Abstract
Parent-of-origin-specific expression of the mouse insulin-like growth factor 2 gene (Igf2) and the closely linked H19 gene located on distal chromosome 7 is regulated by a 2.4-kb imprinting control region (ICR) located upstream of the H19 gene. In somatic cells, the maternally and paternally derived ICRs are hypo- and hypermethylated, respectively, with the former binding the insulator protein CCCTC-binding factor (CTCF) and acting to block access of enhancers to the Igf2 promoter. Here we report on a detailed in vivo footprinting analysis—using ligation-mediated PCR combined with in vivo dimethyl sulfate, DNase I, or UV treatment—of ICR sequences located outside of the CTCF binding domains. In mouse primary embryo fibroblasts carrying only maternal or paternal copies of distal chromosome 7, we have identified five prominent footprints specific to the maternal ICR. Each of the five footprinted areas contains at least two nuclear hormone receptor hexad binding sites arranged with irregular spacing. When combined with fibroblast nuclear extracts, these sequences interact with complexes containing retinoic X receptor alpha and estrogen receptor beta. More significantly, the footprint sequences bind nuclear hormone receptor complexes in male, but not female, germ cell extracts purified from fetuses at a developmental stage corresponding to the time of establishment of differential ICR methylation. These data are consistent with the possibility that nuclear hormone receptor complexes participate in the establishment of differential ICR methylation imprinting in the germ line.
Imprinted genes are a small subset of genes that are expressed from only one of the two alleles, according to parental origin. The two alleles of imprinted genes exhibit differential epigenetic states in the same cell such that one is silenced and the other is active. Most imprinted genes are associated with differentially methylated DNA regions (DMRs) (20). Disruption of DNA methylation by gene targeting of DNA methyltransferases results in loss of monoallelic expression (16). Primary DMRs are those DMRs inherited from the gametes, and their methylation may constitute the epigenetic mark that transmits imprinting information from the gamete to the embryo (23, 29, 30, 43, 44). It is important to identify these DMRs and to understand how the methylation marks are erased and established differentially in the germ lines.
Perhaps the best-studied ICR is found next to the imprinted genes insulin-like growth factor 2 (Igf2), an embryonic mitogen, and H19, which produces an untranslated RNA of unknown function. In the mouse, these genes are located 80 kb apart on distal chromosome 7. They are coordinately expressed in tissues of mesoderm and endoderm origin due to the sharing of enhancers located downstream (3′) of the H19 gene (2, 15, 48). The parent-of-origin-specific expression of the two genes is regulated by an imprinting control region (ICR) located at kb −2 to −4.4 relative to the transcription start site of the H19 gene (14, 26, 41). It has been established that the ICR acts as a chromatin insulator on the maternal chromosome through the binding of the zinc finger protein CCCTC-binding factor (CTCF) (3, 4, 8, 11, 32, 34). Binding of CTCF prevents activation of the maternal Igf2 promoter by the enhancers located downstream of H19.
On the paternal chromosome, the ICR sequence is highly methylated at CpG sequences, is not associated with CTCF, and lacks insulator activity. The paternal Igf2 promoter is therefore able to access the enhancers and is active. The hypermethylated paternal ICR, while lacking insulator activity, inactivates the H19 promoter in cis during early development (32, 41). As this epigenetic information is inherited from sperm, it is likely that this function is dependent on the methylation in the ICR.
The germ line-specific processes that determine differential ICR methylation, which could be viewed as equivalent to the imprinting mechanisms per se, are unknown. It appears that these processes are entirely separate from the later somatic ICR functions of chromatin insulation and H19 promoter silencing. When the CTCF sites were inactivated by site-directed mutagenesis in the mouse, the mutant ICR lacked enhancer-blocking activity and the expression of Igf2 was activated on the mutant maternal chromosome (24, 27, 39a). However, the mutant ICRs were not methylated in ovulated oocytes and blastocysts after subtle mutagenesis of the CTCF binding sites (27) or in oogonia and oocytes after more drastic mutagenesis of the same sites (39a), indicating that binding of CTCF is not required to establish an unmethylated ICR during oogenesis. Upon paternal transmission, the mutant ICR becomes methylated as usual, indicating also that CTCF is not required for de novo methylation of the ICR in the paternal germ line. In addition to its independence of CTCF, the acquisition of differential ICR methylation is a function autonomous to this region, at least when the ICR is moved to another location within the Igf2/H19 domain (10).
