Abstract
mRNA translation is mainly regulated at the level of initiation, a process that involves the synergistic action of the 5′ cap structure and the 3′ poly(A) tail at the ends of eukaryotic mRNA. The eukaryote initiation factor 4G(eIF4G) is a pivotal scaffold protein that forms a critical link between mRNA cap structure, poly(A) tail, and the small ribosomal subunit. There are two functional homologs of eIF4G in mammals, the original eIF4G, renamed eIF4GI, and eIF4GII that functionally complements eIF4GI. To date, biochemical and functional analysis have not identified differential activities for eIF4GI and eIF4GII. In this report, we demonstrate that eIF4GII, but not eIF4GI, is selectively recruited to capped mRNA at the onset of cell differentiation. This recruitment is coincident with a strong and long-lasting phosphorylation of eIF4E and the release of 4E-BP1, a suppressor of eIF4E function, from the cap structure, without a concomitant change in 4E-BP1's phosphorylation. Our data further indicate that cytokines such as thrombopoietin can differentially regulate eIF4GI/II activities. These results provide the first evidence that eIF4GI/II does fulfill selective roles in mammalian cells.
Cell fate specification is achieved by differential gene expression, which can involve regulation at various levels, including transcription, RNA processing, translation, and posttranslation protein modifications. Transcriptional regulation has long been thought to play the central role in that process. However, translational control is also a well-known important determinant of cell proliferation and survival, as well as cell maturation, and generally varies in response to treatment with growth factors, cytokines, hormones, and mitogens.
The regulation of translation is exerted mainly at the level of initiation (31, 36, 39). The critical step during translation initiation is the recruitment of the small 40S ribosomal subunit to an mRNA, a process that involves the synergistic action of the 5′ cap structure and the 3′ poly(A) tail at the end of eukaryotic mRNAs (6, 47). The poly(A) tail is recognized by the poly(A)-binding protein (PABP) while the cap structure (m7GpppN) interacts with a protein complex termed the eukaryotic initiation factor 4F(eIF4F), which consists of three subunits: eIF4E, eIF4G, and eIF4A. The cap-binding subunit of eIF4F, eIF4E, can simultaneously bind to the cap structure and to the N-terminal region of eIF4G. eIF4A is an ATP-dependent RNA helicase that, in conjunction with eIF4B, is thought to unwind the secondary structure in the mRNA 5′ untranslated region; it binds to the central and the C-terminal regions of eIF4G (16). eIF4G functions as a pivotal scaffolding factor: in addition to eIF4E and eIF4A, it also binds to eIF3, a multiprotein complex directly associated with the small ribosomal subunit, and to PABP, allowing a circularization of the mRNA molecule (46) which explains the synergistic effect of the 5′ cap and the 3′-poly(A) tails of mRNA on translation initiation (15, 35). Thus, eIF4G provides a physical link between the mRNA cap structure, the poly(A) tail, and the small ribosomal subunit. eIF4E specifically binds to the cap structure and, through association with eIF4G and eIF4A, allows the cap-proximal region of mRNA to be unwound and rendered accessible to an incoming 43S complex to facilitate ribosomal subunit binding (as reviewed in references 6 and 33). There are two functional homologs of eIF4G in mammals, the original eIF4G, renamed eIF4GI, and eIF4GII. eIF4GII is 46% identical to eIF4GI, exhibits similar biochemical activities, and functionally complements eIF4GI (8, 15).
Under most circumstances, eIF4E is the least abundant of all initiation factors and is a major target for translation control. An important mechanism to regulate eIF4E function in the initiation process is the modulation of its availability to form an active eIF4F complex (reviewed in reference 6). This occurs through modulation of the amount of eIF4E present within the cell or, more often, through regulation of the association of eIF4E with a family of three translational repressors, the eIF4E-binding proteins (4E-BPs) (32). The 4E-BPs do not inhibit eIF4E binding to the cap but, instead, block eIF4F assembly by competing with eIF4Gs for a common binding site on eIF4E (11, 23). The binding of eIF4E to 4E-BPs is regulated through phosphorylation of 4E-BPs, as hyperphosphorylation of 4E-BP1 inhibits the association of 4E-BPs with eIF4E (10, 20, 45).
An additional level of regulation is the phosphorylation of eIF4E itself at serine 209. In mammals, stimulation of mRNA translation by mitogenic growth factors, serum, or nutrients correlates with increased phosphorylation of eIF4E, whereas dephosphorylation of eIF4E strongly correlates with inhibition of cap-dependent mRNA translation during heat shock or nutrient deprivation, metaphase arrest of the cell cycle, and infection with certain viruses (6). Phosphorylation of eIF4E is critical for growth in Drosophila melanogaster (19). Two kinases phosphorylate Ser209 and are targets of the mitogen-activated extracellular-signal-regulated kinases (Erks) and the stress- and cytokine-activated p38 mitogen-activated protein (MAP) kinase pathways. These enzymes (Mnk1 and Mnk2) also associate with eIF4G in vivo; this mode of recruitment is thought to ensure that eIF4E is phosphorylated only as a part of the eIF4F complex (41, 50). However, the key issue of the effect of phosphorylation on the function of eIF4E or eIF4F remains largely unknown.
While much research has focused on the biochemical mechanisms that regulate cell cycle and cell growth in eukaryotes, considerably less attention has focused on the biochemical mechanisms that regulate translation at the onset of cell differentiation and cell fate decisions, particularly in mammalian systems. Platelets are blood particles that fulfill a crucial hemostatic function. They originate from bone marrow megakaryocytes by proplatelet shedding; megakaryocytes derive from hematopoietic stem cells. Thrombopoietin (TPO) is the primary physiological regulator of platelet production, favoring the commitment of hematopoietic stem cell-multipotent hematopoietic progenitors to the megakaryocyte lineage and acting on all the stages of megakaryocyte development (17). By using a TPO-responsive multipotent hematopoietic cell line, we addressed the possibility that a cytokine such as TPO affects the translation machinery coincident with cell commitment to megakaryocyte differentiation. We therefore investigated the mechanism of translational control under these conditions, with emphasis on the early steps of translation initiation. We show here that induction of cell differentiation correlates with the selective recruitment of eIF4GII—but not eIF4GI—to the cap structure, in contrast to mitogen-activated cells. Concomitant with this recruitment, 4E-BP1 dissociates from the cap structure and eIF4E becomes highly phosphorylated. No significant changes in the amount of 4E-BPs and 4E-BP1's phosphorylation were observed. Our data provide strong evidence that eIF4GI and eIF4GII do fulfill different roles within the cell.
MATERIALS AND METHODS
Reagents and antibodies.
