–5 cells/well, washed with PBS containing 2% (v/v) FBS, and covered with a LifterSlip™ (EMS). Given that these cells constitute only ~5% of total PBMCs, each occupied well at this density holds 1 B cell on average. Transmitted-light and fluorescent images were then acquired on an automated epifluoresence microscope (Carl Zeiss) fit with an EM-CCD camera (Hamamatsu). The images were analyzed on a block-by-block basis using custom software [18] to determine the viability and the level of expression for each marker. Data were gated first for size then mean fluorescence intensities were extracted for the different fluorescent channels.
2.7. Microengraving
A protocol outlining this technique has been published [11]. For evaluating human B cells, poly-L-lysine-functionalized glass slides were coated with a polyclonal donkey anti-human (H+L) antibody (25 μg/mL, Jackson Immunoresearch), and microengraving was conducted for 2 h. Microarrays were scanned on a GenePix® 4200AL and analyzed with GenePix® Pro 6.0 (Molecular Devices). Printed elements in the arrays were considered Ig+ with % saturation ≤ 2, signal-to-noise ratio ≥ 1, ≥ 40% of pixels above background + 1 standard deviation and CV ≤ 200, and after compensation for bleed-through of signal from adjacent channels. All antigen-specific events were also confirmed by visual inspection since they were typically <10% of all Ig+ events.
2.8. Variable gene recovery and amplification
Cells were retrieved with an automated micromanipulator (AVISO CellCelector™, ALS GmbH) [19], and deposited into 20 μL 1× First-Strand Buffer (Invitrogen) and 10 U RNasin® (Promega) in the wells of a 96-well PCR plate. Heavy and light chain variable genes were amplified using primer sets and protocols previously described with modifications [20, 21]. Germline V(D)J usage and CDR3 sequences were determined using IMGT [22].
2.9. Antibody expression and validation
Paired heavy and light chain genes were cloned into vectors for expression as human IgG1 (courtesy of M. Nussenzweig, Rockefeller University), based on a published protocol [23]. Cloned antibodies were transiently expressed in human embryonic kidney (HEK) 293T cells (ATCC). Despite successful recovery and sequencing, we note that ~30% of antibodies selected did not clone or express as full-length human IgG1 after transfection in the host cell line, especially with antibodies from non-IgG1 isotypes (Table A1, Appendix). Antibodies in culture supernatants were assayed for binding on a custom protein microarray comprising a cross-clade panel of recombinant HIV envelope (Env) proteins (Table A2, Appendix). Microarrays were scanned on GenePix® 4200AL and analyzed with GenePix® Pro 6.0.
2.10. Neutralization assay
Neutralizing activity of expressed antibodies was tested using a published neutralization assay [24].
3. Results
3.1. Nanowell-based analysis of human antibody-secreting cells
We designed a new analytical process that combines microengraving, epifluoresence microscopy and single-cell RT-PCR to comprehensively characterize primary human B cells involved in antigen-specific responses (Fig. 1). Cells were used either directly or after activation with stimulants – depending on the subset analyzed – and then stained for viability and surface receptors. The labeled cells were deposited into arrays of 84,672 nanowells, to allow the analysis of ~100,000 total cells per array. The arrays were imaged on an epifluoresence microscope to record the expressed phenotypes of every cell and the occupancy of each well. Microengraving was then performed to capture secreted antibodies. The antibody microarrays produced were incubated with solution-phase antigens and then with labeled detection antibodies to highlight antigen-specific antibodies. Cells of interest were recovered with an automated micromanipulator (~60 cells/h), and heavy and light chain variable genes amplified by RT-PCR then sequenced. The efficiency of amplifying genes from cells recovered after microengraving ranged from 30–70%. This combined approach typically yields data for ~103–104 Ig-secreting cells per array and can be completed in 3–4 days.
Figure 1.
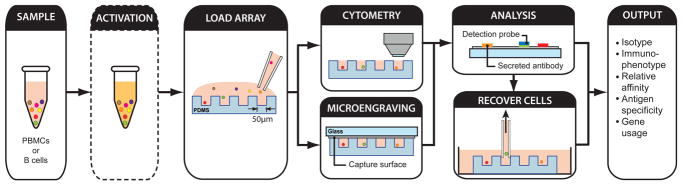
Process schematic for the integrated analysis of B cells using microengraving and on-chip cytometry. Incubation step to activate memory B cells is optional (dashed outline).