The sequences responsible for differential DNA methylation of the ICR occurring in the male and female germ lines are unknown. A major hurdle is in the limited amount of cellular material that can be obtained from these lineages. To gain insight into the mechanisms involved, therefore, we have carried out maternal- and paternal-chromosome-specific in vivo footprinting studies over the 2.4-kb ICR in somatic cells and used extracts prepared from purified male or female fetal germ cells to test whether these new footprint sequences bind to protein factors. In addition to the four CTCF binding sites previously identified (34), we discovered five prominent in vivo footprints on the maternal chromosome. Further, we show that the footprint sequences interact with nuclear hormone receptors (NHRs) in the male, but not in the female, germ line.
MATERIALS AND METHODS
In vivo footprinting.
Mouse primary embryo fibroblasts (PEFs) were derived by a standard method (9) from euploid 13.5-day-postcoitum (13.5-dpc) mouse fetuses which possessed two maternal or paternal copies (or duplication) of distal chromosome 7 (MatDup.d7 or PatDup.d7, respectively). They are derived from an intercross of mice heterozygous for the reciprocal translocation T(7;15)9H and are identified by homozygosity for the albino mutation (Tyrc) (28). Cells were grown in cultures in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 10−4 M β-mercaptoethanol, and nonessential amino acids, l-glutamine, and antibiotics at standard concentrations. Confluent 15-cm-diameter plates provide about 15 to 35 million PEFs. Early passages (up to passage 4) were used in this study.
DNase I treatment was done essentially as described before (25). The cells were rinsed with Dulbecco's phosphate-buffered saline (DPBS) without calcium and magnesium (DPBS−) and permeabilized with 0.05 mg of lysolecithin/ml in 10 ml of solution I (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2) for 2 min. The cells were washed with solution I, and then solution II (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 2 mM CaCl2) containing 10 μg of DNase I (catalog no. 104132; Boehringer Mannheim)/ml was added. The cells were then incubated for 5 min at room temperature to obtain an optimal nicking frequency of one nick every 200 to 800 nucleotides (25). This was assessed by alkaline gel electrophoresis after DNA purification and before ligation-mediated PCR (LMPCR). Cells were trypsinized, washed with DPBS−, pelleted, and resuspended in 2.5 ml of buffer B (150 mM NaCl, 5 mM EDTA, pH 7.8). Then, 2.5 ml of buffer C (20 mM Tris HCl [pH 8], 20 mM NaCl, 20 mM EDTA, 1% sodium dodecyl sulfate) containing 1.2 mg of proteinase K/ml was added and the sample was incubated for 1 h at 37°C. The DNA was phenol-chloroform extracted and ethanol precipitated. As a control, 40 μg of genomic DNA isolated from untreated PEFs as described above was mixed with 0.3 μg of DNase I/ml in a 400-μl reaction volume and incubated at room temperature for 5 min in 40 mM Tris-Cl (pH 7.7)-10 mM NaCl-6 mM MgCl2.
Dimethyl sulfate (DMS) treatment was done as described previously (38). The medium was changed to serum-free medium containing 0.2% DMS, and cells were incubated for 5 min at room temperature. The action of DMS was stopped by washing the cells twice with 40 ml of ice-cold DPBS−. The cells were trypsinized and washed with DPBS− followed by centrifugation at 250 × g for 3 min to pellet the cells. The cell pellet was suspended in 2.5 ml of buffer B. Then, 2.5 ml of buffer C containing 1.2 mg of proteinase K/ml was added and incubated at 37°C for 3 h and the DNA was isolated by phenol-chloroform extraction and ethanol precipitation. As a control, genomic DNA was isolated from untreated PEFs. DNA (50 μg) was treated with 0.2% DMS in 200 μl of DMS buffer (50 mM Na-cacodylate, 1 mM EDTA, pH 8) for 5 min at room temperature. The reaction was stopped with 50 μl of 1.5 M sodium acetate (pH 7)-1 M β-mercaptoethanol. The DNA was precipitated with 750 μl of ethanol, dissolved, precipitated again, and dissolved in 50 μl of Tris-EDTA (TE) buffer.