The TPO mimetic peptide GW395058 (1) was synthesized by Genosys Biotechnologies Ltd. Erythropoietin (EPO) was from Boerhinger Mannheim (Mannheim, Germany). Antibodies directed against eIF4E, phospho-eIF4E (Ser209), and anti-rabbit horseradish peroxidase-conjugated secondary antibody were from Cell Signaling Technology, Inc. Anti-4E-BP1, -4E-BP2, -4E-BP3, -eIF4GI, and -eIF4GII antibodies were previously described (8, 34, 41). Fluorescein isothiocyanate-conjugated CD61 and control immunoglobulin G1 (IgG1) antibody were from Immunotech (Marseille, France). The chemical inhibitors PD98059, SB203580, and rapamycin were obtained from Calbiochem (Germany); LY294002 was from Sigma-Aldrich Corp. (St. Louis, Mo.).
Cell culture.
The human UT7-mpl cells were maintained in alpha minimal essential medium supplemented with 10% fetal calf serum and either 2 U of EPO/ml or 2.5 ng of recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF)/ml, as described previously (33). UT7-mpl cells were treated for the appropriate time with 10 nM TPO mimetic peptide GW395058, a concentration equivalent to 100 ng of the recombinant cytokine per ml for both proliferation and differentiation of UT7-mpl cells and of megakaryocyte progenitors derived from normal CD34+ human cells (5; E. Cramer, unpublished data). Where indicated, UT7-mpl cells were pretreated for 30 min in the presence of the indicated inhibitors, PD98059 (30 μM), SB203580 (20 μM), LY294002 (50 μM), or rapamycin (20 nM), before adding TPO.
Flow cytometric analysis.
Cells, treated or not treated with TPO, were washed in PBA (phosphate-buffered saline containing 0.2% sodium azide and 1% bovine serum albumin) and incubated for 30 min at 4°C with 1 mg of normal goat aggregated IgG per ml to saturate Fc receptors. After washing, fluorescein isothiocyanate-conjugated control IgG1 or CD61 mouse monoclonal antibodies were added, and the incubation continued for 30 min at 4°C. Cells were analyzed for fluorescence on an ELITE flow cytometer.
Electron microscopy (EM) and immuno-EM.
Cells were fixed for 1 h in 3% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), washed, and embedded in Epon. Representative fields of sections performed on the entire samples were chosen for illustration. For immuno-EM, cells were fixed in 1% glutaraldehyde in 0.1 M phosphate buffer for 1 h, washed, and embedded in glycomethacrylate. Postembedding immunolabeling was performed on thin sections, with polyclonal rabbit anti-human antibodies for αIIbβ3 (10 μg/ml; kindly provided by Michael Berndt, Melbourne, Australia), followed by goat anti-rabbit IgG conjugated with 10-nm gold particles. Thin sections were examined with a Philips CM 10 EM (Philips, Eindoven, The Netherlands) after uranyl acetate and lead citrate staining.
Expression vectors and transient transfections.
Vector encoding hemagglutinin (HA)-tagged Mnk1 was previously described (41). UT7-mpl cells (5 × 106) were transiently transfected with the indicated plasmid (20 μg) by means of electroporation, as previously described (2). Immediately following electroporation, cells were resuspended in proliferative medium, split between two flasks, and maintained in culture for 36 h before being stimulated with or without TPO (10 nM) for 30 min and lysed.
Immunoprecipitation and Western blotting.
Total cell extracts were prepared by lysing the cells for 15 min at 4°C in buffer containing 50 mM Tris (pH 7.4), 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 1% Triton X-100 supplemented with protease inhibitor cocktail (Complete EDTA-free; Roche). The homogenate was centrifuged at 15,000 × g for 5 min, and the supernatant was collected. Samples (150 μg) were denatured by boiling in Laemmli lysis buffer for 5 min and analyzed by one- or two-dimensional gel electrophoresis followed by Western blotting. Alternatively, lysates (0.2- to 0.5-mg proteins) were incubated with m7GTP-Sepharose beads (Amersham Inc.) or with the indicated antibodies for 16 h at 4°C on rotating wheels. Protein G-agarose was added to immunoprecipitates, and the reaction was allowed to proceed for an additional 30 min of incubation. Proteins bound to the beads were eluted by boiling in Laemmli lysis buffer and were further identified by Western blotting using the appropriate antibodies. All Western blot experiments were performed at least four times independently. Where indicated, Western blots were quantified by using ECL Plus substrate (Amersham Biosciences), a Typhoon 9400 fluorescent imager, and Imagequant software analysis.
Measurement of protein synthesis.
UT7-mpl cells were treated with TPO for the indicated time. Four hours before the end of the treatment, 106 cells were counted and 100 μCi of [35S]methionine/ml was added to culture medium. At the end, medium was removed, and the cells were intensively washed and then lysed in buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM EDTA, and 100 mM KCl. Incorporation of radioactivity into solubilized proteins was determined by precipitation with trichloracetic acid.
RESULTS
TPO favors early megakaryocyte differentiation.