3.2. On-chip cytometry for immunophenotyping of human B cells
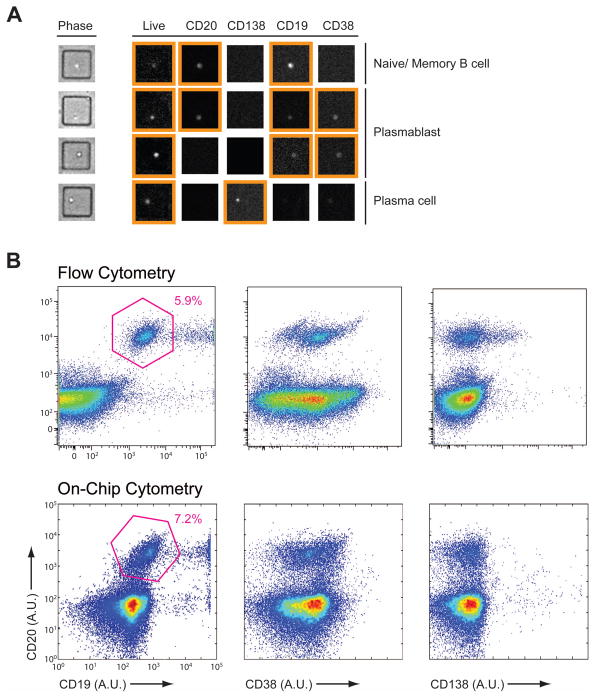
The evolution of B cells from plasmablasts to terminally differentiated plasma cells is accompanied by the increased expression and/or downregulation of CD19, CD20, CD38 and CD138 cell-surface receptors [25]. We validated a panel of detection antibodies against these receptors for analysis of B cells in nanowells by image-based cytometry (Fig. 2A). The distributions of populations enumerated on-chip were similar to those determined by flow cytometry (Fig. 2B). These results indicate that image-based cytometry can sufficiently resolve the major classes of B cells, despite the narrower dynamic range typically available on epifluoresence microscopes equipped with CCDs.
Figure 2.
Comparison of on-chip cytometry and flow cytometry. PBMCs from a healthy donor were stained for viability with Calcein violet AM, and for expression of CD19, CD20, CD38 and CD138. A portion of the labeled cells were loaded onto an array of nanowells and imaged by epifluoresence microscopy. The remaining cells were analyzed on a BD LSRFortessa™ cell analyzer. (A) Representative micrographs from wells with single cells displaying different expression patterns. Signals gated as positive with custom-built analysis script are highlighted with orange boxes for each. (B) Density plots of both flow cytometry (top) and on-chip cytometry (bottom) generated from the same sample.
3.3. Microengraving for the classification of isotypes and antigen-specific antibodies
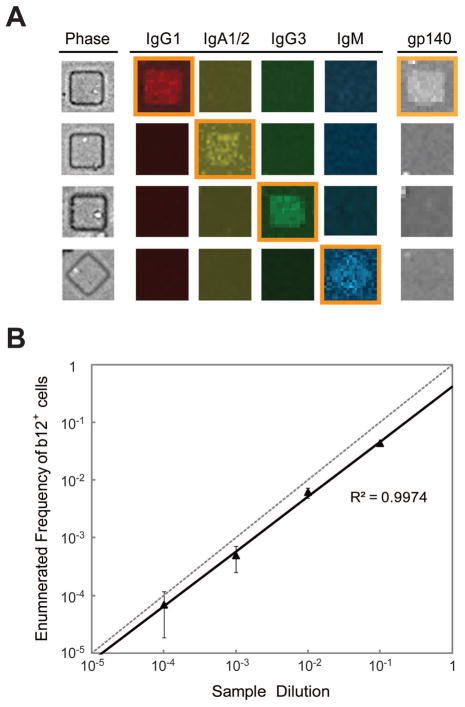
Microengraving is a soft-lithographic technique that allows the capture and analysis of molecules secreted by cells confined in an array of nanowells [10]. To establish the utility of this method for characterizing the antibodies secreted from human B cells, we used cells isolated from HIV-infected subjects. This choice of sample allowed us to test and optimize different configurations of the assay, and to compare our results to well-characterized antibodies (e.g., b12, 2G12, 2F5, VRC01). To analyze the distribution of isotypes of antibodies secreted, we validated a panel of antibodies to detect secreted IgG1, IgA1/2, IgG3, and IgM, as well as specificity for an HIV-related antigen (Fig 3A). These isotypes were chosen based on their prevalence and importance in mediating B cell effector functions. We verified with hand-spotted microarrays [11] that >10 ng/ml of each isotype can be detected in supernatants—a concentration much less than that estimated to result during microengraving for 2 h with ASCs (~10 μg/mL) [26].
