Abstract
Fluorescence-activated cell sorting (FACS) provides a rapid means of isolating large numbers of fluorescently tagged cells from a heterogeneous mixture of cells. Collections of transgenic plants with cell type-specific expression of fluorescent marker genes such as green fluorescent protein (GFP) are ideally suited for FACS-assisted studies of individual cell types. Here we describe the use of Arabidopsis and rice enhancer trap lines with tissue-specific GFP expression patterns in the root to isolate specific cell types of root tissues using FACS. Additionally, protocols are provided to impose a ramped salinity stress for 48 h prior to cell sorting.
Keywords: FACS, Cell type-specific, Salinity, Rice, Arabidopsis, Protoplast, GAL4
1. Introduction
Fluorescence-activated cell sorting (FACS) of plant protoplasts expressing green fluorescent protein (GFP) has enabled numerous cell type-specific gene expression studies in the model dicotyledonous species Arabidopsis (Arabidopsis thaliana). The use of Arabidopsis lines with cell type-specific patterns of GFP expression (typically reporter lines with tissue-specific promoters driving GFP or GAL4 enhancer trap lines) as source material for FACS experiments permits thousands of protoplasts from a specific cell type to be harvested within minutes. Following the initial publication of a gene expression map spanning five cell populations and three developmental zones of the Arabidopsis root (1), FACS was used to examine developmental and spatiotemporal gene expression as well as cell type-specific responses to abiotic stresses and nitrogen uptake in the Arabidopsis root (2– 4). FACS has also been utilized to collect numerically small cell types such as the root quiescent center and sperm cells for transcriptomic analyses (5, 6). Additionally, FACS of nuclei with nuclear-targeted GFP has been employed as an alternative means of harvesting genetic material from specific cell types of Arabidopsis (7). These studies demonstrate that FACS is an efficient means of isolating and analyzing a variety of GFP-labeled Arabidopsis cell types and provides an alternative to manual isolation techniques such as laser-capture microdissection.
While methods for FACS of Arabidopsis protoplasts are well established (8, 9), the use of FACS to examine cell type-specific gene expression in the model monocotyledonous species rice (Oryza sativa) has not been reported. Here, we demonstrate the use of GAL4 enhancer trap lines of rice and Arabidopsis to isolate specific cell types of the root from both species under control and saline conditions. We describe a supported hydroponics setup for Arabidopsis and rice that allows for growth of large numbers of plants under control and saline conditions, followed by high-throughput protoplasting from root tissues of both species. The FACS procedure to collect fluorescent protoplasts is also described in detail. The enhancer trap lines utilized in these experiments originate from large GAL4 collections of Arabidopsis (10) and rice (11) that display a wide variety of highly specific patterns of GFP expression in root tissues and other organs (Fig. 1). In addition to their use as GFP marker lines, the Arabidopsis and rice GAL4 collections have been used in cell type-specific overexpression studies to increase sodium exclusion and salinity tolerance in both model species (12, 13). The following methods show that it is possible to collect specific cell types of rice and Arabidopsis after exposure to salinity stress through use of FACS. While this protocol is geared towards transcriptomic analyses of the harvested cells, the protocol can be easily adapted for metabolic analyses of the cells through use of a suitable collection buffer.
Fig. 1.