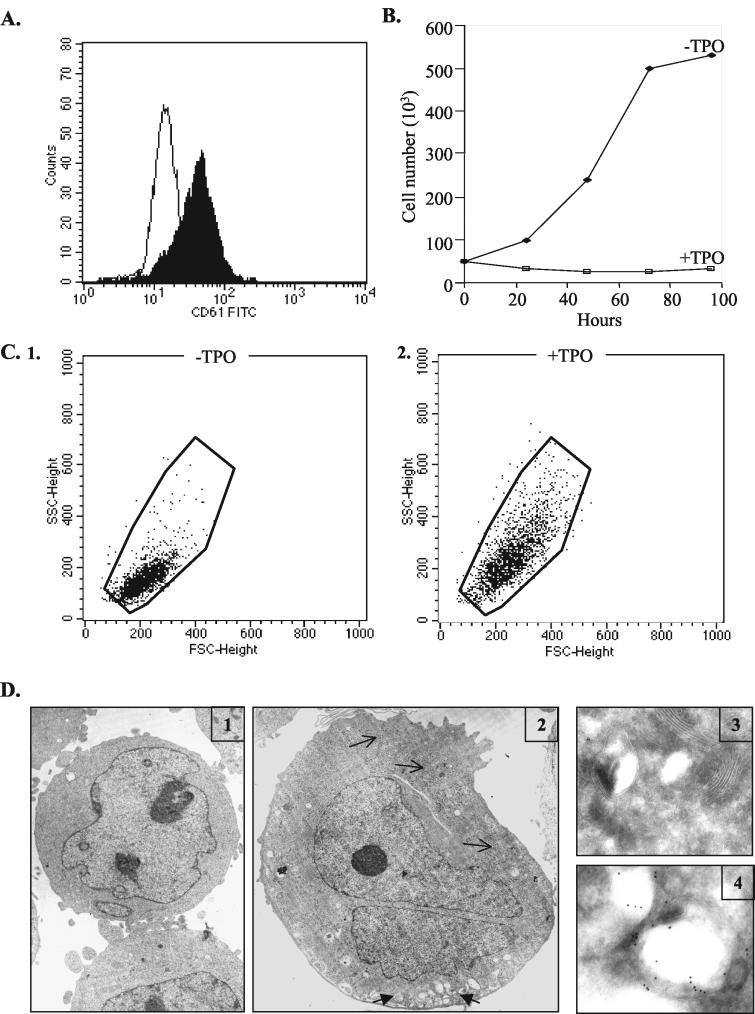
A few multipotent hematopoietic cell lines possess the ability to differentiate in response to selective cytokine treatments. The multipotent human hematopoietic UT7-mpl cells express exogenous TPO receptor and require the presence of growth factors such as EPO or GM-CSF for their growth and survival. Upon TPO addition, UT7-mpl cells undergo megakaryocytic differentiation evidenced by the enhanced expression of differentiation antigens CD61 (GpIIIa; Fig. 1A) and CD41 (GpIIb/IIIa complex; data not shown) on the cell surface and the loss of markers for other lineages (33). Coincidentally, the cell division rate strongly slowed down (Fig. 1B). Since megakaryocyte differentiation is characterized by significant development of cell membranes and granules and an increase in cell size, we also analyzed these characteristic phenotypic changes following TPO treatment of the UT7-mpl cells. TPO induced a large increase in both the size of the cells and the complexity of the cells, as exemplified by their forward- and side-scatter index in flow cytometry (Fig. 1C) and analysis by EM (Fig. 1D1 and D2). After 2 days of TPO treatment, we observed in detail, as detected by EM, that rough endoplasmic reticulum strongly increased (Fig. 1D2) and that cells had started to develop an intracellular membrane network resembling the future demarcation membranes that would divide the megakaryocyte cytoplasm into platelet fields (Fig. 1D2); these membranes expressed large amounts of the typical integrin GpIIb/IIIa at their surface, as detected by immuno-EM (Fig. 1D3 to D4). Therefore, TPO-treated UT7-mpl cells recapitulate a large number of the early differentiation events observed in normal megakaryocyte progenitors. By using this cellular model, we wished to investigate whether TPO affects translation while favoring differentiation.
FIG.1.
UT7-mpl cells recapitulate early differentiation events observed in normal megakaryocyte progenitors. (A, C, and D) UT7-mpl cells were incubated for 48 h with or without TPO (10 nM). At the end of the treatment, the expression of the integrin chain β3 (CD61) (A) or the forward- (FSC) and side-scatter (SSC) light transmission characteristics of the untreated cells (C1) or treated cells (C2) were assayed by fluorescence-activated cell sorter analysis; alternatively, untreated (D1, D3) or treated (D2, D4) cells were processed for EM (D1 and D2; magnification, ×6,600) or for immuno-EM with anti-human αIIbβ3 (D3 and D4). Open arrows, rough endoplasmic reticulum; filled arrows, intracellular membrane network. (B) UT7-mpl cells were diluted at a concentration of 50,000/ml and treated with TPO for the indicated time and counted.
TPO regulates the phosphorylation state of eIF4E in a rapid and sustained manner.
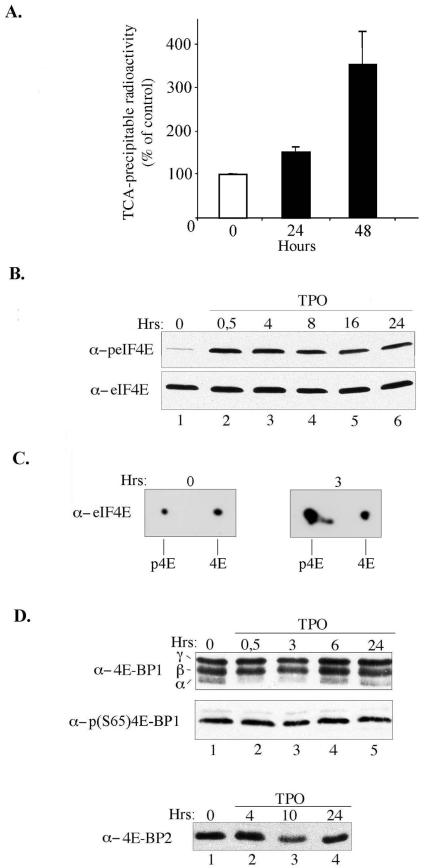
As a first step, we tested whether TPO affects overall protein synthesis. As shown in Fig. 2A, while cells stopped growing and started to differentiate in the presence of TPO, the cells showed a strong increase (i.e., 50% after 1 day of TPO treatment and 300% after 2 days) in the rate of protein synthesis, as assessed by [35S]methionine incorporation into proteins.
FIG. 2.
TPO enhances protein synthesis and induces a robust phosphorylation of eIF4E. (A) UT7-mpl cells were incubated with TPO for the indicated time. Four hours prior the end of the treatment, [35S]methionine was added to cell culture medium of 106 cells. Radioactivity incorporated into protein was determined by trichloracetic acid (TCA) precipitation. (B to D) Cells were incubated with TPO for the indicated time and then lysed. Cell extracts were analyzed by one-dimensional (B and D) or two-dimensional (C) gel electrophoresis followed by Western blotting with specific antibodies as indicated on the left. In the experiment represented in panel C, 60% of the eIF4E was phosphorylated upon TPO treatment while only 30% was phosphorylated from untreated cells.
The initial recruitment of ribosomes to mRNA is the major rate-limiting step of translation in eukaryotes. We thus investigated whether expression of the cap-binding protein, eIF4E, and its inhibitors, 4E-BPs, and/or their phosphorylation states change upon TPO treatment. Western blot analyses were performed with various antibodies directed against total or phosphorylated forms of these factors. As shown in Fig. 2B, TPO did not affect the expression of eIF4E (lower panel). However, TPO induced a rapid increase (less than 30 min) in eIF4E phosphorylation on Ser209. This increase was intense and maintained for at least 1 day of TPO treatment (Fig. 2B, upper panel). We wished to quantify the percentage of eIF4E that was phosphorylated upon TPO treatment. Two-dimensional gel electrophoresis coupled to Western blotting with anti-eIF4E antibody and fluorescent imaging indicated that, depending on the experiments, between 60% (Fig. 2C) and 90% of the whole eIF4E present in the cells was phosphorylated on Ser209 after 3 h of TPO treatment.