Figure 3.
Concurrent profiling of isotype and antigen-specificity by microengraving. (A) Primary antibody-secreting cells were analyzed by microengraving, and the resulting microarray was incubated with solution-phase, fluorescently-labeled recombinant gp140 monomer (YU2 strain), and our optimized panel of anti-human Ig detection antibodies. Representative micrographs of the different isotypes detected are shown, with signals gated as positive for IgG1, IgA1/2, IgG3, IgM and bound antigen highlighted. (B) Plot of number of b12-producing cells enumerated by microengraving as a function of their dilution. Frequencies of CHO-b12 cells in samples were calculated as the ratio of the number of IgG+MV+ spots with CHO-b12 cells present in the wells, and the total number of IgG+ events.
One advantage of microengraving is the flexibility to interrogate the resulting antibody microarrays with different probes to highlight specificities for different antigens. We found that microengraving could identify Env-specific antibodies using a range of probes derived from Env proteins. The probes tested included recombinant proteins (e.g., gp120, gp140), peptides (e.g., MPER-derived peptides 179.4 and 57), and inactivated virus-like particles (BaL microvesicles, MVs; courtesy of J. Lifson, NCI-Frederick). Using cell lines producing antibodies that bind gp120 (b12), gp41 (2F5) and influenza HA (4D20) as a negative control, we determined that sensitivities for antigen-specific detection were all high (monomeric YU2 gp140: 88.4% for b12; BaL MVs: 99.9% for b12; MPER peptides: ≥ 99.7% for 2F5) with thresholds set such that specificities were ≥ 95% (Fig. A1, Appendix). Furthermore, serial dilution of CHO cells producing b12 into populations of cells producing 2F5 allowed us to estimate the lower limit of detection for rare cells in a population to be 1 in 100,000 cells (Fig. 3B). The limit appears to scale linearly with the number of wells/cells analyzed, and is 1–2 orders of magnitude lower than that typically achieved by flow cytometry [27, 28]. These results together demonstrate that microengraving can identify rare cells producing desired antibodies.
3.4. Nanowell-based analysis of primary B cells from HIV-infected subjects
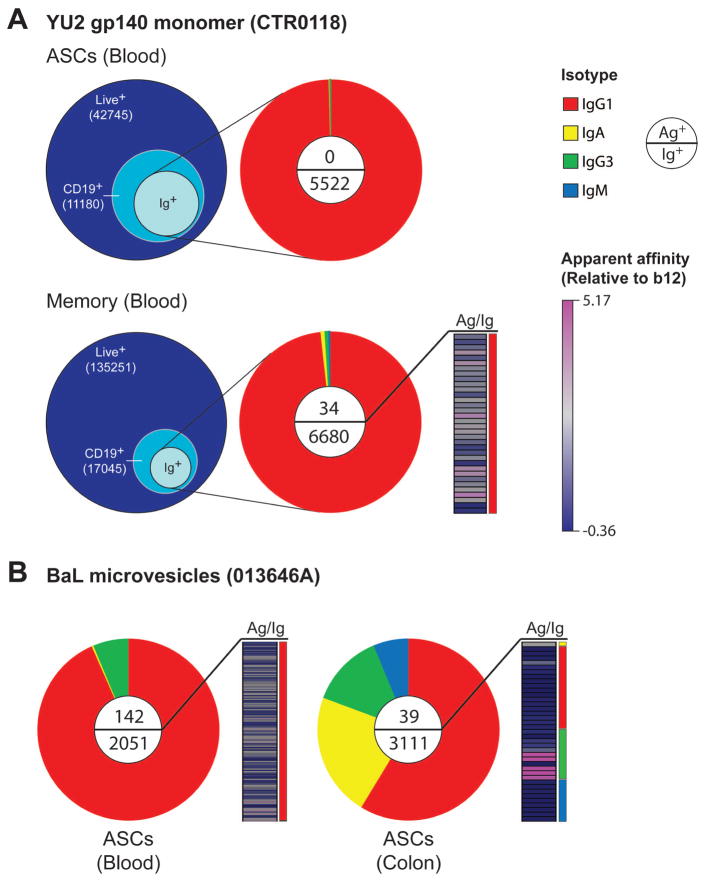
To establish the utility of this nanowell-based approach for examining the human humoral response, we applied our integrated analytical process to cells from HIV-infected subjects. In one example, we compared antibodies produced by plasmablasts/plasma cells and memory B cells circulating in blood from an HIV-infected elite controller (CTR0118). For both populations of cells, we determined the phenotypes of the viable cells, the isotypes of the secreted antibodies and their relative affinities for monomeric YU2 gp140 (Fig. 4A). IgG1 was observed to be the predominant isotype secreted in circulation, as expected [29]. For this subject, no Env-specific events were found in the circulating ASCs, but ~0.5% of Ig+ events were Env-specific among the memory B cells.
