Abstract
Ras family GTPases (RFGs) regulate signaling pathways that control multiple biological processes. How signaling specificity among the closely related family members is achieved is poorly understood. We have taken a proteomics approach to signaling by RFGs, and we have analyzed interactions of a panel of RFGs with a comprehensive group of known and potential effectors. We have found remarkable differences in the ability of RFGs to regulate the various isoforms of known effector families. We have also identified several proteins as novel effectors of RFGs with differential binding specificities to the various RFGs. We propose that specificity among RFGs is achieved by the differential regulation of combinations of effector families as well as by the selective regulation of different isoforms within an effector family. An understanding of this new level of complexity in the signaling pathways regulated by RFGs is necessary to understand how they carry out their many cellular functions. It will also likely have critical implications in the treatment of human diseases such as cancer.
Ras genes (the H-Ras, K-Ras, and N-Ras genes) are found mutated in 20 to 25% of human tumors (4). They code for small GTPases that act as molecular switches, cycling between an inactive GDP-bound state and an active GTP-bound state. They are activated in response to a wide variety of extracellular stimuli and regulate signaling pathways that control multiple biological processes, allowing a cell to respond to its microenvironment. Mutations found in tumors lock the protein in a constitutively active GTP-bound state. The activation state of the proteins is normally tightly regulated by the concerted action of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs catalyze the release of GDP, allowing binding of the more abundant GTP and the activation of the proteins, whereas GAPs greatly stimulate their intrinsic GTPase activity and therefore catalyze their inactivation. The exchange of GDP for GTP induces a conformational change in the proteins that allows them to interact with their downstream effectors and carry out their multiple functions (40, 71).
The best characterized Ras effectors are the Raf kinases, through which Ras activates the mitogen-activated protein kinase (MAPK) cascade, the p110 catalytic subunit of class I phosphoinositide 3-kinases (PI3Ks), and a family of Ral exchange factors (RalGEFs). Other Ras effectors include RIN1, Tiam1, phospholipase Cɛ (PLCɛ), AF6, and Nore1 (14).
Ras effectors interact with Ras through a small region called the Ras binding domain (RBD). The RBDs of Raf, PI3Ks, and RalGEFs, though having considerable structural similarities, have little sequence homology and seem to define three types of distinct domains that are found in other proteins. Tiam1, for example, has a Raf-type RBD (35). The type of RBD found in RalGEFs has been termed the RA (RalGDS/AF6, Ras-associating) domain (50). RA domains are found in a wide variety of proteins, some of which, including Rin1, AF6, Nore1, and PLCɛ, are known to interact with Ras.
Ras proteins were the first members of what is now a large superfamily of small GTPases, comprising more than 150 proteins, to be identified (71). This superfamily is structurally classified into at least five families: the Ras, Rho, Rab, Arf, and Ran subfamilies. The Ras family now includes at least 21 members: H-Ras, K-Ras (A and B), N-Ras, R-Ras, TC21/R-Ras2, R-Ras3/M-Ras, Rap1a, Rap1b, Rap2a, Rap2b, Rap2c, Rit, Rin, Rheb, Noey2, DiRas1/Rig, DiRas2, ERas, RalA, RalB, DexRas/RasD1, and RasD2/Rhes.
Many of these Ras family GTPases (RFGs) remain poorly characterized, and little is known about their properties and functions. Several of these RFGs do share some of the biological properties of Ras in various cell systems. Mutated TC21 has been found in human tumors (2, 7, 27), and activated versions of TC21 transform a variety of cell types. Like Ras, TC21 also induces neurite outgrowth in PC12 rat pheochromocytoma cells and blocks C2 mouse myoblast differentiation (10, 20, 21, 45). Activated versions of R-Ras3 transform NIH 3T3 fibroblasts and induce neurite outgrowth in PC12 cells (16, 31, 32). R-Ras can transform some, but not other, cell types (12, 39, 61) and also has functions clearly different from those of Ras (62, 70, 78). Rap1 proteins, although originally proposed to act by antagonizing Ras function, have functions that are different from those of Ras, some of which may involve the regulation of integrin-mediated adhesion (6). Rit and Rin lack the Ras family characteristic CAAX prenylation signal but instead contain a cluster of basic amino acids and are reported to be membrane localized (36). Activated Rit, but not Rin, can transform NIH 3T3 fibroblasts (60).
Although it is still not well defined, there is some overlap in the way in which at least some of the RFGs are regulated, with both GEFs and GAPs having overlapping specificities and acting on several Ras family members (14, 53). This fact raises important considerations about Ras protein functions previously reported based on the use of certain experimental tools. Many of the crucial cellular functions ascribed to Ras proteins in the literature are based on experiments involving the blocking of Ras function with the use of dominant-negative mutants, such as N17 Ras. N17 Ras is thought to prevent the activation of endogenous Ras by sequestering RasGEFs. However, because RasGEFs also act on other RFGs, their activation is also expected to be inhibited by N17 Ras and therefore this Ras mutant cannot discriminate between contributions from different RFGs. Similarly, the neutralizing monoclonal antibody Y13-259, another tool widely used to block Ras function, cross-reacts with R-Ras3 (16). Therefore, its reported effects upon microinjection, such as the inhibition of cell cycle progression, do not exclude the possibility of a contribution from other RFGs.
In order to understand the biological functions of the different RFGs and their individual contributions to human diseases, such as cancer, it is crucial to understand which effector pathways they can regulate. Different RFGs also overlap in their abilities to activate the different effector pathways. TC21 can activate the PI3K/protein kinase B (PKB) pathway, but there are conflicting results regarding its ability to activate the Raf/MAPK and RalGEF pathways (37, 45, 46, 57-59). R-Ras3 interacts with a variety of Ras effectors in the yeast two-hybrid (Y2H) system, though different groups have reported different results (15, 31, 52). R-Ras activates the PI3K pathway but not the Raf/MAPK pathway (44). Rit and Rin can interact by Y2H with RalGEFs and AF6 but not with Rafs (64, 65), and Rit failed to activate the MAPK or PKB pathway in NIH 3T3 fibroblasts (60) but did induce Erk phosphorylation in PC6 pheochromocytoma cells (68).
Conflicting results reported by different laboratories may have arisen from the use of different experimental systems, including different assays and cell types. In vitro interaction assays with purified proteins most often rely on the use of fragments of the effectors, typically just the RBD. Interactions with these minimal binding regions may not accurately reflect interactions with the full-length proteins. Similarly, in the Y2H assay that has been instrumental in the identification of Ras effectors and in the characterization of their abilities to interact with different RFGs, often only truncated versions of the effectors have been used. Furthermore, the assay relies on the mislocalization of the GTPases. The carboxy-terminal sequences that target these GTPases for posttranslational lipid modifications and membrane localization are deleted in order to allow nuclear localization of the fusion proteins and transcriptional activation of the reporter genes used in the assay. Therefore, any contributions of these posttranslational modifications or the subcellular localizations of the RFGs to the interactions will be lost.
Another important consideration is that there are several isoforms for most Ras effectors. For example, the class I family of PI3K has p110α, p110β, p110δ, and p110γ isoforms and the Raf kinase family comprises Raf-1, A-Raf, and B-Raf genes. RalGEFs now include RalGDS, RGL, RGL2/Rlf, and RGL3. RFGs may differ in their abilities to regulate different isoforms of the same family, and these selective interactions may have important biological consequences, depending on the specific properties and functions of the different isoforms as well as the patterns of expression of the various GTPases and the effector isoforms in different cell types. Furthermore, in many instances the ability of RFGs to activate effector pathways, such as the Raf/MAPK and PI3K pathways, relies on the use of the activation of a downstream target as a readout (e.g., Erk or PKB, respectively). These assays rely on the endogenous Raf or PI3K isoforms expressed within the given cell type, but because these isoforms very often have not been determined, important isoform-specific differences may have been overlooked and may also account for some of the conflicting reports in the literature.
To understand the effector specificity of RFGs, we have set out to compare comprehensively the ability of a large set of RFGs to interact with and directly activate the various effector isoforms of the PI3K, Raf, and RalGEF families. We have found some striking isoform-specific differences and discuss their critical implications. We have also compared the abilities of the different RFGs to interact with a wide array of proteins containing RA domains, some previously described and some recently uncovered by sequencing projects and yet to be characterized. We have identified several new proteins as novel effectors of RFGs with differential binding specificities to the various GTPases of the Ras family.