For AlkA digestion, 10 μg of DMS-modified DNA was incubated in a 100-μl reaction volume in AlkA buffer (70 mM HEPES, pH 7.5, 0.5 mM EDTA, 10 mM β-mercaptoethanol, 5% glycerol) with 10 μg of AlkA enzyme for 30 min at 37°C. Then 100 μl of 2 M piperidine was added, and the DNA was incubated at 37°C for 15 min to produce strand breaks with 5′-terminal phosphates at the resulting abasic sites. After precipitation, the DNA was washed two times with 80% ethanol, resuspended in 100 μl of water, and vacuum dried overnight in a Speed-vac concentrator (Savant Instruments). The pellet was resuspended in 10 μl of TE buffer.
For piperidine treatment, 50 μg of DMS-modified DNA was dissolved in 100 μl of 1 M piperidine and incubated at 88°C for 30 min. After transference into a new tube, the DNA was precipitated, washed two times with 80% ethanol, resuspended in 100 μl of water, and vacuum dried overnight in a Speed-vac concentrator (Savant Instruments). The pellet was resuspended in 50 μl of TE buffer.
For UV treatment (42), plates were washed with DPBS with calcium and magnesium (DPBS+). The DPBS+ was removed, leaving a 1-mm-thick layer, and the cells were irradiated with 2,400 J of UV light/m2 in a UV Stratalinker 2400 apparatus (Stratagene). Cells were trypsinized, and DNA was extracted as described for the DMS protocol. As a control, genomic DNA was extracted from untreated PEFs, dissolved at a 0.5 μg/μl concentration in TE buffer, and irradiated with 4,800 J of UV light/m2 in 10-μl drops on parafilm.
For enzymatic treatment of UV-irradiated DNA, 10 μg of DNA was incubated in a 100-μl reaction volume with T4 endonuclease V in 50 mM Tris-HCl (pH 7.6)-50 mM NaCl-1 mM EDTA-10 mM dithiothreitol (DTT)-100 μg of bovine serum albumin/ml for 1 h at 37°C. Photolyase (Pharmingen) (5 μg) was added under yellow light, and the DNA was incubated for 1 h at room temperature under black lights (42). The DNA was phenol extracted and ethanol precipitated.
Maxam-Gilbert sequencing (21) was done to serve as a position marker.
For alkaline gel electrophoresis, the size of DNA fragments generated by cleavage reactions was checked on an alkaline gel before LMPCR. A 2% agarose gel was made with 100 ml of 50 mM NaCl and a 1 mM EDTA solution presoaked overnight in the running buffer (50 mM NaOH and 1 mM EDTA). DNA (1 μg) was incubated in a 10-μl volume at room temperature for 15 min after the addition of 2 μl of 6× loading buffer (0.15% bromocresol green, 50% glycerol, 300 mM NaOH) and loaded on the gel. The gel was run in fresh running buffer at 50 V until the dye reached two-thirds of the gel length. The gel was neutralized for 30 min in 1 M Tris-HCl (pH 7.6)-1.5 M NaCl, stained for 30 min with 5 μg of ethidium bromide/ml, destained for 30 min with water, and photographed.
LMPCR was done as previously described (37) except that 21 cycles of PCR were used. Briefly, the starting material for LMPCR was total genomic DNA that had undergone a specific modification or cleavage procedure that produces single-strand breaks. In chromatin-probing experiments, these reactions are carried out inside the cell. After the cells had been subjected to the specific footprinting protocol, DNA was purified by standard methods. In the LMPCR reaction, the genomic DNA was heat denatured and a gene-specific oligonucleotide (primer 1) was annealed and used in a primer extension reaction with modified T7 DNA polymerase (Sequenase; United States Biochemicals) to create blunt ends. After the primer extension step, the double-stranded linker oligonucleotide was ligated to the blunt ends to introduce a common sequence at the 5′ ends of all sequence ladder fragments. After ligation, exponential PCR with the longer oligonucleotide of the linker (linker-primer) and a second, nested gene-specific primer (primer 2) generated a ladder of gene-specific PCR products. The length of these PCR products is determined by the initial positions of the DNA breaks in the genomic template relative to that of the gene-specific PCR primer. After PCR, the DNA fragments were separated on a sequencing gel, electroblotted onto a nylon membrane, and hybridized with a gene-specific probe to visualize the sequence ladders. Single-stranded hybridization probes were made from a premade PCR product with primer 3 by multiple runoff polymerizations with Taq polymerase.