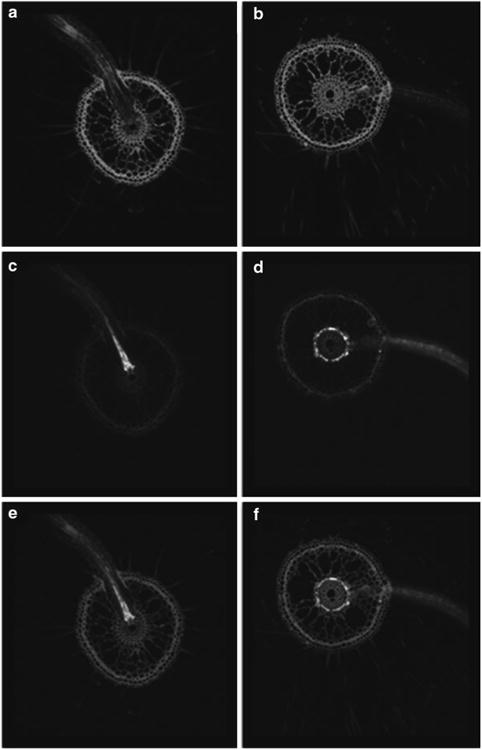
Cell type-specific expression of GFP in roots of two GAL4-GFP rice enhancer trap lines as visualized by confocal laser scanning microscopy. Images of root cross sections are presented as fluorescence of the cell wall stain propidium iodide (a, b), GFP (c, d) and overlay of the propidium iodide and GFP fluorescence patterns (e, f). (a, c, e) Enhancer trap line ASG F03 displays bright GFP fluorescence in xylem parenchyma cells and the pattern is most evident in emerging lateral roots. (b, d, f) Enhancer trap line ASU A03 displays bright GFP fluorescence in cortical cells just outside of the endodermis (note that most cortical cells have been replaced by aerenchyma).
2. Materials
2.1. GAL4-GFP Enhancer Trap Lines
Selected Arabidopsis enhancer trap lines in C24 background from the GAL4 catalogue http://www.plantsci.cam.ac.uk/Haseloff/assembly/page167/index.html.
Selected rice enhancer trap lines in cv. Nipponbare background from the O. sativa GAL4-GFP database http://129.127.183.5.
2.2. Arabidopsis Supported Hydroponics
Phytatray (Sigma).
1 mL tip rack (96 tips per rack) cut to fit inside of the Phytatray (95 × 80 mm). Nitex 03-100/47 mesh (Sefar) cut to fit on top of the tip rack. Use 4 × 1 mL wide-bore tips (cut-off 1 mL tips) to support the tip rack within the Phytatray. Autoclave tip rack and Nitex mesh prior to assembly within the Phytatray.
Growth cabinet set to 14 h light at 22°C (200 μm s-1 m-1) and 10 h dark at 18°C.
100 mL of 0.1% agarose. Autoclaved.
100% Ethanol.
100% bleach solution with 1 drop Tween-20 detergent. Made fresh on day of experiment.
Sterile water (see Note 1).
Arabidopsis growth solution: 1/4× MS basal salt mixture, 1× Gamborg's B5 vitamin solution, 0.5% sucrose, 0.05% MES, pH 5.7 (with 10 M KOH). Prepared in 1 L batches. Autoclaved.
Arabidopsis 25 mM NaCl growth solution: Same composition as growth solution supplemented with 25 mM NaCl and 0.22 mM CaCl2 (see Note 2), pH 5.7 (with 10 M KOH). Autoclaved.
Arabidopsis 50 mM NaCl growth solution: Same composition as growth solution supplemented with 50 mM NaCl and 0.375 mM CaCl2, pH 5.7 (with 10 M KOH). Autoclaved.
2.3. Rice Supported Hydroponics
95 × 95-mm petri dish (BD Falcon).
Filter paper (Whatman) wrapped in foil. Autoclaved.
70% Ethanol
30% Bleach solution with 1 drop Tween-20 detergent. Made fresh on day of experiment.
Elfa® mesh basket (L527 × W327 × H85 nm). Soak in a 50% bleach overnight, rinse well.
20 L Starmaid® plastic container (L558 × W390 × H144 nm) or equivalent.
Growth cabinet set to 12 h light at 28°C (700 μm s-1 m-2) and 12 h dark at 25°C.
Rice growth solution: 1/4× MS basal salt mixture, pH 5.7 (with 10 M KOH). Prepared in 10 L batches (1 L should be autoclaved for rice germination in petri dishes).