TPO did not affect the total expression of 4E-BP1 (Fig. 2D, upper panels) or 4E-BP2 (lower panel), while 4E-BP3 was absent from UT7-mpl cells (data not shown). 4E-BP1 exhibited a shift to low-mobility migrating species on acrylamide gels, suggesting the presence of a high proportion of phosphorylated 4E-BP1 species in TPO-untreated and -treated cells. The high amount of phosphorylated 4E-BP1 species was confirmed by Western blotting with phospho-specific anti-4E-BP1 antibodies (Fig. 2D, middle panel) and was in accordance with the initial intense proliferation of untreated UT7-mpl cells (Fig. 1B). Yet, following TPO treatment, while cells stopped proliferating, the proportion of the remaining hypophosphorylated α form of 4E-BP1 still decreased (Fig. 2D, upper panel, lanes 2 to 3), in a transient but reproducible way. No additional changes to the proportion of the other hypo- or hyperphosphorylated forms of 4E-BP1 were observed in the presence of TPO (Fig. 2D, middle panel; data not shown). These data indicate that commitment to differentiation, with its coincident inhibition of proliferation, does not trigger major changes in the 4E-BP1 phosphorylation state or in 4EBP1-3 expression.
The TPO-dependent phosphorylation of eIF4E requires two different MAP kinase pathways.
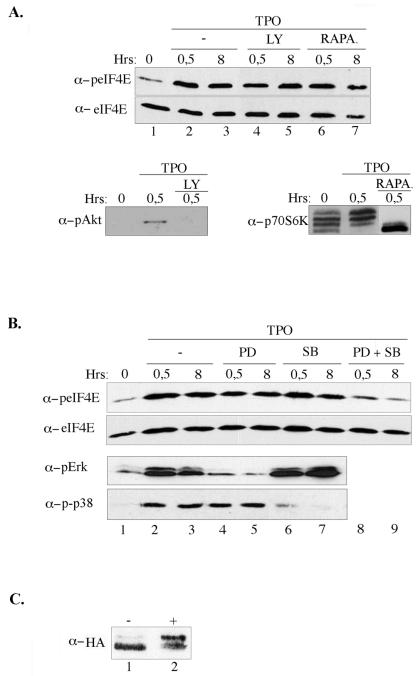
eIF4E activity is thought to be critical for normal cell growth, as overexpression of eIF4E in rodent cells is oncogenic (21), while injection of eIF4E into quiescent NIH3T3 cells induces DNA synthesis (44). Moreover, eIF4E is phosphorylated on Ser209 following treatment of cells with numerous growth factors and mitogens, and this has been correlated with increased cellular proliferation in mammals (45) and shown to be essential for normal growth in Drosophila (19). However, we repeatedly observed a rather weak phosphorylation of eIF4E in proliferating UT7-mpl cells (Fig. 2B, lane 1) and a robust eIF4E phosphorylation when cells committed to differentiate and stopped proliferating (lanes 2 to 6). We thus decided to identify the pathways involved in eIF4E phosphorylation in our model. Protein kinase C and the MAP kinase-activated protein kinase Mnk1 were previously shown to phosphorylate eIF4E on Ser209 in vitro and in vivo following mitogenic stimuli (50, 51). p38 MAP kinase and mTOR (mammalian target of rapamycin) signaling pathways may also influence eIF4E Ser209 phosphorylation following cytokine or stress stimuli, as suggested previously (3, 49). We tested whether any of these pathways participate in the induction of eIF4E phosphorylation at the onset of TPO-dependent cell differentiation. Neither the phosphatidyl inositol 3-kinase (PI3-k) inhibitor LY294002 nor the mTOR inhibitor rapamycin inhibited Ser209 phosphorylation (Fig. 3A), although these two inhibitors did block TPO-dependent Akt and p70S6 kinase phosphorylation in the UT7-mpl cells, respectively, as expected (Fig. 3A, lower panels). Also, these two drugs did not affect TPO-dependent differentiation (data not shown). When UT7-mpl cells were preincubated with the PD98059 inhibitor of the Mek/Erk1/Erk2 signaling pathway or with the SB203580 inhibitor of p38 MAP kinase before adding TPO, the TPO-induced phosphorylation of eIF4E was poorly affected (Fig. 3B, lanes 4 to 7), although these two inhibitors did block the TPO-dependent phosphorylation of Erk1/2 and p38 MAP kinase, respectively, in the UT7-mpl cells (Fig. 3B, lower panels), as already reported (5). However, in the presence of both the MEK inhibitor and the p38 MAP kinase inhibitor, the TPO-dependent phosphorylation of eIF4E was strongly impaired (lanes 8 to 9). To determine whether TPO activated Mnk1 per se, influenza HA-tagged Mnk1 was introduced in UT7-mpl cells. As shown in Fig. 3C, TPO rapidly activated Mnk1, as assessed by its mobility shift on an analytical gel; this activation was maintained for a few hours (data not shown). These data suggest that TPO enhances the phosphorylation of eIF4E by activating the same kinase Mnk1 that is activated by mitogens and that was previously shown to associate with eIF4GI (40). It also suggests that TPO-activated Mnk1 is controlled by the Erk1/2 and the p38 MAP kinase pathways.
FIG. 3.
Erk1/2 and the p38 MAP kinases participate in the TPO-induced phosphorylation of eIF4E. (A) UT7-mpl cells were preincubated without (−, lanes 1 to 3) or with LY294002 (50 μM) (LY, lanes 4 to 5) or rapamycin (20 nM) (RAPA, lanes 6 to 7) for 30 min before adding TPO for 0 to 8 h, and then cells were lysed. Cellular extracts (150 μg) were analyzed for the presence of phospho-Ser209-eIF4E or total eIF4E proteins by Western blotting with the antibodies indicated on the left. As controls for drug activity, Akt and p70S6 kinase phosphorylation blotting were also performed. (B) Cells were pretreated without (−, lanes 1 to 3) or with PD98059 (30 μM) (PD, lanes 4 to 5) or SB203580 (20 μM) (SB, lanes 6 to 7), or with both (lanes 8 to 9), and analyzed as above. As controls for drug activity, Erk and p38 MAP kinase phosphorylation blotting were also performed. (C) HA-tagged Mnk1 expression vector was introduced in UT7-mpl cells following electroporation. Cells were maintained in culture for 36 h, then treated with (+) or without (−) TPO for 30 min before lysis. Lysates (100 μg) were analyzed by Western blotting using an anti-HA antibody.
TPO induces the selective recruitment of eIF4GII to the cap structure.