The LMPCR primers were as follows: for the footprint 1 (FP1) lower strand, primers II1 (5′-CCAGGACTCAAAGGAACATG-3′), II2 (5′-TCAAAGGAACATGCTACATTCACACGA-3′), II3 (5′-CGAGCATCCAGGAGGCATAAGAA-3′), and JJ3 (5′-CCACGAGGTACCAGCCTAGAAAATG-3′); for the FP2 upper strand, DZS1 (5′-TGAGAACGTTTTATCAAGGACTAG-3′), DZS2 (5′-ACGTTTTATCAAGGACTAGCATGAACC-3′), DZS3 (5′-ACTAGCATGAACCCCTGGCCTC-3′), and DZ3 (5′-GTCACAAATGCCACTAGGGGGG-3′); for the FP3 lower strand, X1 (5′-CCAAAGTTCGGGTTCGC-3′), X2 (5′-CACAGCAATGTCCGAAGCCGC-3′), X3, 5′-AGCCGCTATGCCTCAGTGGTCG-3′, and Y3 (5′-CTTACCACCCCTATGAATCCCTATTTGG-3′); for the FP4 lower strand, LY1 (5′-ACTGAACCAGAGAACTTGACTCA-3′), LY2 (5′-TTGACTCATTCCCTACACAGCCCGA-3′), and LY3 (5′-CCCTACACAGCCCGAGATCGTCAG-3′); for the FP4 upper strand, NY1 (5′-CCACCACGCGGCATC-3′), NY2 (5′-GCGGCATCGTCTGTCCATTTAGCT-3′), and NY3 (5′-AATCTGCACAGCGTGGAGAGTGAAC-3′); for the FP5 lower strand, CS1 (5′-CAGGCATAGCATTCAATGATT-3′), CS2 (5′-TTCAATGATTCATAAGGGTCATGGGGT-3′), and CS3 (5′-TAAGGGTCATGGGGTGGTACAACACA-3′); and for the FP5 upper strand, GY1 (5′-CCCGCCTATAACCGATTCT-3′), GY2 (5′-GCCTATAACCGATTCTGTATTGAGTTTGGAT-3′), and GY3 (5′-TTTGGATTGAACAGATCTGGCTAGCTTG-3′).
The first primers were biotinylated.
EMSA.
Whole-cell extract for the binding assays was made from confluent plates of 13.5-dpc normal PEFs as previously described (46), with minor modifications. Cells were trypsinized and washed with cold DPBS−. Aliquots of 107 cells were suspended in 565 μl of extraction buffer (50 mM Tris HCl [pH 8.0], 500 mM KCl, 0.5 mM EDTA [pH 8.0], 0.5 mM EGTA [pH 8.9], 1% Nonidet P-40, 2 mM DTT, 1× protease inhibitor cocktail [P2714; Sigma]) and incubated on ice for 30 min. After centrifugation at 13,000 rpm with a MicroCentaur (MSE) microcentrifuge for 5 min, the supernatant was placed into a new tube and gently mixed with 10% ice-cold glycerol and small aliquots were snap frozen on dry ice. The aliquots were kept at −80°C. To prepare the probe for an electrophoretic mobility shift assay (EMSA), 10 pmol of one strand of the oligonucleotide probe was end labeled and annealed to 20 pmol of the other strand and then diluted to 10 fmol/μl (∼30,000 to 60,000 cpm/μl). For the binding reaction, 2 μl of PEF extract was mixed with 2 μl of antibody (Santa Cruz Biotechnology) or 1 μl of 500 fmol/μl of competitor in 1× binding buffer (20 mM HEPES [pH 7.6], 2 mM MgCl2, 5% glycerol, 1 mM DTT, 1% Nonidet P-40) and incubated on ice for 20 min in the presence of 0.25 μg of sheared herring sperm DNA and 0.2 μg of poly(dI-dC) (Sigma) in a 20-μl reaction volume. A total of 1 μl (10 fmol) of probe was added, the incubation was continued for another 20 min at room temperature, and the bound and unbound fractions were separated on a prerun 4% polyacrylamide gel containing 5% glycerol in 25 mM Tris-borate-EDTA for 3 h at 200 V in a cold room. After drying at 80°C, the gel was analyzed on a PhosphorImager (Molecular Dynamics).