We propose a model in which specificity among the RFGs is achieved by the differential regulation of combinations of effector families as well as by the selective regulation of different isoforms within an effector family. An understanding of this new level of complexity in the Ras field is necessary to understand how RFGs carry out their many cellular functions. It will also help to design strategies for treating diseases in which these pathways are deregulated, such as human cancer.
MATERIALS AND METHODS
Materials.
Anti-myc (A-14) and antihemagglutinin (anti-HA) (Y-11) antibodies were obtained from Santa Cruz Biotechnology, anti-Flag M2 antibody was obtained from Sigma-Aldrich, and phospho-T202/Y204 p44/42 MAPK was obtained from Cell Signaling Technology. Horseradish peroxidase-coupled secondary antibodies and glutathione Sepharose were obtained from Amersham-Pharmacia Biotechnologies.
Cell culture and transfections.
293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Cells (1 × 106) were seeded in six-well dishes and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The total amount of plasmid DNA was always 2 μg, and when several plasmids were used, the ratios were always 1:1, except in the RalGEF assays, in which one-fourth of the amount of RalGDS was used.
Constructs.
All RFGs were of human origins except RalA, which was of a simian origin (18a). All RFGs carried activating mutations (V12 H-Ras, V12 N-Ras, V12 K-Ras4B, V12 Rap1a, V12 Rap2a, V38 R-Ras, V23 TC21, L81 R-Ras3, L79 Rit, L78 Rin, L64 Rheb, and L89 Rab5). V22 R-Ras3, V30 Rit, and V29 Rin were also tested and exhibited no significant differences from their respective L61 versions. Mutations were generated with the QuikChange site-directed mutagenesis kit (Stratagene). All Raf kinases, RalGEFs, class I PI3Ks, and RA domain-containing proteins were of human origins except p110α, which was of a bovine origin. Most were cloned by reverse transcription-PCR from pooled human fetal brain, liver, and lung poly(A) RNAs (Clontech) by using the Superscript One-Step reverse transcription-PCR for long templates (Life Technologies) or from human full-length cDNA (Panomics) by PCR with LA-Taq polymerase (Takara). All genes were full length except APBB1IP/RIAM, which was cloned as a hypothetical protein FLJ20805 and was a truncated version (amino acids 1 to 261) of APBB1IP/RIAM, and PLCɛ, which was a C-terminal fragment (amino acids 1383 to 2303). Genes were cloned into pENTR vectors (Invitrogen) and transferred in frame into cytomegalovirus promoter-driven expression plasmids with N-terminal myc, HA, or glutathione S-transferase (GST) tags by using recombination-mediated Gateway technology (Invitrogen).
Raf and Erk assays.
Two days after transfection, cells were lysed in 350 μl of 1% Triton X-100-TNE (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA) containing protease and phosphatase inhibitor cocktails (Sigma). HA-tagged Rafs or HA-tagged Erk1 was immunoprecipitated with anti-HA antibodies, and after extensive washing, Raf activity was assayed in a coupled MEK/ERK2 kinase assay and Erk activity was measured by using myelin basic protein as a substrate as described previously (1).
Interaction assays.
GST-RFGs were transfected into 293T cells with myc effectors. Two days after transfection, cells were lysed in 350 μl of 1% Triton X-100-TNM containing protease and phosphatase inhibitor cocktails (Sigma). RFGs were pulled down from cleared lysates with glutathione Sepharose, and beads were washed four times with 1% Triton X-100-TNM, drained, and resuspended in sample buffer. Bound effectors were detected by Western blotting with myc antibodies.
Ral activation assays.
Flag-RalA was cotransfected with RalGEFs, RFGs, or empty vector. Two days later, cells were lysed and the GTP-bound form of RalA was specifically pulled down with the GST-tagged RBD of RalBP1 as described previously (73). Cells were lysed and processed as described above for interaction assays. Ral was detected by immunoblotting with an anti-Flag antibody.
PI3K assays.
The p110 catalytic subunits of class I PI3Ks were cotransfected with RFGs into 293T cells. Two days later, cells were labeled in 600 μl of phosphate-free Dulbecco's modified Eagle's medium with 60 μCi of [32P]orthophosphate for 4 h. PI3K activity was assayed by measuring the levels of total 3′ phosphorylated lipids by high-pressure liquid chromatography as described previously (56).
RESULTS
Activation of Raf kinases by RFGs.
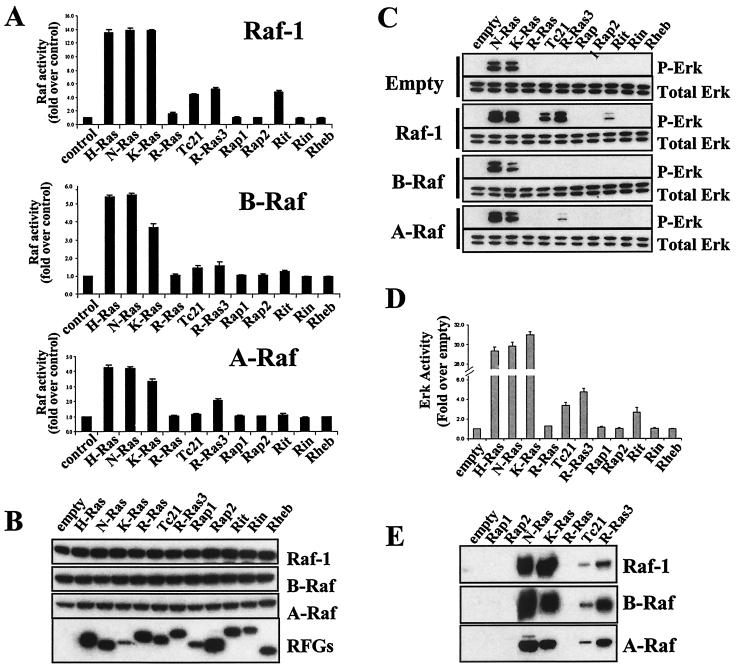
Activation of Raf kinases by H-Ras, N-Ras, and K-Ras has been well established, but the ability of other RFGs to activate the Raf/Erk pathway is controversial, possibly because different experimental systems have been used. In addition, different RFGs have been analyzed mostly individually, making it difficult to make relative comparisons between the various RFGs. We have compared the abilities of a large subset of RFGs to activate the three Raf isoforms in 293T cells. As shown in Fig. 1A, H-Ras, N-Ras, and K-Ras are the strongest activators of Raf-1, with a 12- to 15-fold increase in stimulation compared with the activity seen in immunoprecipitates from cells expressing Raf-1 alone. TC21, R-Ras3, and Rit also activate Raf-1, although less efficiently than Ras proteins (with a four to sixfold increase in stimulation, depending on the experiment). Rap1, Rap2, Rin, and Rheb had no detectable effects. H-Ras, N-Ras, K-Ras, and, to a smaller but significant extent, R-Ras3 can also stimulate A-Raf, whereas only H-Ras, N-Ras, and K-Ras are able to detectably activate B-Raf. Thus, different RFGs vary greatly in their abilities to activate the various Raf isoforms, with differences both in isoform specificities and in the efficiencies with which they activate individual isoforms.
FIG. 1.
Activation of the Raf/Erk pathway by RFGs. (A) Activation of Raf kinases by RFGs. Constructs expressing HA-tagged Raf kinases were cotransfected into 293T cells with constitutively active myc-tagged RFGs or empty vector (control). Two days later, Raf kinase activity on HA immunoprecipitates was measured in a coupled assay for its ability to activate MEK. (B) Expression levels of transfected proteins were measured by Western blotting with anti-HA or anti-myc antibodies. (C) Stimulation of Erk phosphorylation by RFGs. Erk phosphorylation was measured in lysates from transfections as described for panel A by Western blotting with phospho-specific Erk antibodies. (D) Activation of Erk1 activity by RFGs. HA-tagged Erk1 was contransfected with RFGs, and Erk kinase activity was measured in HA immunoprecipitates by using myelin basic protein as a substrate. (E) Interaction of Raf kinases with RFGs. GST-tagged RFGs were cotransfected into 293T cells with myc-tagged Raf kinases, and interactions were measured by pulling down RFGs with glutathione beads and detecting bound Raf in Western blots with anti-myc antibodies.