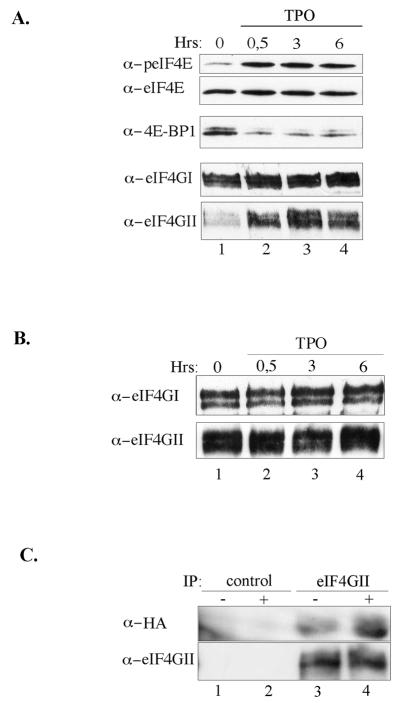
The recruitment of the 40S ribosomal subunit to the mRNA depends on the formation of the subunit complex eIF4F, which consists of three proteins: eIF4E, eIF4A, and an eIF4G member (eIF4GI or eIF4GII). Among these, eIF4E is thought to be the main regulatory factor in most cellular systems (6). Since TPO induced a robust phosphorylation of eIF4E while only marginally affecting the phosphorylation or the amounts of the inhibitory factor 4E-BPs, we tested whether TPO enhanced the assembly of eIF4F. UT7-mpl cells were treated with TPO, and the eIF4F complex was isolated by affinity chromatography on m7GTP-Sepharose 4B; bound factors were solubilized in Laemmli lysis buffer and analyzed by Western blotting. As shown in the upper panels of Fig. 4A, we observed a strong binding of phospho-eIF4E to the m7GTP-Sepharose beads, whereas the overall recruitment of total eIF4E to the affinity matrix did not change with TPO treatment. These data suggest that TPO induced the phosphorylation of eIF4E that is preassociated with the cap structure. Concomitantly, TPO triggered a quick and sustained release of the majority of m7GTP-bound 4E-BP1 from the matrix. When we assayed for the presence of eIF4G members, we found that eIF4GI did associate with the cap matrix in TPO-untreated cells, but we did not observe any reproducible change in the amounts of the cap-associated eIF4GI following TPO treatment. Samples from the same experiments were run on a parallel gel and immunoblotted with anti-eIF4GII antibodies. Surprisingly, while some eIF4GII proteins associated with the affinity matrix from TPO-untreated cell lysates, TPO strongly increased eIF4GII recruitment to the cap affinity matrix. This enhanced recruitment was selective for eIF4GII; it was sustained for hours (Fig. 4A, bottom panel). The levels of expression of either eIF4GI or eIF4GII were unaffected by TPO (Fig. 4B). Quantitation of eIF4GII retained on m7GTP-Sepharose matrix indicated that approximately 5% of total eIF4GII formed eIF4F complexes upon TPO treatment. Knowing that UT7-mpl cells express larger amounts of eIF4GII than other cell lines like 293T cells or Cos cells (unpublished data), this rather low percentage may simply reflect the amounts of eIF4E present within the cells that directly bind m7GTP-Sepharose 4B and are able to recruit eIF4GII.
FIG. 4.
Increased amounts of phospho-eIF4E and eIF4GII bind to the cap structure following TPO treatment. (A) Lysates from cells treated with or without TPO for the indicated time were incubated with m7GTP-Sepharose beads, and proteins bound to the affinity matrix were characterized by Western blotting using the indicated antibodies. (B) Cell lysates were analyzed directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with the indicated antibodies. (C) HA-tagged Mnk1 expression vector was introduced in UT7-mpl cells as described for Fig. 3C. Transfected cells were treated with or without TPO for 30 min before lysis. Lysates were immunoprecipitated (IP) with control (lanes 1 to 2) or anti-eIF4GII (lanes 3 to 4) antibodies and analyzed by Western blotting using an anti-HA antibody.
The above data indicated that TPO enhanced the phosphorylation of eIF4E at a time when eIF4GII was recruited to the eIF4F complex. eIF4E phosphorylation was previously demonstrated to depend on the activation and association of Mnk1 to eIF4GI in various cell contexts. We therefore wondered whether Mnk1 similarly associated with eIF4GII in our cellular context. Influenza HA-tagged Mnk1 was introduced in UT7-mpl cells as described above; transfected cells were activated in the presence or absence of TPO and eIF4GII protein was immunoprecipitated. As shown in Fig. 4C, Mnk1 coprecipitated with eIF4GII in TPO-treated and -untreated cells. Similarly, we observed that Mnk1 was recruited to the m7GTP-Sepharose affinity matrix upon TPO treatment (data not shown). These experiments suggest that eIF4GII-containing eIF4F functions in a manner similar to the eIF4GI-containing complex and does associate with activated Mnk1.
The phosphorylation of eIF4E and the recruitment of eIF4GII to the cap correlate with cell differentiation.
Our above data indicate that commitment of the UT7-mpl cells to megakaryocyte differentiation coincides with enhanced recruitment of eIF4GII to the cap structure and the long-lasting phosphorylation of eIF4E. These data suggest that these two biochemical events are linked to the differentiation process. We wished to further substantiate these results. UT7-mpl cells require growth factors such as EPO or GM-CSF for their growth and survival (18). Removing EPO or GM-CSF from the culture medium for a few hours eliminates DNA synthesis and cell proliferation, while adding back EPO or GM-CSF allows the cells to reenter the cell cycle. Therefore, we first analyzed the eIF4F complex status following growth factor deprivation of the UT7-mpl cells at the time cells reenter the cell cycle. Figure 5A shows that growth factor stimulus induced a strong phosphorylation of eIF4E that quickly declined thereafter, whereas the overall level of eIF4E was unaffected. Also, stimulation of the growth factor-deprived cells increased recruitment of eIF4GI (Fig. 5B, top panel, lane 2) but not obvious recruitment of eIF4GII to the cap affinity matrix (Fig. 5B, middle panel). As a control, stimulation of the growth arrest cells did not affect eIF4E levels (Fig. 5B, bottom panel; note that the increased amounts of eIF4E bound to the affinity matrix observed in lane 3 only reflects a difference in protein loading). These observations suggest that the eIF4F complex that forms at the onset of cell proliferation selectively includes eIF4GI.
FIG. 5.