For collection of germ cells for EMSA and reverse transcription-PCR (RT-PCR), male mice of the homozygous transgenic line B6;CBA-Tg(Pou5f1-EGFP)2Mnn (stock no. 004654; Jackson Laboratory, Bar Harbor, Maine), which express the enhanced green fluorescent protein (EGFP) reporter gene specifically in germ cells (35), were mated to wild-type CF1 females (Charles River, Wilmington, Mass.). A MoFlo flow cytometer (Cytomation, Fort Collins, Colo.) was used for flow cytometry to purify germ cells from the resulting female and male 15.5- and 18.5-dpc fetuses as previously described (35). Spermatogonia were purified from newborns by the same method, while pachytene spermatocytes of adult testes were sorted by flow cytometry on the basis of physical characteristics (the “R2” subpopulation) (19). Germ cell extracts were prepared from purified germ cells in similarity to PEF treatment except that the volumes were scaled down to have the same number of cells per volume (20,000 cells/μl) for germ cells as was used for PEFs.
For Rxrα−/− PEF extracts, Rxrα+/− mice (33) were intercrossed to obtain Rxrα−/− embryos, which were identified at dpc 13.5 by the small size of their livers. The corresponding placentas were typed by PCR. Cell extract was prepared from 2 million cells in a procedure similar to that used for normal PEFs.
The oligonucleotides used were as follows (upper strands are shown; lowercase characters signify point mutations introduced): FP1U (5′-ATTCACAAATGGCAATGCTGTGGGTCACCCAAGTTCAGTACCTCAGGGGGGTCACAAATG CCACTAGGGG-3′), FP1m (5′-ATTCACAAATGGCAATGCTGTGGaTCcCCCAAGTTCAGTACCTCAGGGaGcTCACAAATGCCACTAGGGG-3′), FP2U (5′-CTATCACCATCTATGATCCCATAGTCATGGGCTTCATGAGGCCAGGGGTTCATGCTAGTCCTTGATAAAACGTTCTCAAGAGCTATCTCAGGTATCTGACTTATAGGGTT-3), FP2m (5′-CTATCACCATCTAGATCCCATAGTCcATGGGCTTCATGAGGCCAGGGGTgCATGCTAGTCCTTGATAAAACGTTCTCAAGAGCTATCTCAGGTAgTCGACTTATAGGGTT-3′), FP3 (5′-CTTGGGGGGAGCGATTCATTCCCAGCAATATCCCAGGGTCACCCAAATAGGGATTCATAGGGGTGGTAAGATGTGTGCACCTCTGGAATGGTTCCCTTACACACTGAACCAGAGAACTTGACTCATTCCCTACAC-3′), FP3m (5′-CTTGGGGGGAGCGATgCATTCCCAGCAATATCCCAGGGTACCcCAAATAGGGATTaATAGGGGTGGTAAGATGTGTGCACCTCTGGAATGGTTCCCTTACACACTtAAgCAGAGAgCTcGACgTCTTCCCTACAC-3),FP4 (5′-GGTGACCAAAATTGCGGTTCACCTATGGCAAACTCATGGGTCACTCAGGC ATAGCATTCAATGATTCATAAGGGTCATGGGGTGGTACAACACACATTTC TTGGGTAGCT-3′), FP4m (5′-GGTGACCAAAATTGCGGaTCcCCTATGGCAAACTCATGGGTacCTCAGGCATAGCATTCAAgaATTCATAAGctTCATGGGGTGGTACAACACACATTTCTTGGGTAGCT-3′), FP5 (5′-AAGCTTTGAGTACCCCAGGTTCAACAAAGGGATCAGGCATTTGTGCACTTAGG-3′), and FP5del (5′-AAGCTTTGAGTACCCCAGGccTGTGCACTTACGG-3′).