To assess whether the differential stimulation of Raf kinase activity correlates with activation of the downstream Erk pathway, we measured the phosphorylation state of the endogenous Erks with phospho-specific antibodies. Figure 1C shows that when RFGs are expressed by themselves, only N-Ras and K-Ras can stimulate Erk phosphorylation. Surprisingly, we did not detect any Erk phosphorylation in response to TC21, R-Ras3, or Rit, despite the fact that 293T cells express Raf-1 and that TC21, R-Ras3, and Rit can activate Raf-1 kinase activity in Raf assays (Fig. 1A). To address the possibility that a weaker level of Erk phosphorylation may have been below the threshold of detection of our Western blots, we cotransfected a tagged Erk1 construct with the RFGs and measured Erk activity on the Erk immunoprecipitates. As shown in Fig. 1D, in this assay, TC21, R-Ras3, and Rit could all activate Erk1 activity, although considerably less efficiently (6- to 10-fold less) than the Ras proteins.
When Raf-1 is coexpressed, phosphorylation of endogenous Erks by TC21, R-Ras3, and Rit becomes detectable (Fig. 1C). When A-Raf is coexpressed, R-Ras3 is able to weakly but detectably stimulate Erk phosphorylation. Therefore, stimulation of the kinase activity of the Raf isoforms by the various RFGs correlates well with Erk activation within the same cells.
We also studied the ability of the RFGs and Raf kinases to interact within the cell in coimmunoprecipitation experiments. As shown in Fig. 1E, N-Ras and K-Ras bind strongly to all three Raf isoforms. H-Ras behaved in a fashion identical to that of N-Ras and K-Ras (data not shown). Raf-1, A-Raf, and B-Raf were also detected in association with TC21 and R-Ras3, but these interactions were considerably weaker than those with the Ras proteins. Contrary to previous reports, we did not detect any association of any Raf isoforms with R-Ras or Rap1.
Activation of RalGEFs by RFGs.
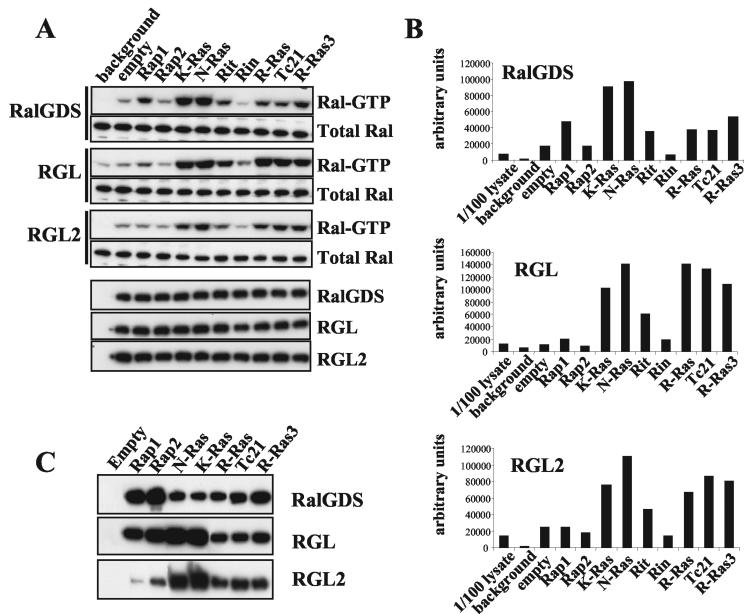
We analyzed the ability of the RFGs to activate three members of the RalGEF family, RalGDS, RGL, and RGL2/Rlf. RalGEF activity was measured by using an activation-specific RBD pulldown assay to measure Ral-GTP levels. Upon cotransfection of all three RalGEFs, N-Ras, K-Ras, R-Ras, TC21, R-Ras3, and Rit can all increase the levels of Ral-GTP, though to slightly different extents (Fig. 2A and B). H-Ras behaves in a fashion identical to that of N-Ras and K-Ras (data not shown). Rheb failed to activate any of the three RalGEFs tested (data not shown). Ras proteins are the strongest activators of RalGDS, whereas R-Ras, TC21, and R-Ras3 are as potent as Ras proteins in the activation of RGL and RGL2. Rap1 (but not Rap2 or Rin) can also activate RalGDS. In some experiments, we detected a small effect of Rap1 on RGL activity (see Discussion).
FIG. 2.
Activation of RalGEF pathway by RFGs. (A) Flag tagged-RalA was cotransfected into 293T cells with empty vector (background), HA-tagged RalGEFs, and empty vector or constitutively active myc-tagged RFGs. Two days after transfection, the levels of GTP-bound flag-RalA were measured in total cell lysates in pulldown assays with GST-RalBP1-RBD. In each gel, 1/100 of the lysate used was run. Expression levels of transfected proteins were measured by Western blotting with anti-Flag or anti-HA antibodies. Results shown are representative of at least three independent experiments. (B) The levels of Ral-GTP shown in panel A were quantified with a STORM phosphorimager. (C) Interaction of RalGEFS with RFGs. GST-tagged RFGs were cotransfected into 293T cells with myc-tagged RalGEFs, and interactions were measured by pulling down RFGs with glutathione beads and detecting bound RalGEFs with anti-myc antibodies in Western blots.
Figure 2C shows that all RFGs tested are able to interact with all three RalGEFs. However, the ability to associate with the protein in vivo does not correlate with the ability to stimulate its activity. Rap1 and Rap2 display the strongest association with RalGDS, but Rap1 is weaker than Ras proteins in activating RalGDS activity, whereas Rap2 has no effect. Similarly, Rap1 and Rap2 interact with RGL more strongly than R-Ras, TC21, and R-Ras3, with little or no effect on their activity.
Activation of class I PI3Ks by RFGs.
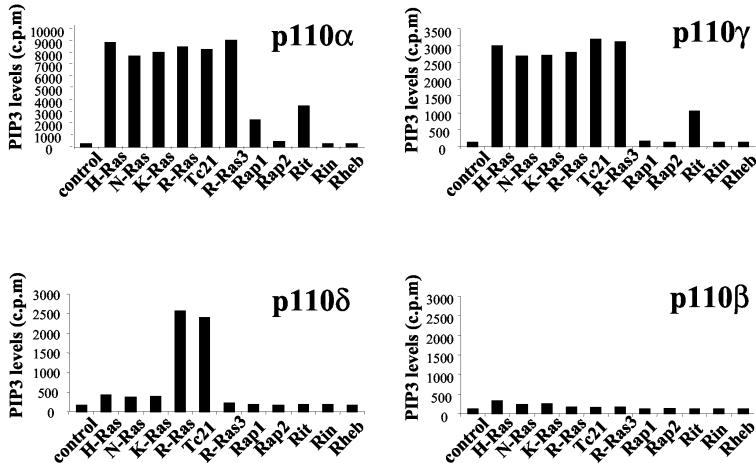
We assessed the ability of the RFGs to stimulate the lipid kinase activity of the four class I PI3K isoforms, p110α, p110β, p110δ, and p110γ, by measuring the levels of their 3′ phosphorylated lipid products in intact cells (Fig. 3). H-Ras, N-Ras, K-Ras, R-Ras, TC21, and R-Ras3 potently activate p110α and p110γ to similar extents; when R-Ras is coexpressed, the levels of PIP3 are 30- to 40-fold or 25- to 30-fold higher than those in cells expressing only p110α or p110γ, respectively. Rap1 and Rit expression also significantly stimulate p110α activity, with a 10- to 15-fold increase in 3′ lipid levels compared with those for p110α alone. Rit can also stimulate p110γ activity. The ability of the different RFGs to stimulate p110α activity correlates well with their ability to stimulate Akt activation in a variety of cell lines tested, such as NIH 3T3 fibroblasts and PAE cells (data not shown).
FIG. 3.
Activation of class I PI3Ks by RFGs. Constitutively active RFGs were cotransfected into 293T cells with PI3K isoforms. Two days after transfection, cells were labeled with [32P]orthophosphate, and total cellular PIP3 levels were measured by high-pressure liquid chromatography. The levels of PI(4,5)P2 were standardized to 200,000 cpm. Results shown are representative of at least three independent experiments.
Remarkably, only R-Ras and TC21 are able to activate the p110δ isoform. p110β failed to undergo activation by any of the RFGs tested. It is possible that p110β activity may be selectively inhibited in our cells. However, immunoprecipitates of p110β had readily detectable lipid kinase activity in vitro (data not shown). Similarly, the ability of this isoform to associate with the p85α and p85β regulatory subunits was undistinguishable from that of p110α or p110δ (data not shown). p110β has been reported to interact with Rab5 (9), suggesting that p110β may be regulated by Rab family GTPases. We were, however, unable to detect any effect of activated Rab5 on p110β activity (data not shown).