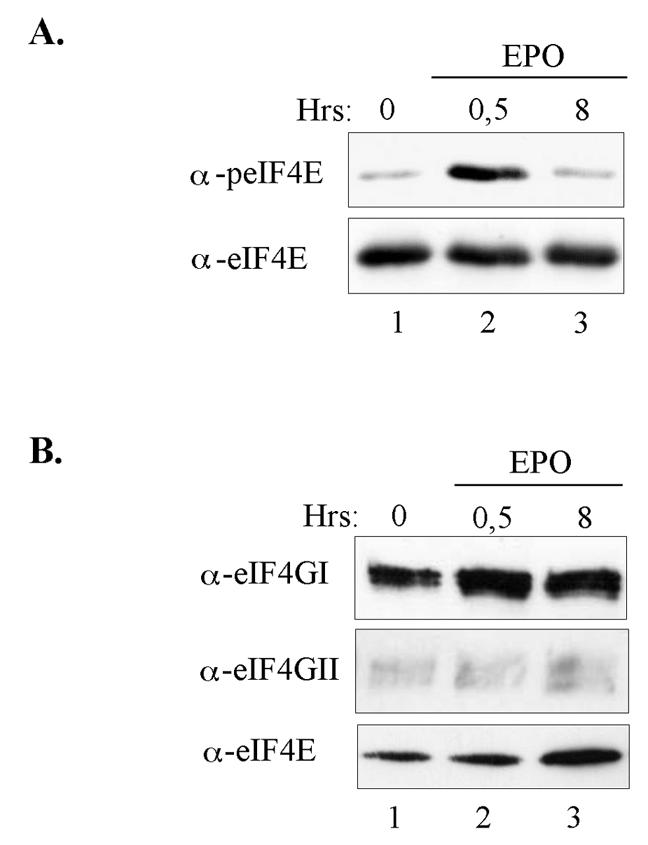
Mitogenic and survival stimuli do not induce a prolonged eIF4E phosphorylation and a selective recruitment of eIF4GII to the cap affinity matrix. UT7-mpl cells were made quiescent by a 15-h starvation in EPO-deprived culture medium. Cells were then stimulated to proliferate by adding EPO back to the culture medium for the indicated time (lanes 1 to 3). Cell lysates were prepared and either directly analyzed by Western blotting with anti-phospho-specific or total eIF4E antibodies (A) or incubated with m7GTP affinity matrix (B). Cap-bound proteins were analyzed by Western blotting using the indicated antibodies.
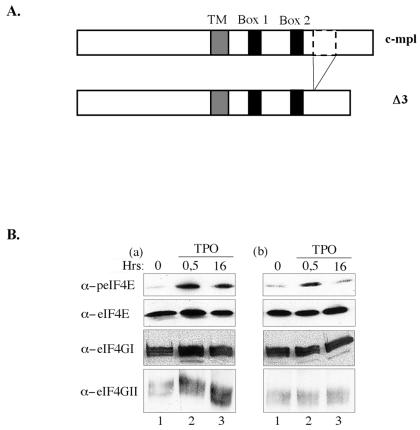
We also made use of another UT7 transfectant which expressed a truncated version of the TPO receptor. These cells, named UT7-mplΔ3, express a TPO receptor mutant which lacks a 25-amino-acid-long sequence in the intracellular domain of the receptor (Fig. 6A). As reported earlier, UT7-mplΔ3 are unable to commit to differentiation in the presence of TPO, as exemplified by the absence of enhanced integrin GpIIb/IIIa expression on the cell surface (33). TPO failed to induce enhanced cell size and complexity, as revealed by fluorescence-activated cell sorter analysis, and did not allow the development of an intracellular membrane network visible in EM, as opposed to TPO-treated UT7-mpl cells (data not shown). Yet, UT7-mplΔ3 cells express an active TPO receptor which allows them to highly proliferate in the presence of TPO, as reported previously (33). By using such cells, we tested the action of TPO on eIF4E phosphorylation and eIF4F formation. As shown in Fig. 6B, while TPO induced a strong and sustained phosphorylation of eIF4E in UT7-mpl cells (top left panel), it only induced a weak and transient phosphorylation of eIF4E in the highly proliferating UT7-mplΔ3 (top right panel, lane 3). This suggests that a robust phosphorylation of eIF4E is linked to the differentiation process. Similarly, while TPO induced the selective recruitment of eIF4GII on the cap affinity matrix in UT7-mpl cells (Fig. 6Ba, bottom left panel), it did not enhance the recruitment of eIF4GII in TPO-treated UT7-mplΔ3 cells (Fig. 6Bb, bottom right panel) nor that of eIF4GI, even though a weak but nonreproducible increase may be detected in some experiments. These data indicate that cell differentiation coincides with the enhanced recruitment of eIF4GII to the cap structure and a long-lasting phosphorylation of eIF4E.
FIG. 6.
Sustained phosphorylation of eIF4E and enhanced recruitment of eIF4GII to the cap correlate with cell differentiation. (A) Schematic representation of the wild-type TPO receptor and its mplΔ3 mutant; TM stands for the transmembrane domain, and box 1 and box 2 refer to conserved regions (33). (B) UT7-mpl cells (a) or UT7-mplΔ3 (b) were treated with TPO for the indicated time, cell extracts were prepared and incubated with m7GTP beads. Cap-bound proteins were analyzed by Western blotting using the antibodies indicated on the left.
DISCUSSION
An important and central conclusion of our work is that the two scaffolding factors eIF4GI and eIF4GII, which functionally complement each other and exhibit similar protein-RNA and protein-protein interactions, exhibit differential properties, since they can be differentially recruited to the mRNA cap structure coincident with the onset of cell differentiation. These observations have quite important implications regarding the regulation of translation initiation per se and also regarding the existence of possible selectivity toward the mRNA populations translated.
The selective and additive recruitment of eIF4GII to the cap structure at the onset of cell differentiation was observed in the absence of a concomitant increase in the amounts of eIF4GII. It is conceivable that some intracellular signals target eIF4GII and enhance—or stabilize—its association with the cap structure. Consistent with their central role in initiation, eIF4Gs are targets for translational control (reviewed in reference 37). eIF4GI and eIF4GII are phospho-proteins, and their phosphorylation states were reported to vary following serum stimulation (eIF4GI) or also, most probably, at the onset of mitosis (eIF4GII) (40, 42). However, these changes do not enhance the selective recruitment of eIF4GI or eIF4GII to the cap structure (40, 42). eIF4G proteins cannot bind to the cap structure directly. Their binding requires interaction with the cap-binding protein eIF4E. Previous reports have already indicated that the amount of eIF4G associated with eIF4E is highly variable, but these variations are most often correlated with the amounts of hypophosphorylated 4E-BP since 4E-BPs mimic eIF4Gs and bind to the eIF4G-binding motif of eIF4E, therefore blocking direct eIF4E-eIF4G interactions (24, 41). In the present report, however, the recruitment of eIF4GII to the cap structure did not correlate with a change in the overall amounts or phosphorylation state of the 4E-BPs. Therefore, our data indicate the existence of signaling pathways that specifically affect eIF4GII-eIF4E interaction in addition to the 4E-BP-eIF4E interactions.