Figure 4.
(A) Integrated analysis of humoral responses from actively secreting cells or memory B cells in an HIV-infected sample. Bulk mononuclear cells from the blood were profiled for viability, surface-expressed phenotypes, isotype distribution and specificity of secreted antibodies for recombinant monomeric gp140 (YU2 strain, 25 μg/mL). Values in Venn diagrams represent the total numbers of cells on each array of nanowells. Distributions in pie-charts include events from wells with ≤ 5 live cells (~ 1 B cell/well). Apparent relative affinities are presented compared to median values observed from binding of b12 from CHO cells to primary bait. (B) Similar analysis was performed on samples of blood and disaggregated colon tissue from an HIV controller for the comparison of responses in both sites. HIV Env-specific antibodies were identified by screening with gp160-presenting MVs (BaL strain, 50 μg/mL).
The ability to isolate small numbers of cells using arrays of nanowells makes this process well-suited to characterize B cells recovered from other anatomical sites such as the colon, small bowel or reproductive tracts. To demonstrate this aspect of the technology, we analyzed mononuclear cells from the blood and colon of another HIV-infected elite controller (013646A) (Fig 4B). In this example, antibodies captured by microengraving were assayed for reactivity to BaL MVs. A large fraction of the cells isolated from the colon produced IgA1/2, as expected [29, 30]. For this subject, the frequency of antigen-specific events within this isotype was small (≤ 0.14%). Overall, the enumerated frequency of Env-specific antibodies was higher in the blood (7%) than in the colon (1.2%), with a wide range of apparent affinities observed.
Cells from the different samples were recovered for subsequent amplification by RT-PCR and the resulting sequences analyzed (Table 1). The average mutation rate (considering only mutations in V genes) for heavy chains of Env-specific monoclonals was 6.9% nucleotides and for light chains, 3.5% nucleotides. CDR3 lengths ranged from 11–25 amino acids for heavy chains, and from 7–13 amino acids for light chains. Together, the data from these two elite controllers demonstrate the nanowell-based approach can easily accommodate the comparative analyses of B cells from different cellular and anatomical compartments.
Table 1.
Sequence analysis for recovered clones
| Clone No. |
Patient ID | Bait | Isotype | V Region | D Region | J Region | Mutation Rate (%) |
CDR3 | CDR3 Length |
V Region | J Region | Mutation Rate (%) |
CDR3 | CDR3 Length |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIT 01 | 013646A | MVs | IgG1 | IGHV4-30-4*01 | IGHD4-11*01 | IGHJ5*02 | 4.1 | CARDLADSYNRLDPW | 15 | IGKV1D-33*01 | IGKJ3*01 | 0.6 | CQQYDNLPRGFTF | 13 |
| MIT 02 | 013646A | MVs | IgM | IGHV3-23*04 | IGHD3-3*01 | IGHJ4*02 | 4.4 | CAGPEGDSWSGRTFTLFDYW | 20 | IGKV3-20*01 | IGKJ4*01 | 4.2 | CQLYGSSLTF | 10 |