Interaction of RFGs with RA domain-containing proteins.
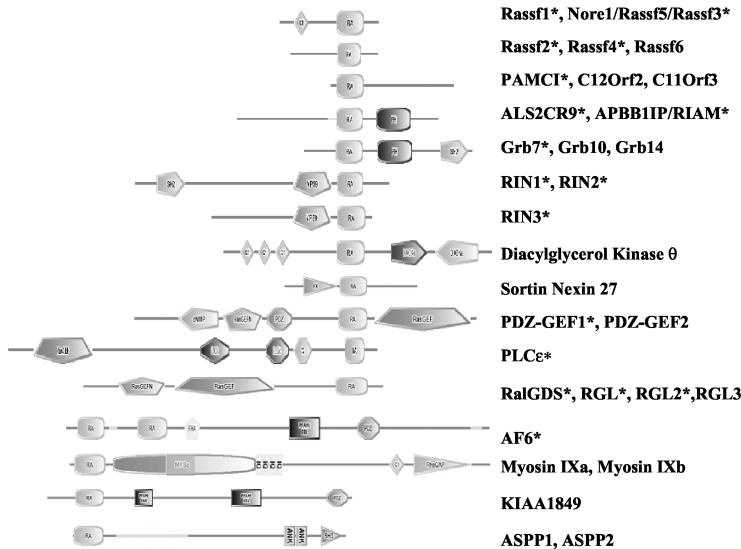
Data presented above and elsewhere indicate that RFGs interact with effector proteins with a remarkable combination of selectivity and promiscuity. In addition to Raf, PI3Ks, and RalGEFs, RFGs are known to interact with other effectors. The type of RBD found in RalGEFs, termed the RA domain, is also found in other proteins known to interact with RFGs, such as RIN1, Nore1, PLCɛ, and AF6. In addition, as shown in Fig. 4, RA domains are found in many other proteins. The abilities of many of these proteins to interact with RFGs have not been determined.
FIG. 4.
Proteins with RA domains. Domain structures of RA domain-containing protein families according to the SMART protein domain database (http://smart.embl-heidelberg.de). Proteins are not drawn to scale. Note that some genes have splice variants that give rise to proteins lacking some of the depicted domains. Asterisks indicate proteins that have been analyzed in this study.
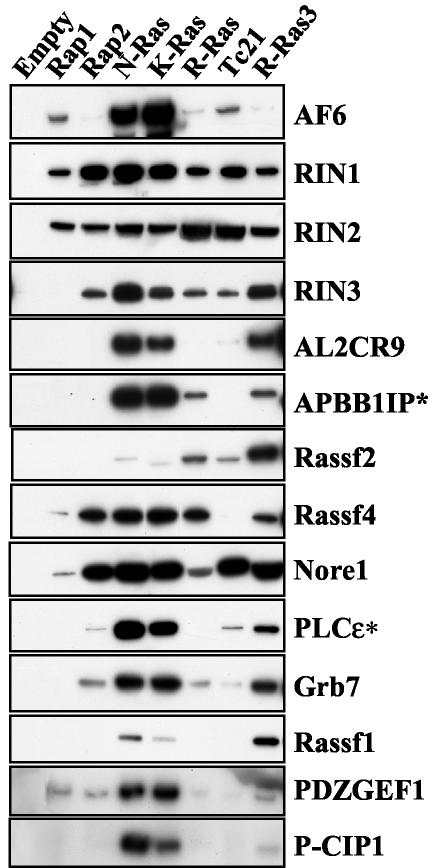
To better characterize the scope of interactions between RFGs and other potential effectors, we tested the binding of 14 other RA domain-containing proteins in coimmunoprecipitation assays. As shown in Fig. 5, all of the RA domain-containing proteins that we tested interacted with at least some RFGs, showing promiscuous as well as selective interactions. Some proteins, such as RIN1, RIN2, and Nore1, interacted with all RFGs, though with variable efficiencies. Many others showed differential interactions with the various RFGs. Some, like PLCɛ, AF6, PDZ-GEF, APBB1IP, and P-CIP1, showed preferential binding to N-Ras and K-Ras but also bound more weakly to other RFGs. APBB1IP, for example, also interacted with R-Ras and R-Ras3. PLCɛ, on the other hand, could also interact with R-Ras3 and, weakly, with TC21 but not with R-Ras. AF6 bound most strongly to N-Ras and K-Ras and considerably more weakly, but detectably, to the other RFGs. Weak binding of R-Ras3, but of no other RFG, was also detected for P-CIP1. Rassf4/ADO37 could interact with all of the RFGs except TC21. In some cases, like those for Rassf1 and Rassf2, R-Ras3 displayed the strongest interaction. Rassf1 also bound to N-Ras and K-Ras, whereas Rassf2 interacted more weakly with R-Ras, TC21, N-Ras, and K-Ras. In conclusion, there was clear promiscuity in the interactions between RFGs and RA domain-containing proteins, with several RFGs binding to the same proteins. There were, however, overlapping but distinct sets of interactions, with different RFGs binding to some RA domain-containing proteins but not others and doing so with various relative efficiencies.
FIG. 5.
Interaction of RFGs with RA domain-containing proteins. Constitutively active GST-tagged RFGs were cotransfected into 293T cells with myc-tagged RA domain-containing proteins, and interactions were measured by pulling down RFGs with glutathione beads and detecting bound proteins with anti-myc antibodies in Western blots. All proteins were full length except those shown with asterisks (APBB1IP/RIAM [amino acids 1 to 261] and PLCɛ [amino acids 1383 to 2303]).
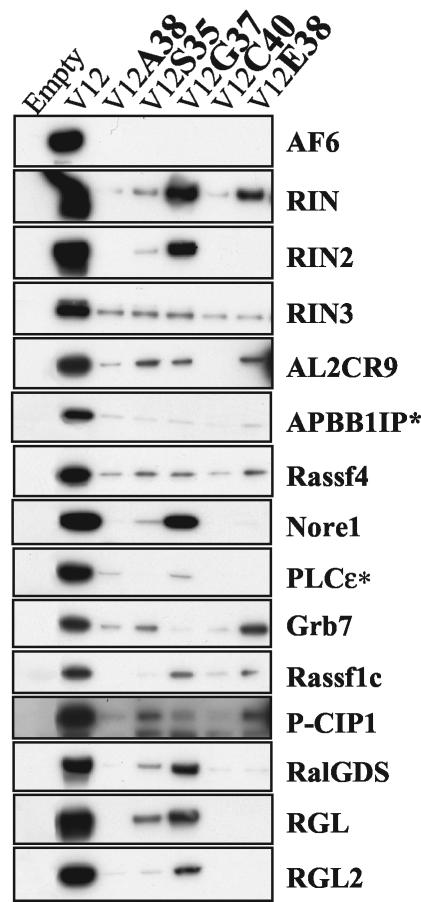
We next analyzed whether the interactions required the effector domain of Ras and determined their differential sensitivity to known partial-loss-of-function Ras mutants. As shown in Fig. 6, the interactions of RFGs with all of the RA domain-containing proteins tested were severely impaired by at least some of the mutations in the effector domain of Ras. This finding highlights the specificity of the interactions and is consistent with these proteins behaving as effectors.
FIG. 6.
Interaction of H-Ras effector mutants with RA domain-containing proteins. GST-tagged H-Ras effector mutants in a V12 backbone were cotransfected into 293T cells with myc-tagged RA domain-containing proteins, and interactions were measured in GST pulldown assays and with anti-myc Western blots.
DISCUSSION
Activation of Raf kinases.
RFGs differ greatly in their abilities to regulate the three Raf isoforms. H-Ras, N-Ras, and K-Ras activate all three Raf isoforms; R-Ras3 can active Raf-1 and A-Raf, whereas TC21 and Rit are able to detectably activate only Raf-1. Raf-1 displays the more promiscuous regulation by RFGs, but the efficiencies with which the various GTPases stimulate its kinase activity vary greatly. H-Ras, N-Ras, and K-Ras have much stronger effects than TC21, R-Ras3, or Rit. We propose that the detection of these weaker effects of TC21, R-Ras3, and Rit may depend on the sensitivities of the assays used by different groups and that differences in these sensitivities may account for previously reported conflicting results.