A number of reports indicate that eIF4E binding to the cap is strongly enhanced in the presence of eIF4G (12, 48). In addition, the effect of binding of full-length eIF4G on the cap affinity of eIF4E can be further enhanced through binding of the PABP to eIF4G (4, 9, 12, 48). PABP can also modulate cap-dependent translation in the absence of a direct interaction with eIF4G, raising the possibility that PABP forms contacts with other factors at the 5′ end of mRNA (30). Therefore, signaling pathways may selectively enhance eIF4GII recruitment to the cap structure either by acting directly on the eIF4GII protein or by modulating eIF4E, PABP, or other specific eIF4GII partners or even by acting on some selective PABP partners, which would strengthen eIF4G-cap interaction.
In addition, our work indicates that commitment to differentiation is coincident with a long-lasting phosphorylation of eIF4E. eIF4E is strongly phosphorylated at Ser209 at the onset of cell differentiation while cells stop proliferating. This phosphorylation persists during the first 3 days of differentiation (S. Caron, unpublished data), whereas the phosphorylation of eIF4E induced by mitogenic stimuli in the same cells lasts only for a few hours. These data are quite surprising in light of previous reports where phosphorylation of Ser209 was most often correlated with cell growth in mammalian cells and was also shown to be critical for growth in Drosophila in toto (19). Although it was first reported that phosphorylation of eIF4E enhanced its affinity for the cap (25), reconstitution experiments of eIF4F complexes with purified molecules indicated that phosphorylation of eIF4E was not a prerequisite for complex formation with eIF4G (28), it was not required for translation (27), and it even reduced cap binding in vitro (43). Nevertheless, phosphorylation of Ser209 may still modulate a number of other protein-protein interactions that can affect the multiprotein 43S or 48S preinitiation complex, since the binding of eIF4E to eIF4G per se is known to change eIF4GI conformation, rendering it accessible to proteolytic cleavage (13, 28). These changes may thus similarly expose domains of eIF4GII involved in interactions with specific partners. These conformational changes may differ depending on the phosphorylation state of eIF4E.
We observed that TPO-dependent phosphorylation of Ser209 depends on the activation of either of two signaling pathways, the Erk and the p38 MAP kinase pathways, that seemed to play a redundant role in this process. TPO was already reported to activate these two pathways, and TPO-dependent differentiation was previously shown to depend on the integrity of the Erk pathway (5). We showed that Mnk1 was indeed activated at the onset of cell differentiation and was associated with eIF4GII. Mnk kinases associate with the C-terminal domain of eIF4G and phosphorylate its substrate eIF4E (41). Therefore our data suggest that phosphorylation of eIF4E is a consequence of the increased association of eIF4GII with eIF4E at the onset of differentiation. However, one cannot exclude that additional signaling pathways (kinases or phosphatases) may also be activated that sustain the long-lasting phosphorylation of eIF4E observed along the differentiation process.
The selective recruitment of eIF4GII to the cap moiety , coincident with the onset of cell differentiation, raises the interesting possibility that the eIF4GII-dependent initiation complexes actually favor translation of specific mRNAs. eIF4G proteins contain a central region that interacts with specific RNA domains such as internal ribosomal entry sites of viral RNAs and also short motifs that exhibit general RNA-binding activities that are critical for ribosome scanning (7, 22, 38). Also, yeast eIF4G interacts with eIF1 and eIF5, and this interaction is required for the optimal AUG selection, indicating that yeIF4G participates in the selection of the initiation codon (14). These observations lead us to propose that eIF4GII may similarly possess selective intrinsic RNA-binding activities or may associate with specific, but still unknown, partners that contribute to the selection of particular mRNAs to be translated. Such function would fit with the intense expression of the appropriate proteins that are required for platelet function. Alternatively, recruitment of eIF4GII to the mRNA cap structure may simply induce an increase of the absolute activity of eIF4F within the cells under selective conditions.
Indirect evidence for nonredundant roles of the two mammalian eIF4Gs was recently reported. eIF4GI and eIF4GII are subjected to regulation of their activities through limited proteolysis during apoptosis or certain viral infections, which most often results in the inhibition of cap-dependent host protein synthesis and the stimulation of internal ribosome entry site-mediated translation (26, 37). The human immunodeficiency virus type 1 protease, HIV-Pr, cleaves eIF4GI but not eIF4GII. This processing of eIF4GI was recently shown to inhibit translation of various RNAs despite the presence of uncleaved eIF4GII, suggesting differential mRNA selectivity for eIF4GI and eIF4GII (29). More importantly, in Saccharomyces cerevisiae two genes were identified (TIF4631 and TIF4632) as encoding eIF4G proteins. Although TIF4631-disrupted strains exhibited a slow-growth phenotype, disruption of TIF4632 failed to show any phenotype, while a double disruption of these genes was lethal (7). These last observations also emphasized some functional difference between the two forms of eIF4G, which fits well with our data. In accordance with these reports, we suggest that eIF4GI may, like TIF4631, direct the translation of survival and growth-related genes, while eIF4GII may be dedicated to the translation of genes linked to the differentiation process.
In summary, our data provide the first evidence that eIF4GI and eIF4GII can be selectively regulated by cytokines, which suggests that they may fulfill different roles in mammalian cells.
Acknowledgments
We thank J. M. Massé, J. Guichart, P. Mayeux, and L. Haddaoui for helpful technical assistance with EM (J.M.M. and J.G.) and with two- dimensional gel electrophoresis (P.M. and L.H.). We thank T. Ohlmann for helpful discussions and P.-H. Romeo for valuable comments on the manuscript.
This work was supported by a grant from the Ligue Contre le Cancer. S.C. is a recipient of a fellowship from the Ligue Contre le Cancer.