| MIT 03 | 013646A | MVs | IgG3 | IGHV4-34*01 | IGHD2-2*01 | IGHJ5*02 | 4.1 | CASGIGYCGSSSCLGW | 16 | IGLV2-14*01 | IGLJ3*02 | 4.1 | CVSSTTTNTRVF | 12 |
| MIT 21 | 013646A | MVs | IgG3 | IGHV1-18*01 | IGHD1-1*01 | IGHJ4*02 | 1.5 | CARSWRNGDDLGGGYW | 16 | IGKV2-24*01 | IGKJ1*01 | 0.0 | CMQATQFPQTF | 11 |
| MIT 34 | 013646A | MVs | IgG1 | IGHV1-69*01 | IGHD2-15*01 | IGHJ6*02 | 3.6 | CAGQDIVVVMATDADFYYYGLDDW | 24 | IGLV2-14*01 | IGLJ3*02 | 2.7 | CSSHASGDTLVF | 12 |
| MIT 35 | 013646A | MVs | IgG1 | IGHV1-2*02 | IGHD3-9*01 | IGHJ5*02 | 4.9 | CARVLSGLNWFDPW | 14 | IGKV1D-33*01 | IGKJ4*01 | 6.5 | 0 | |
| MIT 41 | 013646A | MVs | IgG1 | IGHV1-24*01 | IGHD3-22*01 | IGHJ5*02 | 7.1 | CTTTRGQKDYYDGSGVPGIYFDPW | 24 | IGKV1D-12*02 | IGKJ4*01 | 5.0 | CQQANSLLALSF | 12 |
|
| ||||||||||||||
| MIT 44 | CTR0118 | gp140 | N.D. | IGHV1-69*02 | IGHD2-15*01 | IGHJ4*02 | 10.2 | CARDAGLPQSFDFW | 14 | IGKV3-20*01 | IGKJ1*01 | 3.3 | CQQYGRSPRTF | 11 |
| MIT 45 | CTR0118 | gp140 | N.D. | IGHV1-69*02 | IGHD2-15*01 | IGHJ4*02 | 9.6 | CARDAGLPQSFDFW | 14 | IGKV3-20*01 | IGKJ1*01 | 3.3 | CQQYGRSPRTF | 11 |
| MIT 46 | CTR0118 | gp140 | N.D. | IGHV1-8*01 | IGHD2-15*01 | IGHJ5*02 | 6.4 | CARGRLLMQWPPRGGFDPW | 19 | IGKV3-20*01 | IGKJ2*02 | 3.5 | CQYYGSSPPSTF | 12 |
| MIT 48 | CTR0118 | gp140 | N.D. | IGHV1-18*01 | IGHD4-23*01 | IGHJ4*02 | 4.5 | CARDWGDDYGGSSDYFDYW | 19 | IGKV4-1*01 | IGKJ1*01 | 2.5 | CQQYYSPPRTF | 11 |
| MIT 61 | CTR0118 | gp140 | IgG1 | IGHV1-69*10 | IGHD5-24*01 | IGHJ5*02 | 5.4 | CASPGDGYYTW | 11 | IGKV2-30*01 | IGKJ2*01 | 1.4 | CMQGTHWPHTF | 11 |
| MIT 62 | CTR0118 | gp140 | IgM | IGHV3-23*04 | IGHD1-26*01 | IGHJ4*02 | 2.6 | CAKGGSVVGTTTRRFDCW | 18 | IGKV3-15*01 | IGKJ5*01 | 2.6 | CQQYNDWPITF | 11 |
| MIT 63 | CTR0118 | gp140 | IgA | IGHV3-23*04 | IGHD3-22*01 | IGHJ4*02 | 3.2 | CAKSLDGYYLFHYW | 14 | IGKV3-11*01 | IGKJ4*01 | 1.7 | CQQRSDWPTF | 10 |
| MIT 64 | CTR0118 | gp140 | IgG1 | IGHV1-8*01 | IGHD3-3*01 | IGHJ5*02 | 6.4 | CARGRLFMQWPPQGGFDPW | 19 | IGKV3-20*01 | IGKJ2*01 | 2.6 | CQEYGRSPPYPF | 12 |
| MIT 65 | CTR0118 | gp140 | IgG1 | IGHV3-23*04 | IGHD3-3*01 | IGHJ6*03 | 5.1 | CAKAAGHNFWSGYQAASYYYYMDVW | 25 | IGKV1-5*03 | IGKJ2*02 | 1.4 | CQQYNSYPYTF | 11 |
| MIT 66 | CTR0118 | gp140 | IgG1 | IGHV4-61*02 | IGHD3-3*01 | IGHJ5*02 | 12.4 | CARDQLTFWSVEYVNRFDPW | 20 | IGKV3-15*01 | IGKJ4*01 | 7.7 | CQSRLVF | 7 |
| MIT 69 | CTR0118 | gp140 | IgG1 | IGHV4-4*07 | IGHD6-13*01 | IGHJ5*02 | 0.0 | CARVGIAAAGTGVGWFDPW | 19 | |||||
| MIT 70 | CTR0118 | gp140 | IgG1 | IGHV1-18*01 | IGHD4-17*01 | IGHJ4*02 | 0.6 | CARDRNDYGVLLYW | 14 | IGKV3-11*01 | IGKJ4*01 | 0.6 | CQQRSNWRLTF | 11 |
| MIT 71 | CTR0118 | gp140 | IgG1 | IGHV4-30-2*01 | IGHD2-15*01 | IGHJ4*02 | 0.0 | CARGGYCSGGSCPSLFDYW | 19 | IGKV1-33*01 | IGKJ4*01 | 2.7 | CHQYYNFPLTF | 11 |