There is a good correlation between the abilities of the various RFGs to interact with Raf-1 and A-Raf and their abilities to stimulate their kinase activities, with the strongest interactors (Ras proteins) being the strongest activators. TC21 and R-Ras3 bound weakly to B-Raf and TC21 bound weakly to A-Raf, but we could not detect the stimulation of their kinase activity. A low level of activation may, however, be below the sensitivity of our assays. Consistent with this possibility, another group has reported the activation of B-Raf by TC21 (58).
Nonetheless, it is clear that different RFGs vary greatly in their abilities to activate the three Raf isoforms, and these differences could have important physiological implications. Raf isoforms have distinct biochemical properties and could potentially have different functions (43, 51, 77). Furthermore, different magnitudes of activation of the Erk pathway are known to have strikingly different consequences (51, 63, 76). It is thus likely that the differential efficiencies with which the different RFGs activate the Erk pathway (in a Raf isoform-specific way) may have critical physiological consequences.
Surprisingly, we did not see any interaction of R-Ras or Rap1 with any of the Raf isoforms. Both R-Ras and Rap1 have previously been reported to interact with Raf-1 in vitro without being able to activate it (11, 44, 55, 67). In the case of Rap1, this fact was used to support a model in which Rap1 would function as a Ras antagonist by competing for Ras effectors. There are also reports in the literature of the inhibition of Erk activation by Rap1 that would be in agreement with this model (69). It should be noted, however, that the previously reported interactions between Rap1 and Raf1 (as well as those between R-Ras and Raf-1) were carried out by using the Raf1 RBD, not full-length Raf. We also saw binding of Rap1 and R-Ras to the Raf RBD (data not shown). However, we could not detect any interaction when the full-length proteins were expressed in cells, even under conditions of overexpression, for which we readily detected interactions of Raf-1 with other RFGs and interactions of Rap1 and R-Ras with other proteins. Other studies have also failed to detect interactions between Rap1 and full-length Raf proteins (48, 75). Our data thus argue against a model of direct competition of Rap1 and Ras for Raf-1 and is consistent with reports that Rap1 activation does not antagonize Ras-dependent Erk signaling (6, 17, 79).
Rap1 has also been reported to activate the Erk pathway in some cell types through direct activation of B-Raf (69). This finding is also controversial, however, and there are reports that Rap1 was unable to activate Erks, even in cells expressing B-Raf (6, 17). As was the case for Raf-1, we failed to detect any interaction between Rap1 and full-length B-Raf in our assays. Furthermore, whereas coexpression of the Raf isoforms results in increased phosphorylation of the endogenous Erks by RFGs in some cases, we did not detect any increase in Erk phophorylation by Rap1, even when B-Raf was overexpressed. It should be noted that we did see effects of Rap1 on the activities of other proteins, such as RalGDS or p110α. Therefore, our results argue against a role of Rap1 in directly regulating B-Raf kinase activity. It is possible, however, that Rap1 could indirectly modulate Erk activation in some cell types. Rap1 has been implicated in regulating integrin-mediated cell adhesion (5). Through changes in integrin function, Rap1 could possibly indirectly modulate Erk activation by extracellular agonists in a cell type-dependent manner.
The differences seen between results obtained with the isolated RBD and those obtained with the full-length proteins highlight the possibility that interactions between RFGs and isolated RBD fragments do not accurately mimic interactions with full-length proteins and therefore should be interpreted with caution. It is likely that effector regions other than the RBD may contribute directly or indirectly to the interaction with the GTPases. Raf-1, for example, is known to have two domains that interact with Ras, the higher-affinity RBD and the lower-affinity cysteine-rich domain (CRD) (8). It is possible that the CRD may contribute structural and/or functional specificity to the interactions with different RFGs that are lost when interactions with the RBD are analyzed alone. Consistent with this possibility, it has been proposed that the differential ability to displace 14-3-3 from conserved region 2 may account for the differences in the regulation of Raf-1 kinase activity by RFGs (37). Furthermore, the interaction with the CRD is dependent on the farnesylation of Ras (26, 41, 74). Assays in which the interactions are carried out by using RFGs that are not posttranslationally modified (in vitro interactions with bacterially expressed proteins or Y2H analysis) will overlook any contribution of posttranslational modification of the RFGs to the interactions with the effectors. Our data strongly suggest that interactions previously reported on the basis of the use of isolated RBDs or truncated proteins may not accurately reflect the differential binding specificities to the various RFGs that occur in vivo.
Similar considerations apply to interactions with other effectors. We saw, for example, detectable binding of Rap1, R-Ras, TC21, and R-Ras3 to AF6, but this binding was much weaker than that of Ras proteins. This finding is in stark contrast to reports in the literature in which Rap1 was reported to bind with higher affinity than Ras to the RBD of AF6 both in vitro and in the Y2H assay (3, 38). Issues concerning RBD versus full-length, proper posttranslational processing and/or subcellular localization may account for these differences. Interestingly, AF6, like Raf-1, has two RBD domains at the N terminus. Ras has been reported to bind to the first, but not the second, RA domain of AF6 in the Y2H assay (3), although a lower-affinity interaction may have been below the sensitivity of the assay or lost because of a lack of posttranslational processing. PLCɛ also has a second low-affinity RA domain that is required for the activation of PLC activity by Ras (29). A similar requirement for dual RBDs has been reported for Ras activation of yeast adenylyl cyclase (33, 66). Interestingly, in this case, the second RBD does not lie entirely within the adenylyl cyclase but is created by the binding of a second protein, adenylyl cyclase-associated protein, to adenylyl cyclase.
It is possible that interaction with a second RBD may be a common feature conserved among RFG effectors, with a second lower-affinity site being involved in modulating specificity to different RFGs and dependent on posttranslational processing of the RFG. This second RBD may lie within the effector protein (e.g., Rafs, AF6, and PLCɛ) or could potentially also be provided through binding to other molecules (like adenylyl cyclase-associated protein and adenylate cyclase in yeast). Also, in the case of p110γ, which has been cocrystallized in a complex with Ras, Ras was shown to establish direct contact with regions other than the RBD (49).
Activation of RalGEFs.
RFGs showed considerable promiscuity in their interactions with the RalGEFs tested; all of the RFGs bound to all three RalGEFs. H-Ras, N-Ras, K-Ras, R-Ras, TC21, R-Ras3, and Rit all activated the RalGEFs, although with some differences in the magnitude of the activation. It is clear, however, that interaction even with the full-length effector does not necessarily directly correlate with the ability to stimulate the enzymatic activity of the RalGEF. We observed that the binding of Rap1 and Rap2 to RalGDS was stronger than that of the other RFGs, but Rap1 had a smaller effect on RalGDS activity than other RFGs did, whereas Rap2 had no effect at all. We sometimes saw a barely detectable effect of Rap1 on RGL. This observation raises the possibility that we may be at the limit of sensitivity of our assay and that experimental considerations should be borne in mind. For example, cell type-dependent localizations of the GTPases in discrete subcellular compartments may stimulate the activation of the effector pathway in localized cellular regions, and these spatially restricted effects may be missed when the readout for the activation of the pathway (i.e., Ral-GTP formation) measures the total cellular pool of Ral. It is possible that the small effects observed on the total Ral protein may be greatly amplified in localized subcompartments. It is also possible that specific RFG-RalGEF interactions may differentially target RalA versus RalB.
It is worth noting, for example, that Rap1 localization seems to be remarkably diverse. Rap1 has been detected at the plasma membrane, in the Golgi apparatus, in the perinuclear region, and in the endocytic-phagocytic and exocytic vesicles in different cell types (6). It could be speculated that the ability of Rap1 (and other RFGs) to stimulate Ral-GTP formation upon binding and recruitment of RalGEFs will depend on the colocalization of Rap1 with Ral, highlighting the importance of cell type-dependent spatial considerations (47).
It is possible that some specificity in the activation of RalGEFs by RFGs may be provided by the putative differential compartmentalization of the various RFGs. Ral proteins have been implicated in functions as diverse as cell cycle control through the regulation of cyclin D and p27 levels and membrane trafficking events both endocytic and exocytic in nature (18). It is tempting to speculate that spatially restricted pools of Ral may be differentially activated by the various RFGs to carry out selective functions.
Activation of PI3Ks.