Rice 25 mM NaCl growth solution: Same composition as growth solution supplemented with 25 mM NaCl and 0.22 mM CaCl2 (see Note 2), pH 5.7 (with 10 M KOH). Prepared in 10 L batches.
Rice 50 mM NaCl growth solution: Same composition as growth solution supplemented with 50 mM NaCl and 0.375 mM CaCl2, pH 5.7 (with 10 M KOH). Prepared in 10 L batches.
2.4. Protoplasting and FACS Materials and Solutions
Water bath at 60°C.
Clinical centrifuge.
Vacuum chamber.
BD FACSAria cell sorter or equivalent.
RNeasy plant mini kit (Qiagen)
40 μm cell strainer (BD falcon).
50 mL centrifuge tubes.
No. 10 surgical blades (Feather)
1 M KCl. Autoclaved.
1 M CaCl2. Autoclaved.
1 M Tris pH 8. Autoclaved.
Arabidopsis protoplasting solution (10 mL): directly before collecting roots prepare 400 mM mannitol (0.73 g), 20 mM MES hydrate (39 mg), 20 mM KCl (200 μL of 1 M KCl), 1.25% Cellulase R10 (125 mg), 0.3% Macerozyme R10 (30 mg) in 10 mL water. Adjust pH to 5.7 with 1 M Tris pH 8. Heat 10 min in 60°C water bath (solution should clear), cool to room temperature while stirring. Add 10 mM CaCl2 (100 μL of 1 M CaCl2), 0.1% BSA (10 mg), and 1.79 μL of β-mercapoethanol. Make fresh on day of protoplasting.
Rice protoplasting solution (30 mL): directly before collecting roots prepare 400 mM mannitol (2.2 g), 20 mM MES hydrate (117 mg), 20 mM KCl (600 μL of 1 M KCl), 1.25% Cellulase R10 (375 mg), 1.25% Cellulase RS (375 mg), 0.3% Macerozyme R10 (90 mg), 0.12% Pectolyase (36 mg) in 30 mL water. Adjust pH to 5.7 with 1 M Tris pH 8. Heat 10 min in 60°C water bath (solution should clear), cool to room temperature while stirring. Add 10 mM CaCl2 (300 μL of 1 M CaCl2), 0.1% BSA (30 mg), and 5.38 μL of β-mercapoethanol. Make fresh on day of protoplasting.
3. Methods
3.1. Arabidopsis Germination, Growth, and Application of Salinity Stress
All work should be carried out under sterile conditions. Perform each procedure as early as possible in the morning.
Weigh 40 mg of Arabidopsis seed (approximately 2,000 seed) into a 2 mL tube (40 mg is sufficient for two Phytatrays, scale up as required; also see Note 3).
Add 1.5 mL of 100% ethanol and mix for 1 min. Pipette off the ethanol.
Add 1.5 mL of 100% bleach solution and mix on an orbital shaker for 5 min. Pipette off the bleach.
Rinse multiple times with sterile water until clean. Pipette off final rinse.
Add 1.5 mL of 0.1% agarose solution and mix well by pipetting up and down. Store in the dark for 3 days at 4°C to ensure uniform germination.
Place a sterile tip rack covered with Nitex mesh into two Phytatrays; add 240 mL of sterile Arabidopsis growth solution (Fig. 2a). The growth solution should just touch the base of the Nitex mesh.
Pipette 750 μL of the agarose seed mixture (approximately 1,000 seed) onto the Nitex mesh of each Phytatray in an even distribution.
Close the lid of the Phytatrays and leave for 4 days in the growth cabinet.
On the morning of day 5, replace the growth solution in the Phytatrays with 240 mL of sterile Arabidopsis 25 mM NaCl growth solution (replace the growth solution of any control plants with 240 mL of sterile Arabidopsis growth solution; also see Note 4). Return Phytatrays to the growth cabinet for 24 h.
On the morning of day 6, replace the growth solution in the Phytatrays with 240 mL of sterile Arabidopsis 50 mM NaCl growth solution (replace the growth solution of any control plants with 240 mL of sterile Arabidopsis growth solution; also see Note 4). Return Phytatrays to the growth cabinet for 24 h.