| MIT 75 | CTR0118 | gp140 | IgG1 | IGHV4-34*01 | IGHD1-7*01 | IGHJ6*02 | 0.0 | CARGLNWNYFPISYGMDVW | 19 | IGKV3-20*01 | IGKJ4*01 | 5.0 | CQKYGSSLTF | 10 |
| MIT 78 | CTR0118 | gp140 | IgG1 | IGHV1-58*02 | IGHD6-13*01 | IGHJ5*02 | 11.3 | CSARGHSFSLPFDSW | 15 | IGKV3-20*01 | IGKJ4*01 | 6.3 | CQKYGGSLTF | 10 |
| MIT 79 | CTR0118 | gp140 | IgG1 | IGHV3-30*18 | IGHD3-22*01 | IGHJ6*02 | 5.1 | CAKEEGYYDNSGYSWNYYYYGVDAW | 25 | IGKV3-20*01 | IGKJ3*01 | 0.0 | CQKYGGSLTS | 10 |
| MIT 80 | CTR0118 | gp140 | IgG1 | IGHV1-58*02 | IGHD6-13*01 | IGHJ5*02 | 11.6 | CSARGHSFSLPFDSW | 15 | IGKV3-20*01 | IGKJ4*01 | 7.4 | CQKYGGSLTF | 10 |
| MIT 81 | CTR0118 | gp140 | IgG1 | IGHV4-4*07 | IGHD2-21*02 | IGHJ4*02 | 12.0 | CGTRIDSSSEGLDFW | 15 | IGKV3-20*01 | IGKJ4*02 | 0.0 | CQEYGTSPRKF | 11 |
| MIT 82 | CTR0118 | gp140 | IgG1 | IGHV4-4*07 | IGHD2-21*02 | IGHJ4*02 | 11.7 | CARGIASSSEGLDFW | 15 | IGKV2-30*01 | IGKJ2*01 | 1.4 | CMQGTHWPHTF | 11 |
| MIT 83 | CTR0118 | gp140 | IgG1 | IGHV1-8*02 | IGHD3-3*01 | IGHJ5*02 | 8.2 | CARGRLFVQWPPQGGFDPW | 19 | IGKV3-20*01 | IGKJ2*01 | 6.2 | CQEYGRAPPYPF | 12 |
| MIT 84 | CTR0118 | gp140 | IgG1 | IGHV4-4*07 | IGHD2-21*02 | IGHJ4*02 | 11.4 | CARGIASSSEGLDFW | 15 | IGKV3-20*01 | IGKJ1*01 | 3.0 | CQLYGSSLWTF | 11 |
| MIT 85 | CTR0118 | gp140 | IgG1 | IGHV1-18*01 | IGHD2-15*01 | IGHJ4*02 | 5.5 | CARGCSGGSCYFDSW | 15 | IGLV3-9*02 | IGLJ4*01 | 0.0 | ||
Highlighted variable region analyses are from antibodies identified as negatives (Ag−) in primary screen. N.D., not determined
3.5. Validation of identified antibodies
To confirm the antibodies identified by microengraving are specific, we cloned and transiently expressed a subset of antibodies recovered in screens of samples from infected patients. Of 8 Env-reactive antibodies expressed, 3 were identified with BaL MVs, and 5 with monomeric YU2 gp140. Dilutions of the supernatants containing expressed Ig, and a set of well-characterized antibodies as controls, were applied to protein microarrays comprising a cross-clade panel of HIV antigens and an anti-Ig capture antibody (for normalization of the data) (Fig. 5A). Of the supernatants analyzed, only MIT 02 (derived from IgM) did not bind to any of the proteins. Some of the antibodies exhibited breadth in binding for a number of gp120 variants across all clades (MIT 44, 45, 64, 65), and another exhibited preference for gp41 (MIT 78). Similar to b12 and VRC01, two of the antibodies (MIT 34 and 41) demonstrated selective binding for the wildtype monomeric YU2 gp120, but not a mutant form (D368R) that has the CD4 binding site abrogated, and thus can neutralize the virus. We found that these two antibodies did, indeed, exhibit neutralizing activity (Fig. 6B; Table A3, Appendix). These results demonstrate that microengraving can identify antigen-specific antibodies from human B cells, and that it is feasible to recover neutralizing antibodies from HIV-infected subjects.
Figure 5.