The following NHR consensus competitors were purchased from Santa Cruz Biotechnology: RAR (DR-5) (catalog no. sc-2559); RARm (DR-5) (sc-2560); RXR (DR-1) (sc-2547); RXRm (DR-1) (sc-2548); TR (DR-4) (sc-2563); TRm (DR-4) (sc-2564); ER (sc-2585); and ERm (sc-2586). The following antibodies were purchased from Santa Cruz Biotechnology: RARα (C-20) (catalog no. sc-551); RARβ (C-19) (sc-552); RARγ (C19) (sc-550); RXRα (D-20) (sc-553); RXRβ (C-20) (sc-831); RXRγ (Y-20) (sc-555); ERα (MC-20) (sc-542); ERβ (H-150) (sc-8974); TRα1 (FL-408) (sc-772); and TRβ1 (J51) (sc-737).
ICR targeting vector.
The same gene targeting vector as previously described (39) was used except that the ICR (defined as a 2.4-kb BglII fragment) was mutagenized. A total of 57 and 107 bases were deleted from FP1 and FP4 binding sites, respectively. Before insertion into the targeting construct, the 2.4-kb BglII fragment was subcloned into the BglII site of pSL1180. Site-directed mutagenesis was done using the Transformer site-directed mutagenesis kit (Clontech). The FP1 deletion created a new EcoRV site and destroyed an EcoRI site. The FP2 deletion created a new SacI site and destroyed a BstEII restriction site. Mutagenic primers were FP1del (5′-CCCCTGGTATTGGATATCCACTAGGGGGGCAGG-3′) and FP4del (5′-GTCCCACATACTTTATCATAGAGCTCCTTCAGTCTTGCG-3′) (boldface characters indicate newly created restriction sites in place of the deleted footprint sequences).
The selection oligonucleotide was SalI-HpaI (5′-CACTATAGGGCGTGCTCTAGATCTAGCTC-3′). Mutant clones were identified by their EcoRI, EcoRV, and SacI digestion patterns. DNA sequence analysis of the entire 2.4-kb BglII fragment confirmed that no bases other than the ones at the FP1 and FP4 sites had been altered.
Production of mice carrying the ICR FP1 and FP4 site mutations.
Gene targeting in mouse embryonic stem cells was performed using a replacement vector containing a neomycin and diphtheria toxin A-chain-positive and -negative selection cassette, respectively. The vector was identical to that previously described (39) except for presence of the mutant 2.4-kb BglII ICR. Homologous recombination was detected by PCR amplification across the short arm of the vector to yield a 1.3-kb band with the following primers: upper primer (in the genomic sequence just outside the end of the short arm), 5′-CCTATGCCCATGCCCCATACAAATGACACC-3′; lower primer (in the polyadenylation sequence of the neomycin selection cassette), 5′-GCTGGGGCTCGACTAGAGCTTGCGGAAC-3′. Recombinant clones identified in this PCR assay were examined further by Southern blotting for conservative recombination of the short and long arms as previously described (39) and for retention of FP1 and FP2 deletions. Excision of the neo cassette was achieved by mating male chimeras with females heterozygous for a X-linked Cre recombinase gene—FVB/NJ.129/SvImJ(N7)-Hprtcre/0; eggs of these females excise floxed sequences without mosaicism and regardless of Cre inheritance (40).
Breeding of fetuses carrying the mutation for analysis.
Female mice heterozygous for the mutation and negative for the Cre allele that were obtained from mating male chimeras with Cre/0 females (see above) were mated with males homozygous for the Mus musculus castaneus form of distal chromosome 7 derived from strain CAST/Ei (CS) (The Jackson Laboratory); these males were of strain FVB/NJ.CAST/Ei(N7). The use of this mating and of its reciprocal allowed for allele-specific analysis of expression. The wild-type allele (+) was derived from strain CS, while the mutant allele (−) was derived from strain 129SI/ImJ (129). Hereafter, heterozygous fetuses maternally and paternally inheriting the mutation are designated −(M)/+ and +/−(P), respectively.