Fig. 2.
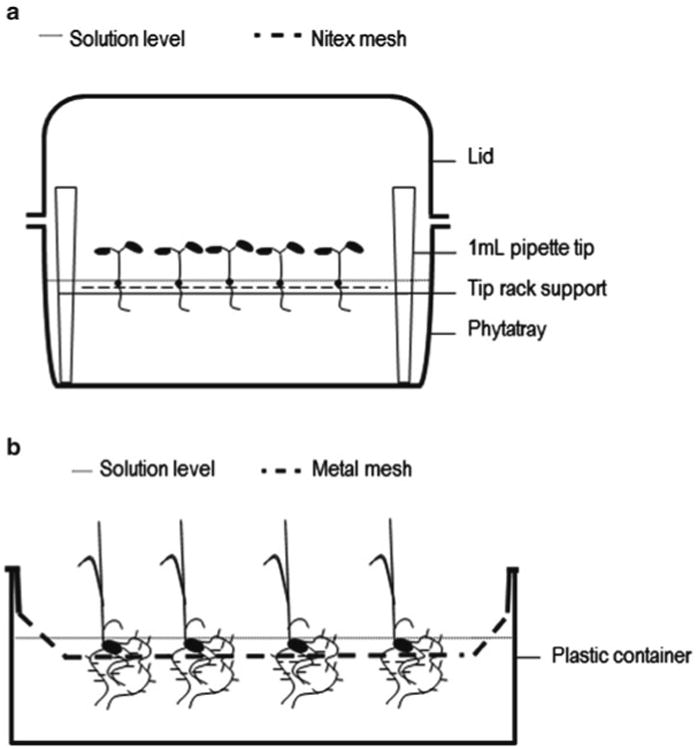
Assembly of the Arabidopsis and rice supported hydroponic systems. (a) Arabidopsis seeds are pipetted directly onto the Nitex mesh (250 μm) that rests on top of the tip rack support. The plant roots grow through the mesh and are easily harvested by scrapping a surgical blade across the lower side of the mesh. (b) Germinated rice seedlings are placed directly into the mesh baskets; 5 mm holes within the mesh allow the rice roots to grow down into the solution. Plastic containers with an inner ridge should be used to hold the mesh basket approximately 5 cm above floor of the container.
3.2. Arabidopsis Protoplasting Protocol
This procedure has been modified from the original publication (7) to increase protoplast yield.
On the morning of day 7, prepare 10 mL of the Arabidopsis protoplasting solution and pour into a 50 mL flask (10 mL is sufficient for digestion of roots harvested from two Phytratrays, scale up as required).
Open Phytatrays and peel the Nitex mesh from the tip rack support, turning over to expose the roots. Quickly harvest the roots by running a surgical blade across the mesh. The roots should collect as a clump at the tip of the blade.
Tap the roots into 10 mL of Arabidopsis protoplasting solution and incubate for 1–2 h at 26°C on a benchtop orbital shaker set at 100 rpm.
Filter the solution two times through a 40 μm cell strainer fitted on top of a 50 mL centrifuge tube to remove debris.
Centrifuge the filtrate for 5 min at 500 × g in a clinical centrifuge. Pour off most of the solution and resuspend the protoplast pellet in a remaining 1–2 mL of protoplasting solution (see Note 5).
Proceed with FACS as soon as possible (see Note 6).
3.3. Rice Germination, Growth, and Application of Salinity Stress
All work until day 5 should be carried out under sterile conditions. Perform each procedure as early as possible in the morning.
Place approximately 75 dehusked rice seed into a 50 mL centrifuge tube (75 seedlings are sufficient to fill one mesh basket, scale up as required).
Add 40 mL of 70% ethanol and mix for 1 min. Pour off the ethanol.
Add 40 mL 30% bleach solution and mix on a benchtop orbital shaker set at 100 rpm for 30 min. Pour off the bleach.