REFERENCES
- 1.Cwirla, S. E., P. Balasubramanian, D. J. Duffin, C. R. Wagstrom, C. M. Gates, S. C. Singer, A. M. Davis, R. L. Tansik, L. C. Mattheakis, C. M. Boytos, P. J. Schatz, D. P. Baccanari, N. C. Wrighton, R. W. Barrett, and W. J. Dower. 1997. Peptide agonist of the thrombopoietin receptor as potent as the natural cytokine. Science 276:1696-1699. [DOI] [PubMed] [Google Scholar]
- 2.Doubeikovski, A., G. Uzan, Z. Doubeikovski, M. H. Prandini, F. Porteu, S. Gisselbrecht, and I. Dusanter-Fourt. 1997. Thrombopoietin-induced expression of the glycoprotein IIb gene involves the transcription factor PU.1/Spi-1 in UT7-Mpl cells. J. Biol. Chem. 272:24300-24307. [DOI] [PubMed] [Google Scholar]
- 3.Fraser, C. S., V. M. Pain, and S. J. Morley. 1999. Cellular stress in xenopus kidney cells enhances the phosphorylation of eukaryotic translation initiation factor (eIF)4E and the association of eIF4F with poly(A)-binding protein. Biochem. J. 342:519-526. [PMC free article] [PubMed] [Google Scholar]
- 4.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20:1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia, J., J. de Gunzburg, A. Eychene, S. Gisselbrecht, and F. Porteu. 2001. Thrombopoietin-mediated sustained activation of extracellular signal-regulated kinase in UT7-Mpl cells requires both Ras-Raf-1- and Rap1-B-Raf-dependent pathways. Mol. Cell. Biol. 21:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 7.Goyer, C., M. Altmann, H. S. Lee, A. Blanc, M. Deshmukh, J. L. Woolford, Jr., H. Trachsel, and N. Sonenberg. 1993. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 13:4860-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, N. K., J. M. Coller, K. S. Dickson, and M. Wickens. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19:4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grolleau, A., N. Sonenberg, J. Wietzerbin, and L. Beretta. 1999. Differential regulation of 4E-BP1 and 4E-BP2, two repressors of translation initiation, during human myeloid cell differentiation. J. Immunol. 162:3491-3497. [PubMed] [Google Scholar]
- 11.Haghighat, A., S. Mader, A. Pause, and N. Sonenberg. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haghighat, A., and N. Sonenberg. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272:21677-21680. [DOI] [PubMed] [Google Scholar]
- 13.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, H., T. von der Haar, C. R. Singh, M. Ii, B. Li, A. G. Hinnebusch, J. E. McCarthy, and K. Asano. 2003. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell. Biol. 23:5431-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17:6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaushansky, K. 1999. Thrombopoietin and hematopoietic stem cell development. Ann. N. Y. Acad. Sci. 872:314-319. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu, N., H. Nakauchi, A. Miwa, T. Ishihara, M. Eguchi, M. Moroi, M. Okada, Y. Sato, H. Wada, Y. Yawata, et al. 1991. Establishment and characterization of a human leukemic cell line with megakaryocytic features: dependency on granulocyte-macrophage colony-stimulating factor, interleukin 3, or erythropoietin for growth and survival. Cancer Res. 51:341-348. [PubMed] [Google Scholar]
- 19.Lachance, P. E., M. Miron, B. Raught, N. Sonenberg, and P. Lasko. 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, J. C., Jr., and R. T. Abraham. 1997. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem. Sci. 22:345-349. [DOI] [PubMed] [Google Scholar]
- 21.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 22.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951-961. [DOI] [PubMed] [Google Scholar]
- 25.Minich, W. B., M. L. Balasta, D. J. Goss, and R. E. Rhoads. 1994. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl. Acad. Sci. USA 91:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morley, S. J., P. S. Curtis, and V. M. Pain. 1997. eIF4G: translation's mystery factor begins to yield its secrets. RNA 3:1085-1104. [PMC free article] [PubMed] [Google Scholar]
- 27.Morley, S. J., and S. Naegele. 2002. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J. Biol. Chem. 277:32855-32859. [DOI] [PubMed] [Google Scholar]
- 28.Ohlmann, T., V. M. Pain, W. Wood, M. Rau, and S. J. Morley. 1997. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 16:844-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlmann, T., D. Prevot, D. Decimo, F. Roux, J. Garin, S. J. Morley, and J. L. Darlix. 2002. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 318:9-20. [DOI] [PubMed] [Google Scholar]
- 30.Otero, L. J., M. P. Ashe, and A. B. Sachs. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18:3153-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pain, V. M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236:747-771. [DOI] [PubMed] [Google Scholar]
- 32.Pause, A., G. J. Belsham, A. C. Gingras, O. Donze, T. A. Lin, J. C. Lawrence, Jr., and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 33.Porteu, F., M. C. Rouyez, L. Cocault, L. Benit, M. Charon, F. Picard, S. Gisselbrecht, M. Souyri, and I. Dusanter-Fourt. 1996. Functional regions of the mouse thrombopoietin receptor cytoplasmic domain: evidence for a critical region which is involved in differentiation and can be complemented by erythropoietin. Mol. Cell. Biol. 16:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulin, F., A. C. Gingras, H. Olsen, S. Chevalier, and N. Sonenberg. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002-14007. [DOI] [PubMed] [Google Scholar]
- 35.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 36.Preiss, T., and M. W. Hentze. 1999. From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev. 9:515-521. [DOI] [PubMed] [Google Scholar]
- 37.Prevot, D., J. L. Darlix, and T. Ohlmann. 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell 95:141-156. [DOI] [PubMed] [Google Scholar]
- 38.Prevot, D., D. Decimo, C. H. Herbreteau, F. Roux, J. Garin, J. L. Darlix, and T. Ohlmann. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22:1909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proud, C. G., and R. M. Denton. 1997. Molecular mechanisms for the control of translation by insulin. Biochem. J. 328:329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyronnet, S., J. Dostie, and N. Sonenberg. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raught, B., A. C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheper, G. C., B. van Kollenburg, J. Hu, Y. Luo, D. J. Goss, and C. G. Proud. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277:3303-3309. [DOI] [PubMed] [Google Scholar]
- 44.Smith, M. R., M. Jaramillo, Y. L. Liu, T. E. Dever, W. C. Merrick, H. F. Kung, and N. Sonenberg. 1990. Translation initiation factors induce DNA synthesis and transform NIH 3T3 cells. New Biol. 2:648-654. [PubMed] [Google Scholar]
- 45.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 46.Tarun, S. Z., Jr., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 47.Tarun, S. Z., Jr., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 48.von Der Haar, T., P. D. Ball, and J. E. McCarthy. 2000. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J. Biol. Chem. 275:30551-30555. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., A. Flynn, A. J. Waskiewicz, B. L. Webb, R. G. Vries, I. A. Baines, J. A. Cooper, and C. G. Proud. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273:9373-9377. [DOI] [PubMed] [Google Scholar]
- 50.Waskiewicz, A. J., J. C. Johnson, B. Penn, M. Mahalingam, S. R. Kimball, and J. A. Cooper. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19:1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whalen, S. G., A. C. Gingras, L. Amankwa, S. Mader, P. E. Branton, R. Aebersold, and N. Sonenberg. 1996. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J. Biol. Chem. 271:11831-11837. [DOI] [PubMed] [Google Scholar]