Gene expression.
Transcription levels of Rxrα and Erβ were assessed by RT-PCR (36). RNA was prepared from 200,000 purified germ cells or somatic cells of 15.5-dpc female or male gonads with the RNeasy-Micro kit (Qiagen). RT was done on total RNA equivalent to that of 30,000 cells. The subsequent PCR was divided into equal aliquots, and the aliquots were subjected to increasing numbers of PCR cycles in the linear range of amplification as indicated (see Fig. 7). Oligonucleotides were ErβU (5′-TTCCTCCTATGTAGAGAGCCGTCACG-3′), ErβL (5′-CCCTCTTGGCGCTTGGACTAGTAA-3′), RxrαU (5′-TCCAACGGGTCGAGGCTCCA-3′), and RxrαL (5′-AGGAACCTTGAGGACGCCATTGAG-3′).
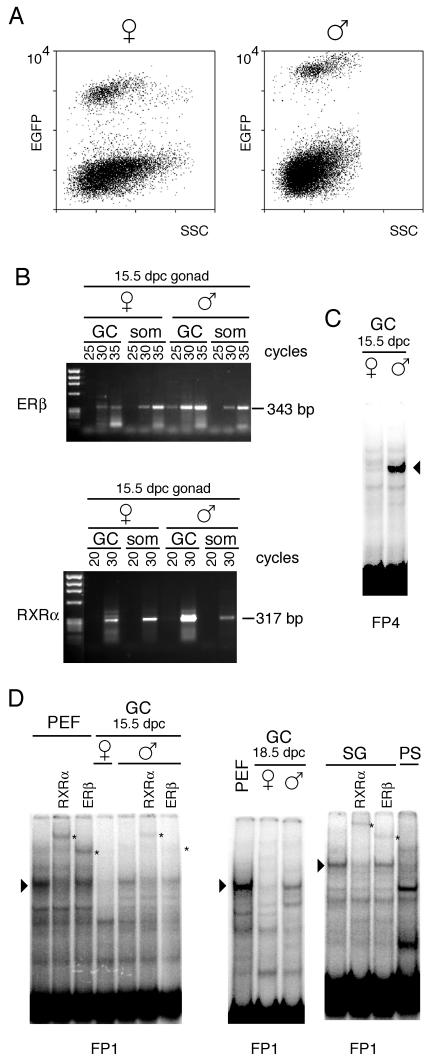
FIG. 7.
Protein binding at the FP sequences in germ cell extracts. (A) Germ cells from male (♂) and female (♀) 15.5-dpc fetuses were obtained by flow sorting of EGFP-expressing germ cells. The populations shown higher in the graphs are comprised of germ cells, while the lower populations are comprised of gonadal somatic cells. SSC, side scatter. (B) Assessment by semiquantitative RT-PCR of relative Rxrα and Erβ transcript levels in germ cells. RNA prepared from equal numbers of purified germ cells (GC) or somatic cells (som) of 15.5-dpc female or male gonads was subjected to RT. Aliquots of the PCR were subjected to increasing number of cycles in the linear amplification range. A representative experiment is shown. (C) Male germ cell-specific complex at the FP4 sequence. (D) Protein-DNA complexes of identical levels of mobility are present in nuclear extracts from PEF and in extracts from 15.5-dpc male germ cells. Female germ cells from the same stage of development do not contain this DNA binding activity. The complex is also present in 18.5-dpc male germ cells and in spermatogonia (SG) but not in pachytene spermatocytes (PS). Arrowheads point to the specific gel shifts. Antibody supershift assays indicate the presence of RXRα and ERβ in the complex in male germ cells (indicated by asterisks).
Allele-specific expression was determined using RT-PCR single-nucleotide primer extension assays as previously described (36, 39). Each assay relies on a single known sequence difference between allelic RNAs. Each sample represents an individual embryo.