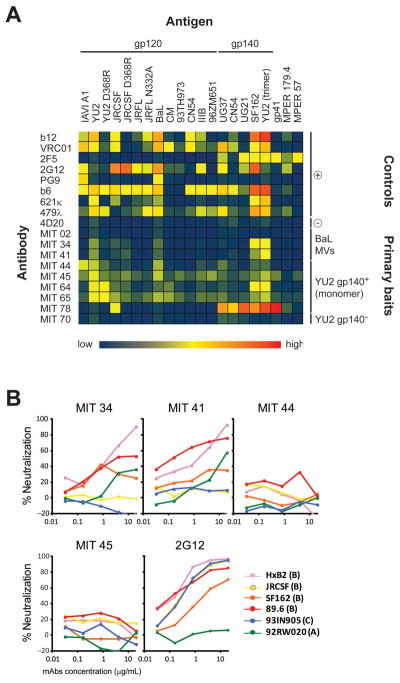
(A) Supernatants from cultures of HEK 293T cells expressing recombinant antibodies from screens were collected and incubated over custom antigen microarrays, along with dilutions of control antibodies (gp120-specific: b12, VRC01, b6, PG9, 2G12, 621κ and 479λ; gp41-specific: 2F5 and 4E10; and HA-specific 4D20). Binding to spotted antigens and anti-Ig capture species was detected with a fluorescently labeled goat anti-human antibody. Relative affinities (Ag/Ig) of the different antibodies tested are presented as a heatmap. (B) Neutralizing activity was tested on 4 antibodies against a panel of tier 1 and tier 2 HIV strains.
4. Discussion
We present here an integrated process for generating detailed profiles of human humoral responses that highlight characteristics of both antigen-specific B cells and their antibodies. The approach extends the capabilities of the nanowell platform beyond the characterization of mouse B cells and hybridomas [12], and incorporates the ability to recover the genes encoding human antibodies of interest by single-cell RT-PCR. The combination of on-chip cytometry, microengraving, and RT-PCR yields comprehensive data on the immunophenotypes of B cells, the isotypes and relative affinities of their antibodies for particular antigens, as well as the genes of the variable regions of the antibodies. We showed that this approach produces detailed profiles of the diversity of HIV Env-specific B cells present in blood and mucosal tissue, and yielded new antibodies with neutralizing activity.
The integrated process for profiling the breadth of antigen-specific human B cells has three advantages over methods relying on FACS and other microtechnologies. First, the method is well-suited to characterize primary B cells from any anatomical site from which a biopsy can be obtained. The example shown here with cells from a colonic biopsy underscores the ability to isolate and interrogate single cells from samples when the total numbers of cells are limited (~105 cells). This capability should enable detailed profiling of the cells contributing to humoral responses at sites of infection and those raised in mucosal tissues following vaccination. Second, compared to flow cytometry, microengraving has better sensitivity for rare cells and allows characterization of the specificity of antibodies from plasmablasts/plasma cells without compulsory recombinant expression. Third, compared to other microtechnologies demonstrated for antibody screening [9, 31], the ability to separate secreted antibodies physically from the cells producing them allows for easy multiplexed characterization and repeated analysis of the antibodies without potential damage or loss of source cells. Here we have shown that antibody microarrays produced by microengraving can be probed by inactivated virions, but other modalities of screening that use intact pathogens (e.g., fungi or bacteria) are also possible (J.C.L., unpublished results), highlighting the applicability of the method for identifying antibodies of interest in other disease models.
In conclusion, our technique for the integrated analysis of ASCs and memory B cells from primary human clinical samples should facilitate the discovery of cells producing antigen-reactive antibodies against targets such as HIV, or immunogens used in vaccination. The routine application of this technology in the analysis of tissue and blood samples would provide, at the single-cell level, a snapshot of the antigen-specific humoral response elicited following infection or vaccination. The further integration of this technology for direct enumeration of antigen-specific B cells with next-generation sequencing technologies would provide a comprehensive view of the B cell repertoire, and improve the understanding and design of vaccines targeting elusive pathogens like HIV.