Rinse multiple times with sterile water until clean, allowing the seed to remain in the final rinse for 20 min on the shaker. Pour off the final rinse.
Place sterile filter papers into the 95 × 95-mm petri dishes and place ten sterilized seed per dish. Add 20 mL of autoclaved rice growth solution to each dish and seal with parafilm.
Leave petri dishes for 4 days in the growth cabinet.
On the morning of day 5, place up to 75 rice seedlings into an Elfa® mesh basket supported within a plastic container holding 10 L of 25 mM NaCl growth solution (or standard growth solution for controls). The solution should just cover the base of the mesh basket (Fig. 2b). Place the container into the growth cabinet for 24 h.
On the morning of day 6, replace the solution with 10 L of 50 mM NaCl growth solution (or standard growth solution for controls). Return the container to the growth cabinet for 24 h.
3.4. Rice Protoplasting Protocol
On the morning of day 7, prepare 30 mL of the rice protoplasting solution into a 100 mL flask (30 mL is sufficient for digestion of roots harvested from one mesh basket, scale up as required).
Manually remove groups of rice plants from the mesh basket and use a razor blade to separate the roots from the shoot and seed (groups of 15–20 plants can be handled easily). Chop the roots on the lid of petri dish and scrape the root tissue into a few milliliter of rice protoplasting solution to keep the root tissue moist.
Once all of the roots have been harvested, use the razor blade to gently mince the root tissue for 30 s and then transfer the tissue to 30 mL of rice protoplasting solution.
Apply a vacuum for 7 min at 25 Hg to facilitate infiltration of the enzymes. Stop the pump and let the chamber come back to normal pressure.
Incubate for approximately 2 h at 26°C on a benchtop orbital shaker set at 100 rpm. Protoplasting should be monitored under the light microscope every 30 min until protoplasts are seen to be releasing from the root tissue (Fig. 3; also see Note 7).
Filter the solution two times through a 40 μm cell strainer fitted on top of a 50 mL centrifuge tube to remove debris.
Centrifuge the filtrate for 5 min at 500 × g in a clinical centrifuge. Pour off most of the solution and resuspend the protoplast pellet in a remaining 1–2 mL of protoplasting solution (see Note 5).
Proceed with FACS as soon as possible (see Note 6).
Fig. 3.
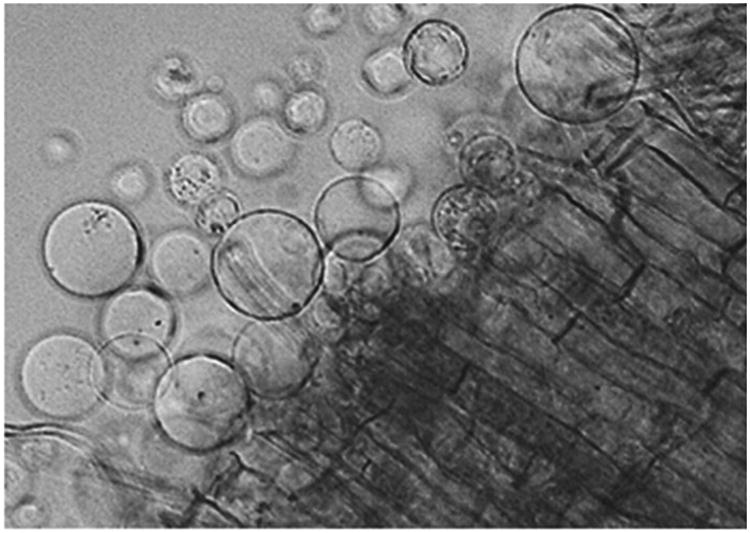
Rice protoplasts being released from the cut edge of a root, indicating that the digestion reaction has proceeded sufficiently to begin filtration and centrifugation to collect the protoplasts for FACS.