Perhaps the most striking isoform-specific differences were seen in the regulation of class I PI3Ks. H-Ras, N-Ras, K-Ras, R-Ras, TC21, and R-Ras3 were equally potent in activating the p110α and p110γ isoforms. Rit had a more modest, but significant, effect on p110α and p110γ activity, whereas Rap1 could modestly stimulate p110α. Strikingly, only R-Ras and TC21 were able to activate the p110δ isoform. We failed to see any effect of any of the RFGs tested on the activity of p110β. We cannot rule out the possibility that this result is due to the presence of some inhibitory signal specific to p110β that may be present in the cell system used. p110β immunoprecipitates, though, have readily detectable lipid kinase activity in vitro. Alternatively, p110β may be regulated by different mechanisms independent of RFGs. In support of this possibility, there are reports that p110β (but not p110α or p110δ) can be activated by Gβγ subunits in vitro (34, 42). p110β has also been shown to directly interact with Rab5 (9), suggesting that GTPases of a family other than the Ras family may regulate the p110β isoform.
The different class I PI3K isoforms may have distinct cell type-specific functions, as suggested by the different phenotypes of knockout mice and by microinjection studies with neutralizing antibodies (28). The remarkable effector isoform specificity displayed by RFGs raises several important considerations. For example, the expression profile of the various isoforms will play a critical role in both the physiological role and the oncogenic properties of the different RFGs in any given cell type; the consequences of activation of RFGs in response to extracellular signals or by mutation will be different, depending on whether the cell expresses the α, β, δ, or γ p110 isoform, Raf-1, B-Raf, A-Raf, or any combination thereof. Considering the strong selection for PI3K activation in human cancer and the fact that the PI3K pathway has been found to be upregulated by various means in different tumor types (e.g., mutations in Ras and PTEN, overexpression of p110α and protein kinase B [28]), it will be interesting to address the possibility that tumors arising from cell types expressing p110δ may have a stronger predisposition to have acquired activating mutations in R-Ras and/or TC21 genes.
Our findings on the remarkable differential specificity of RFGs for the various p110 isoforms also highlight the importance of designing and using isoform-specific inhibitors, a point that is of special relevance because of the pleiotropic functions of PI3Ks. In addition to their roles in cell proliferation, survival, and migration, which make them attractive targets for inhibition in the treatment of cancer, PI3Ks also play roles in the metabolic functions of insulin, in the regulation of endothelial homeostasis, and in inflammatory and immune responses, among other processes (28). Based on our observations that the Ras proteins activated p110α but not p110β or p110δ, we propose that isoform-specific inhibitors of the p110α isoform would be ideal candidates for the inhibition of the PI3K pathway in tumor cells harboring Ras mutations, avoiding any potentially toxic effects of the unnecessary inhibition of p110β and p110δ.
Interaction with other effectors.
In addition to regulating the Raf, PI3K, and RalGEF pathways, it is clear that RFGs have many other effectors. Some were already known to interact with Ras and other RFGs. In addition, there is an incredibly diverse array of proteins with RBDs and thus the potential to behave as effectors of RFGs that have not been characterized yet. In this study, we have analyzed the ability of many proteins containing RA domains to interact with the various RFGs and have identified several of them as novel putative effectors of RFGs. All of the RA domain-containing proteins we have tested in this study interact with at least some RFGs, although with various strengths. We therefore predict that other RA domain-containing proteins that have not been tested will also bind to at least some RFGs.
It is likely that the interactions between the RFGs and their many effectors reported in this study are subject to a more complex set of factors that still need to be characterized. For example, posttranslational modifications of the effectors or interaction with other proteins in response to extracellular agonists may differentially modulate their interactions with the various RFGs. This possibility is illustrated by the recent report that phosphorylation of AF6 by Bcr kinase increases the binding of Ras to AF6 (54). However, it is clear that in the same cell type, under the same conditions, there is a differential pattern of interactions among RFGs and their various effectors, with some, but not all, RFGs binding to some effectors but not others.
Partial-loss-of-function mutants of Ras have been characterized for their ability to discriminate between the Raf, PI3K, and RalGEF pathways and have proved to be useful tools in elucidating the contribution of these pathways to various biological effects of Ras in different cell types. With the realization that RFGs interact with many other proteins, it is clear, however, that the ability of these mutants to interact with other effectors needs to be taken into consideration when interpretations of their biological effects are made. Recently, for example, the E37G Ras mutant (but not the Y40C and T35D mutants or constitutively active PI3K or Raf) was shown to selectively induce anchorage-independent growth in human cells (22). This effect was only partially mimicked by a constitutively active RalGEF and only partially blocked by a dominant-negative Ral, suggesting that the E37G mutant regulates other effector pathways that are making important contributions to malignant transformation of human cells. As previously reported by other groups (24, 29, 30, 49) and shown in this study, the E37G mutant does indeed interact with several other effectors. This finding further highlights the importance of understanding the functions of all of the new effector molecules and their contributions to the functions of the RFGs.
Little is known about most RA domain-containing proteins, and more work is thus needed to understand their contributions to the functions of the various RFGs. RIN1 has been reported to act as a GEF for Rab5 and to regulate endocytosis (72). Some RFGs may also regulate the activation of Rap1 and Rap2 through PDZGEF-1 and PDZGEF-2. R-Ras3 has been previously reported to activate PDZGEF2 activity (19). We found that PDZGEF1 interacts most strongly with N-Ras and K-Ras. This finding suggests that Rap1 proteins, originally proposed to function by antagonizing Ras function, may actually act downstream of Ras under some circumstances.
It is becoming apparent that RFGs can also interact with a large number of proteins without any catalytic domain. These proteins may function as scaffolding proteins, localizing active RFGs to specialized protein complexes. AF6, for example, interacts with cell-cell adhesion molecules like ZO-1 and JAM and may localize RFG signaling to sites of cell-cell contact. The Grb7/10/14 family of adaptors is known to interact with many receptor tyrosine kinases and other signaling proteins (23). Many other adaptor molecules, such as Grb2 and Shc, are known to function upstream of RFGs, linking growth factor receptors to GEFs for RFGs. The ability of RFGs to interact directly with Grb7 suggests the striking possibility that the Grb7/10/14 adaptor proteins may function downstream of RFGs.
Some splice variants of Rassf1 and Nore1/Rassf5 are selectively downregulated by promoter methylation in human tumors, and their reexpression inhibits cell growth (13, 25). This finding suggests the remarkable possibility that RFGs may directly regulate proteins with tumor suppressor properties. We have now identified several other proteins as putative novel effectors of RFGs, some not yet described and of as yet unknown function. The future identification of proteins that interact with these new scaffold-type proteins will shed more light on their function.
Further work is needed to fully understand and characterize this exciting and intriguing new level of complexity in the effector pathways regulated by RFGs. A picture emerges in which there is promiscuity when the interactions between RFGs and individual effectors are considered but specificity when the whole array of effectors is considered; each RFG has its individual blueprint of effector interactions. We propose that specificity in the signaling properties and biological functions of the various RFGs arises from the specific combination of effector pathways they regulate in each cell type. The activation of any RFG by extracellular signals (or aberrantly in cancer) will lead to the activation of a specific set of effector pathways, depending on the expression of the various effector isoforms they can regulate in the cell in question. Furthermore, the consequences of activation of any given pathway should in turn be considered in the context of the wide but specific set of effector pathways being regulated at the same time.
A better understanding of the functions of the different effectors is required to assess how these effectors may contribute to the many cellular functions of the various RFGs in different cell types. It will also likely have important implications both in the identification of new targets of therapeutic intervention and in the design and use of isoform-specific drugs for the treatment of human diseases such as cancer.
Acknowledgments
We thank Deborah Morrison, Takashi Endo, Paul Worley, Doug Anders, John Colicelli, Kozo Kaibuchi, and Marcel Spaargaren for constructs and Benoit Bilanges, Jay Gump, Clodagh O'Shea, David Stokoe, and Martin McMahon for critical reading of the manuscript.