Supplementary Material
Acknowledgments
We thank Dr. Elizabeth Bradshaw, Kenneth M. Law and Tomoyuki Hongo for helpful discussions and assistance with optimization of conditions for memory B cell activation and single-cell RT-PCR. This research was supported by the W.M. Keck Foundation, an IAVI Innovation award, and the NIH/NIAID (U19AI090970). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy And Infectious Diseases or the National Institutes of Health. A.O.O. was supported by UNCF/Merck Graduate Research Dissertation Fellowship. J.C.L. is a Camille Dreyfus Teacher-Scholar and Latham Family Career Development Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2(9):706–13. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 2.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 3.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 4.Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi As, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010 Aug 13;329(5993):811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protocols. 2009;4(3):372–84. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science. 2011 Aug 12;333(6044):850–6. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 8.Zwick MB, Gach JS, Burton DR. A welcome burst of human antibodies. Nat Biotech. 2008;26(8):886–7. doi: 10.1038/nbt0808-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin A, Ozawa T, Tajiri K, Obata T, Kondo S, Kinoshita K, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. 2009;15(9):1088–92. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- 10.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotech. 2006;24(6):703–7. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 11.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat Protoc. 2009;4(5):767–82. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Story CM, Papa E, Hu C-CA, Ronan JL, Herlihy K, Ploegh HL, et al. Profiling antibody responses by multiparametric analysis of primary B cells. Proceedings of the National Academy of Sciences. 2008 Nov 18;105(46):17902–7. doi: 10.1073/pnas.0805470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International HIV Controllers Study. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science. 2010 Dec 10;330(6010):1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shacklett BL, Critchfield JW, Lemongello D. Isolating mucosal lymphocytes from biopsy tissue for cellular immunology assays. Methods Mol Biol. 2009;485:347–56. doi: 10.1007/978-1-59745-170-3_23. [DOI] [PubMed] [Google Scholar]
- 15.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, et al. Cytokine-Mediated Regulation of Human B Cell Differentiation into Ig-Secreting Cells: Predominant Role of IL-21 Produced by CXCR5+ T Follicular Helper Cells. J Immunol. 2007 Dec 15;179(12):8180–90. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455(7212):532–6. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, et al. Rapid, efficient functional characterization and recovery of HIV-specific human CD8+ T cells using microengraving. Proceedings of the National Academy of Sciences. 2012 Feb 21; doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love KR, Panagiotou V, Jiang B, Stadheim TA, Love JC. Integrated single-cell analysis shows Pichia pastoris secretes protein stochastically. Biotechnology and Bioengineering. 2010;106(2):319–25. doi: 10.1002/bit.22688. [DOI] [PubMed] [Google Scholar]
- 19.Choi JH, Ogunniyi AO, Du M, Du M, Kretschmann M, Eberhardt J, et al. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnology Progress. 2010;26(3):888–95. doi: 10.1002/btpr.374. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Stollar BD. Human immunoglobulin variable region gene analysis by single cell RT-PCR. Journal of Immunological Methods. 2000;244(1–2):217–25. doi: 10.1016/s0022-1759(00)00260-x. [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw EM, Orihuela A, McArdel SL, Salajegheh M, Amato AA, Hafler DA, et al. A Local Antigen-Driven Humoral Response Is Present in the Inflammatory Myopathies. The Journal of Immunology. 2007 Jan 1;178(1):547–56. doi: 10.4049/jimmunol.178.1.547. [DOI] [PubMed] [Google Scholar]
- 22.Brochet X, Lefranc M-P, Giudicelli Vr. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Research. 2008 Jul 1;36(suppl 2):W503–W8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of Immunological Methods. 2008;329(1–2):112–24. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. Journal of Virology. 2005 Aug;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Andres M, Paiva B, Nieto WG, Caraux A, Schmitz A, Almeida J, et al. Human peripheral blood B-cell compartments: A crossroad in B-cell traffic. Cytometry Part B: Clinical Cytometry. 2010;78B(S1):S47–S60. doi: 10.1002/cyto.b.20547. [DOI] [PubMed] [Google Scholar]
- 26.Hendershot LM, Sitia R. Molecular Biology of B Cells. London: Elsevier Science Ltd; 2003. [Google Scholar]
- 27.Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123(3):305–13. doi: 10.1111/j.1365-2567.2007.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Tute RM, Jack AS, Child JA, Morgan GJ, Owen RG, Rawstron AC. A single-tube six-colour flow cytometry screening assay for the detection of minimal residual disease in myeloma. Leukemia. 2007;21(9):2046–9. doi: 10.1038/sj.leu.2404815. [DOI] [PubMed] [Google Scholar]
- 29.Kindt TJ, Goldsby RA, Osbourne BA. Kuby’s Immunology. 6. New York: Wn. Freeman and Company; 2007. [Google Scholar]
- 30.Scamurra RW, Nelson DB, Lin XM, Miller DJ, Silverman GJ, Kappel T, et al. Mucosal Plasma Cell Repertoire During HIV-1 Infection. The Journal of Immunology. 2002 Oct 1;169(7):4008–16. doi: 10.4049/jimmunol.169.7.4008. [DOI] [PubMed] [Google Scholar]
- 31.Debs BE, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Functional single-cell hybridoma screening using droplet-based microfluidics. Proceedings of the National Academy of Sciences. 2012 Jul 17;109(29):11570–5. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.