3.5. FACS Collection of GFP-Expressing Protoplasts
The sorting ability of the FACS instrument is based on its capacity to excite and detect specific fluorescence wavelengths emitted by cells in suspension. Cell populations emitting relatively more fluorescence at a specific wavelength (i.e., GFP fluorescence; 488 nm excitation and 530/30 nm emission) can be isolated and collected. However, the autofluorescent properties of protoplast suspension interfere with a distinction of GFP-positive cells based on emission at 530/30 nm alone. Autofluorescent emission at 610/20 nm and a 488 nm excitation is therefore taken as a com-pensational measure of autofluorescence allowing for the identification of sorting events with GFP-specific fluorescence.
Load the protoplast solution into the FACS. The loading tube should be under constant agitation to maintain a homogenous solution.
Run protoplasts through the FACS using a 70- μm (or 100 μm) nozzle (PBS buffer for sheath fluid) at event rates up to 20,000 events/s and a system pressure up to 70 psi (events/s and system pressure will depend on the nozzle utilized). Laser excite the cells at 488 nm and set a green channel to measure GFP emission at 530/30 nm and a red channel to measure auto fluorescence at 610/20 nm. Use a scatter plot with red emission intensity (autofluorescence) vs. green emission intensity (GFP fluorescence) (Fig. 4a, c).
A non- fluorescent control protoplast solution (protoplasts prepared from wild-type plants not expressing GFP) should initially be run to determine gate settings for capture of GFP fluorescent protoplasts (Fig. 4a, c).
When sorting protoplasts from the enhancer trap lines, a clear tail of protoplasts exhibiting proportionally high green fluorescence should be visible (Fig. 4b, d; also see Note 8). Make sure the gate is well positioned to collect GFP positive protoplasts while avoiding as many false positives as possible. (In this study, BDFACSDiva software version 6.1.3 was used with the BD FACSAria cell sorter set to sort precision mode “Purity” to ensure a stringent sorting of GFP protoplasts.) Sort into a desired collection buffer (for our purposes, we used RNA extraction buffer).
The number of cells to collect by FACS is dependent on the type of study and RNA extraction kit used. For rice transcriptomic studies using microarrays, we sorted between 3,000 and 17,000 GFP positive cells into RLT buffer from the RNeasy Plant Micro Kit (see Note 9). Maintain a ratio of 100 μL cell suspension to 350 μL RLT buffer to ensure efficient RNA extraction (refer to manufacturer's instructions). For RT-PCR studies, more cells may be needed.
Fig. 4.
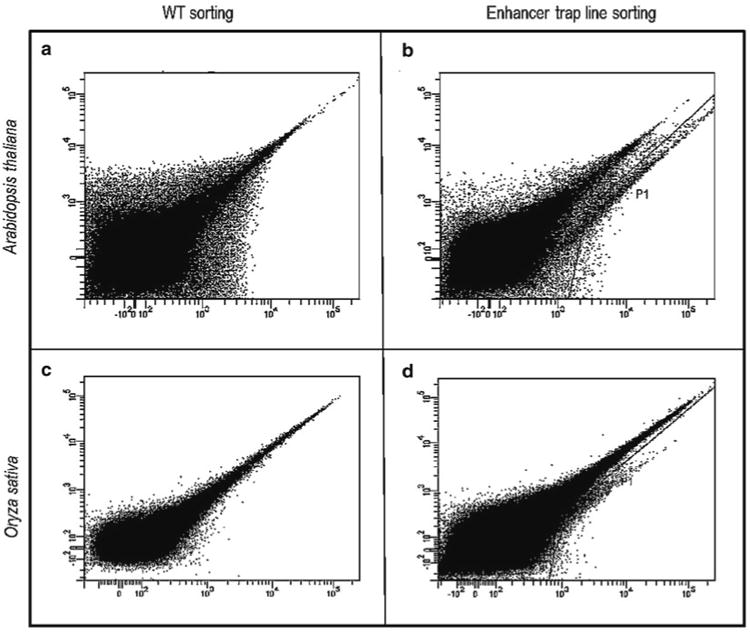
Scatter plots of green fluorescence intensity on the x-axis (arbitrary units; excitation 488 nm/emission 530/30 nm) and red fluorescence intensity on the y-axis (arbitrary units; excitation 488 nm/emission 576/26 nm) from representative Arabidopsis and rice FACS experiments. (a) Scatter plot of protoplasts from wild-type Arabidopsis plants not expressing GFP with most cells having a typical autofluorescence pattern of equal green and red fluorescence. (b) Sort of protoplasts from an Arabidopsis GAL4-GFP enhancer trap line with a gate (P1) set to collect the GFP tail where cells display relatively more green than red fluorescence. (c) Sort of protoplasts from wild-type rice plants not expressing GFP with most cells having a typical autofluorescence pattern of equal green and red fluorescence. (d) Sort of protoplasts from a rice GAL4-GFP enhancer trap line, with a gate (P1) set to collect the GFP tail where cells display relatively more green than red fluorescence.