REFERENCES
- 1.Alessi, D. R., P. Cohen, A. Ashworth, S. Cowley, S. J. Leevers, and C. J. Marshall. 1995. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 255:279-290. [DOI] [PubMed] [Google Scholar]
- 2.Barker, K. T., and M. R. Crompton. 1998. Ras-related TC21 is activated by mutation in a breast cancer cell line, but infrequently in breast carcinomas in vivo. Br J. Cancer 78:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettner, B., C. Herrmann, and L. Van Aelst. 2001. Ras and Rap1 interaction with AF-6 effector target. Methods Enzymol. 332:151-168. [DOI] [PubMed] [Google Scholar]
- 4.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 5.Bos, J. L., K. De Bruyn, J. Enserink, B. Kuiperij, S. Rangarajan, H. Rehmann, J. Riedl, J. De Rooij, F. Van Mansfeld, and F. Zwartkruis. 2003. The role of Rap1 in integrin-mediated cell adhesion. Biochem. Soc. Trans. 31:83-86. [DOI] [PubMed] [Google Scholar]
- 6.Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2:369-377. [DOI] [PubMed] [Google Scholar]
- 7.Chan, A. M., T. Miki, K. A. Meyers, and S. A. Aaronson. 1994. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc. Natl. Acad. Sci. USA 91:7558-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong, H., H. G. Vikis, and K. L. Guan. 2003. Mechanisms of regulating the Raf kinase family. Cell Signal 15:463-469. [DOI] [PubMed] [Google Scholar]
- 9.Christoforidis, S., M. Miaczynska, K. Ashman, M. Wilm, L. Zhao, S. C. Yip, M. D. Waterfield, J. M. Backer, and M. Zerial. 1999. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1:249-252. [DOI] [PubMed] [Google Scholar]
- 10.Clark, G. J., M. S. Kinch, T. M. Gilmer, K. Burridge, and C. J. Der. 1996. Overexpression of the Ras-related TC21/R-Ras2 protein may contribute to the development of human breast cancers. Oncogene 12:169-176. [PubMed] [Google Scholar]
- 11.Cook, S. J., B. Rubinfeld, I. Albert, and F. McCormick. 1993. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 12:3475-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, A. D., T. R. Brtva, D. G. Lowe, and C. J. Der. 1994. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene 9:3281-3288. [PubMed] [Google Scholar]
- 13.Dammann, R., C. Li, J. H. Yoon, P. L. Chin, S. Bates, and G. P. Pfeifer. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 25:315-319. [DOI] [PubMed] [Google Scholar]
- 14.Ehrhardt, A., G. R. Ehrhardt, X. Guo, and J. W. Schrader. 2002. Ras and relatives—job sharing and networking keep an old family together. Exp. Hematol. 30:1089-1106. [DOI] [PubMed] [Google Scholar]
- 15.Ehrhardt, G. R., C. Korherr, J. S. Wieler, M. Knaus, and J. W. Schrader. 2001. A novel potential effector of M-Ras and p21 Ras negatively regulates p21 Ras-mediated gene induction and cell growth. Oncogene 20:188-197. [DOI] [PubMed] [Google Scholar]
- 16.Ehrhardt, G. R., K. B. Leslie, F. Lee, J. S. Wieler, and J. W. Schrader. 1999. M-Ras, a widely expressed 29-kD homologue of p21 Ras: expression of a constitutively active mutant results in factor-independent growth of an interleukin-3-dependent cell line. Blood 94:2433-2444. [PubMed] [Google Scholar]
- 17.Enserink, J. M., A. E. Christensen, J. de Rooij, M. van Triest, F. Schwede, H. G. Genieser, S. O. Doskeland, J. L. Blank, and J. L. Bos. 2002. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat. Cell Biol. 4:901-906. [DOI] [PubMed] [Google Scholar]
- 18.Feig, L. A. 2003. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 13:419-425. [DOI] [PubMed] [Google Scholar]
- 18a.Frech, M., I. Schlichting, A. Wittinghofer, and P. Chardin. 1990. Guanine nucleotide binding properties of the mammalian RalA protein produced in Escherichia coli. J. Biol. Chem. 265:6353-6359. [PubMed] [Google Scholar]
- 19.Gao, X., T. Satoh, Y. Liao, C. Song, C. D. Hu, K. Kariya Ki, and T. Kataoka. 2001. Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras. J. Biol. Chem. 276:42219-42225. [DOI] [PubMed] [Google Scholar]
- 20.Graham, S. M., A. D. Cox, G. Drivas, M. G. Rush, P. D'Eustachio, and C. J. Der. 1994. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 14:4108-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, S. M., S. M. Oldham, C. B. Martin, J. K. Drugan, I. E. Zohn, S. Campbell, and C. J. Der. 1999. TC21 and Ras share indistinguishable transforming and differentiating activities. Oncogene 18:2107-2116. [DOI] [PubMed] [Google Scholar]
- 22.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, D. C., T. L. Shen, and J. L. Guan. 2001. The Grb7 family proteins: structure, interactions with other signaling molecules and potential cellular functions. Oncogene 20:6315-6321. [DOI] [PubMed] [Google Scholar]
- 24.Han, L., D. Wong, A. Dhaka, D. Afar, M. White, W. Xie, H. Herschman, O. Witte, and J. Colicelli. 1997. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc. Natl. Acad. Sci. USA 94:4954-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesson, L., A. Dallol, J. D. Minna, E. R. Maher, and F. Latif. 2003. NORE1A, a homologue of RASSF1A tumour suppressor gene is inactivated in human cancers. Oncogene 22:947-954. [DOI] [PubMed] [Google Scholar]
- 26.Hu, C. D., K. Kariya, M. Tamada, K. Akasaka, M. Shirouzu, S. Yokoyama, and T. Kataoka. 1995. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J. Biol. Chem. 270:30274-30277. [DOI] [PubMed] [Google Scholar]
- 27.Huang, Y., R. Saez, L. Chao, E. Santos, S. A. Aaronson, and A. M. Chan. 1995. A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene 11:1255-1260. [PubMed] [Google Scholar]
- 28.Katso, R., K. Okkenhaug, K. Ahmadi, S. White, J. Timms, and M. D. Waterfield. 2001. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 17:615-675. [DOI] [PubMed] [Google Scholar]
- 29.Kelley, G. G., S. E. Reks, J. M. Ondrako, and A. V. Smrcka. 2001. Phospholipase Cε: a novel Ras effector. EMBO J. 20:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khokhlatchev, A., S. Rabizadeh, R. Xavier, M. Nedwidek, T. Chen, X. F. Zhang, B. Seed, and J. Avruch. 2002. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12:253-265. [DOI] [PubMed] [Google Scholar]
- 31.Kimmelman, A., T. Tolkacheva, M. V. Lorenzi, M. Osada, and A. M. Chan. 1997. Identification and characterization of R-ras3: a novel member of the RAS gene family with a nonubiquitous pattern of tissue distribution. Oncogene 15:2675-2685. [DOI] [PubMed] [Google Scholar]
- 32.Kimmelman, A. C., N. Nunez Rodriguez, and A. M. Chan. 2002. R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade. Mol. Cell. Biol. 22:5946-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda, Y., N. Suzuki, and T. Kataoka. 1993. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science 259:683-686. [DOI] [PubMed] [Google Scholar]
- 34.Kurosu, H., T. Maehama, T. Okada, T. Yamamoto, S. Hoshino, Y. Fukui, M. Ui, O. Hazeki, and T. Katada. 1997. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110beta is synergistically activated by the βγ subunits of G proteins and phosphotyrosyl peptide. J. Biol. Chem. 272:24252-24256. [DOI] [PubMed] [Google Scholar]
- 35.Lambert, J. M., Q. T. Lambert, G. W. Reuther, A. Malliri, D. P. Siderovski, J. Sondek, J. G. Collard, and C. J. Der. 2002. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat. Cell Biol. 4:621-625. [DOI] [PubMed] [Google Scholar]
- 36.Lee, C. H., N. G. Della, C. E. Chew, and D. J. Zack. 1996. Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of ras proteins. J. Neurosci. 16:6784-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Light, Y., H. Paterson, and R. Marais. 2002. 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell. Biol. 22:4984-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linnemann, T., M. Geyer, B. K. Jaitner, C. Block, H. R. Kalbitzer, A. Wittinghofer, and C. Herrmann. 1999. Thermodynamic and kinetic characterization of the interaction between the Ras binding domain of AF6 and members of the Ras subfamily. J. Biol. Chem. 274:13556-13562. [DOI] [PubMed] [Google Scholar]