Acknowledgments
This work was supported by grants from the Australian Research Council (DP0770966 to M.T. and A.A.T.J.) and the U.S. National Institutes of Health (NIH grant R01 GM078279 to K.D.B.)
Footnotes
Use ultrapure water for all rinses and to prepare solutions (2 MΩ at room temperature). Store solutions at room temperature unless specified otherwise.
The addition of CaCl2 to the 25 and 50 mM NaCl growth solutions ensures that Ca activity remains constant upon addition of salt, so that any measured effects of salinity are due to the addition of NaCl and not NaCl-induced decreases in Ca activity of the external solution (14).
If more Arabidopsis protoplasts are required, it is recommended to increase the number of Phytatrays. Overloading Phytatrays with more than 1,000 seed leads to poor plant growth and greater contamination rates.
Work in sterile conditions when changing hydroponics Arabidopsis solutions to avoid problems with bacterial contamination. The rice-supported hydroponics setup does not need to be kept sterile.
A cell pellet may not always be visible after centrifugation and resuspension. It is recommended to observe a drop of the resuspended pellet under the microscope to check protoplast density and condition before proceeding with FACS.
Minimize holding times of the protoplasts to avoid transcriptional changes related to the protoplasting procedure itself. If necessary, protoplasts can be kept for approximately 60 min at 15–20°C prior to beginning of FACS.
We suggest monitoring digesting rice roots under the microscope to better assess how enzymatic digestion is proceeding. Digestion time can be reduced if protoplasts are visible.
The absence of GFP positive protoplasts as a GFP tail on the sorting scatter plot may be due to poor efficiency of protoplasting. Check pH of the protoplasting solution and temperature for any errors and monitor digestion via microscopic examination.
Quickly vortex or vigorously shake the sorted protoplasts in RLT buffer to ensure cell lysis. Either proceed with RNA extraction directly or freeze the lysed cells in liquid nitrogen and store at −80°C.
References
- 1.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 2.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 3.Dinneny JR, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 4.Gifford ML, et al. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nawy T, et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borges F, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, et al. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 2008;147:30–40. doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnbaum K, et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- 9.Bargmann BOR, Birnbaum KD. Fluorescence activated cell sorting of plant protoplasts. J Vis Exp. 2010;(36):e1673. doi: 10.3791/1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haseloff J. GFP variants for multispectral imaging of living cells. Methods Cell Biol. 1999;58:139–151. doi: 10.1016/s0091-679x(08)61953-6. [DOI] [PubMed] [Google Scholar]
- 11.Johnson AAT, et al. Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J. 2005;41:779–789. doi: 10.1111/j.1365-313X.2005.02339.x. [DOI] [PubMed] [Google Scholar]
- 12.Møller IS, et al. Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis. Plant Cell. 2009;21:2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plett D, et al. Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS One. 2010;5:e12571. doi: 10.1371/journal.pone.0012571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tester M, Davenport R. Na + tolerance and Na + transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]