- 39.Lowe, D. G., and D. V. Goeddel. 1987. Heterologous expression and characterization of the human R-ras gene product. Mol. Cell. Biol. 7:2845-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowy, D. R., B. M. Willumsen. 1993. Function and regulation of RAS. Annu. Rev. Biochem. 62:851-891. [DOI] [PubMed] [Google Scholar]
- 41.Luo, Z., B. Diaz, M. S. Marshall, and J. Avruch. 1997. An intact Raf zinc finger is required for optimal binding to processed Ras and for ras-dependent Raf activation in situ. Mol. Cell. Biol. 17:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier, U., A. Babich, and B. Nurnberg. 1999. Roles of non-catalytic subunits in Gβγ-induced activation of class I phosphoinositide 3-kinase isoforms β and γ. J. Biol. Chem. 274:29311-29317. [DOI] [PubMed] [Google Scholar]
- 43.Marais, R., Y. Light, H. F. Paterson, C. S. Mason, and C. J. Marshall. 1997. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J. Biol. Chem. 272:4378-4383. [DOI] [PubMed] [Google Scholar]
- 44.Marte, B. M., P. Rodriguez-Viciana, S. Wennstrom, P. H. Warne, and J. Downward. 1997. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 7:63-70. [DOI] [PubMed] [Google Scholar]
- 45.Movilla, N., P. Crespo, and X. R. Bustelo. 1999. Signal transduction elements of TC21, an oncogenic member of the R-Ras subfamily of GTP-binding proteins. Oncogene 18:5860-5869. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, G. A., S. M. Graham, S. Morita, S. E. Reks, K. Rogers-Graham, A. Vojtek, G. G. Kelley, and C. J. Der. 2002. Involvement of phosphatidylinositol 3-kinase, but not RalGDS, in TC21/R-Ras2-mediated transformation. J. Biol. Chem. 277:9966-9975. [DOI] [PubMed] [Google Scholar]
- 47.Ohba, Y., K. Kurokawa, and M. Matsuda. 2003. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortiz-Vega, S., A. Khokhlatchev, M. Nedwidek, X. F. Zhang, R. Dammann, G. P. Pfeifer, and J. Avruch. 2002. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 21:1381-1390. [DOI] [PubMed] [Google Scholar]
- 49.Pacold, M. E., S. Suire, O. Perisic, S. Lara-Gonzalez, C. T. Davis, E. H. Walker, P. T. Hawkins, L. Stephens, J. F. Eccleston, and R. L. Williams. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103:931-943. [DOI] [PubMed] [Google Scholar]
- 50.Ponting, C. P., and D. R. Benjamin. 1996. A novel family of Ras-binding domains. Trends Biochem. Sci. 21:422-425. [DOI] [PubMed] [Google Scholar]
- 51.Pritchard, C. A., M. L. Samuels, E. Bosch, and M. McMahon. 1995. Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol. Cell. Biol. 15:6430-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quilliam, L. A., A. F. Castro, K. S. Rogers-Graham, C. B. Martin, C. J. Der, and C. Bi. 1999. M-Ras/R-Ras3, a transforming ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J. Biol. Chem. 274:23850-23857. [DOI] [PubMed] [Google Scholar]
- 53.Quilliam, L. A., J. F. Rebhun, and A. F. Castro. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71:391-444. [DOI] [PubMed] [Google Scholar]
- 54.Radziwill, G., R. A. Erdmann, U. Margelisch, and K. Moelling. 2003. The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Mol. Cell. Biol. 13:4663-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rey, I., P. Taylor-Harris, H. van Erp, and A. Hall. 1994. R-ras interacts with rasGAP, neurofibromin and c-raf but does not regulate cell growth or differentiation. Oncogene 9:685-692. [PubMed] [Google Scholar]
- 56.Rodriguez-Viciana, P., and J. Downward. 2001. Ras activation of phosphatidylinositol 3-kinase and Akt. Methods Enzymol. 333:37-44. [DOI] [PubMed] [Google Scholar]
- 57.Rong, R., Q. He, Y. Liu, M. S. Sheikh, and Y. Huang. 2002. TC21 mediates transformation and cell survival via activation of phosphatidylinositol 3-kinase/Akt and NF-κB signaling pathway. Oncogene 21:1062-1070. [DOI] [PubMed] [Google Scholar]
- 58.Rosario, M., H. F. Paterson, and C. J. Marshall. 1999. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 18:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosario, M., H. F. Paterson, and C. J. Marshall. 2001. Activation of the Ral and phosphatidylinositol 3′ kinase signaling pathways by the ras-related protein TC21. Mol. Cell. Biol. 21:3750-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rusyn, E. V., E. R. Reynolds, H. Shao, T. M. Grana, T. O. Chan, D. A. Andres, and A. D. Cox. 2000. Rit, a non-lipid-modified Ras-related protein, transforms NIH3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways. Oncogene 19:4685-4694. [DOI] [PubMed] [Google Scholar]
- 61.Saez, R., A. M. Chan, T. Miki, and S. A. Aaronson. 1994. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene 9:2977-2982. [PubMed] [Google Scholar]
- 62.Self, A. J., E. Caron, H. F. Paterson, and A. Hall. 2001. Analysis of R-Ras signalling pathways. J. Cell Sci. 114:1357-1366. [DOI] [PubMed] [Google Scholar]
- 63.Sewing, A., B. Wiseman, A. C. Lloyd, and H. Land. 1997. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol. Cell. Biol. 17:5588-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao, H., and D. A. Andres. 2000. A novel RalGEF-like protein, RGL3, as a candidate effector for Rit and Ras. J. Biol. Chem. 275:26914-26924. [DOI] [PubMed] [Google Scholar]
- 65.Shao, H., K. Kadono-Okuda, B. S. Finlin, and D. A. Andres. 1999. Biochemical characterization of the Ras-related GTPases Rit and Rin. Arch. Biochem. Biophys. 371:207-219. [DOI] [PubMed] [Google Scholar]
- 66.Shima, F., T. Okada, M. Kido, H. Sen, Y. Tanaka, M. Tamada, C. D. Hu, Y. Yamawaki-Kataoka, K. Kariya, and T. Kataoka. 2000. Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation. Mol. Cell. Biol. 20:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spaargaren, M., G. A. Martin, F. McCormick, M. J. Fernandez-Sarabia, and J. R. Bischoff. 1994. The Ras-related protein R-ras interacts directly with Raf-1 in a GTP-dependent manner. Biochem. J. 300:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spencer, M. L., H. Shao, and D. A. Andres. 2002. Induction of neurite extension and survival in pheochromocytoma cells by the Rit GTPase. J. Biol. Chem. 277:20160-20168. [DOI] [PubMed] [Google Scholar]
- 69.Stork, P. J. 2003. Does Rap1 deserve a bad rap? Trends Biochem. Sci. 28:267-275. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki, J., Y. Kaziro, and H. Koide. 2000. Positive regulation of skeletal myogenesis by R-Ras. Oncogene 19:1138-1146. [DOI] [PubMed] [Google Scholar]
- 71.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 72.Tall, G. G., M. A. Barbieri, P. D. Stahl, and B. F. Horazdovsky. 2001. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1:73-82. [DOI] [PubMed] [Google Scholar]
- 73.van Triest, M., J. de Rooij, and J. L. Bos. 2001. Measurement of GTP-bound Ras-like GTPases by activation-specific probes. Methods Enzymol. 333:343-348. [DOI] [PubMed] [Google Scholar]
- 74.Williams, J. G., J. K. Drugan, G. S. Yi, G. J. Clark, C. J. Der, and S. L. Campbell. 2000. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 275:22172-22179. [DOI] [PubMed] [Google Scholar]
- 75.Winkler, D. G., J. C. Johnson, J. A. Cooper, and A. B. Vojtek. 1997. Identification and characterization of mutations in Ha-Ras that selectively decrease binding to cRaf-1. J. Biol. Chem. 272:24402-24409. [DOI] [PubMed] [Google Scholar]
- 76.Woods, D., D. Parry, H. Cherwinski, E. Bosch, E. Lees, and M. McMahon. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 17:5598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, X., S. J. Noh, G. Zhou, J. E. Dixon, and K. L. Guan. 1996. Selective activation of MEK1 but not MEK2 by A-Raf from epidermal growth factor-stimulated Hela cells. J. Biol. Chem. 271:3265-3271. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, Z., K. Vuori, H. Wang, J. C. Reed, and E. Ruoslahti. 1996. Integrin activation by R-ras. Cell 85:61-69. [DOI] [PubMed] [Google Scholar]
- 79.Zwartkruis, F. J., R. M. Wolthuis, N. M. Nabben, B. Franke, and J. L. Bos. 1998. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J. 17:5905-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]