RESULTS
In vivo footprints at the H19-Igf2 ICR.
In vivo genomic footprinting by LMPCR was performed to analyze the chromatin structure of the Igf2 and H19 ICR in mouse PEFs that carry only maternally or paternally inherited copies of the distal chromosome 7 region on which these genes reside—MatDup.d7 or PatDup.d7 fibroblasts, respectively. In MatDup.d7 fibroblasts, on each of the two distal chromosome 7 regions Igf2 is silent whereas H19 is active; in PatDup.d7 embryos, the opposite is true (22). The modification of DNA by DNase I, DMS, or UV light is sensitive to bound protein, and areas of protein-DNA interaction appear as footprints (protection or enhancements) in LMPCR genomic sequencing ladders. LMPCR analysis of naked DNA from PEFs treated with these reagents in vitro served as a control.
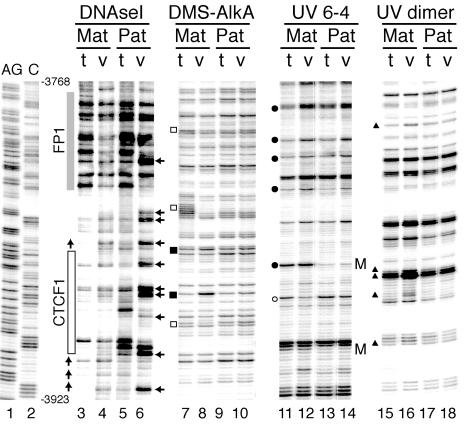
Using these in vivo treatments previously, we revealed the occupation of the four CTCF binding sites on the maternally derived chromosomes (34). In the present LMPCR experiments we obtained evidence for additional protein binding next to all four CTCF sites on maternally derived ICR sequences. Following DNase I treatment, MatDup.d7 PEFs displayed a prominent DNase I-protected area spanning about 70 bp and located about 45 bp 3′ to the CTCF site most distal relative to the H19 promoter (Fig. 1). By contrast, PatDup.d7 PEFs did not show areas of such strong protection. Instead, they displayed periodically spaced strong DNase I hyperreactive sites in the same region indicative of nucleosomes that occupy preferred rotational positions. Following DMS treatment, MatDup.d7 PEFs displayed a G residue of lower relative intensity within the DNase I-protected area (Fig. 1; compare lanes 7 and 8) whereas no footprints were observed for PatDup.d7 PEFs (compare lanes 9 and 10). DMS treatment also indicated the presence of a string of protected G residues between the FP1 and CTCF footprints (Fig. 1, lanes 7 and 8). Following treatment with UV light, MatDup.d7 PEFs showed a number of C and T bands of a much higher or lower relative intensity level within the FP1 footprint area (for 6-4 photoproducts, compare lanes 11 and 12; for cyclobutane pyrimidine dimers, compare lanes 15 and 16).
FIG. 1.
In vivo genomic footprinting of CTCF site 1 and surrounding sequences in the H19-Igf2 ICR. Fibroblasts from 13.5-dpc Matdupd7 (Mat) and Patdupd7 (Pat) embryos were treated with the footprinting agent DNase I or DMS or with 254-nm-wavelength UV light. The DMS-modified DNA bases were converted into strand breaks with the AlkA DNA glycosylase. The UV-induced DNA modifications were converted into strand breaks by heating in 1 M piperidine at 95°C to reveal 6-4 photoproducts (UV 6-4) or by using T4 endonuclease V to reveal cyclobutane pyrimidine dimers (UV dimer). The positions of the resulting strand breaks in the H19-Igf2 ICR were mapped by LMPCR. For controls, DNAs from maternal or paternal duplication fibroblasts were treated with the same agents in vitro and the in vitro patterns (t) were compared with the in vivo patterns (v). The AG and C lanes show position markers obtained by chemical DNA sequencing. The DNase I in vivo footprints produced by binding of CTCF (site 1) and the adjacent footprint (FP1) on the maternal chromosomes are indicated by boxes. The alternating DNase I hyporeactivity and hyperreactivity (horizontal arrows) are characteristic of positioned nucleosomes on the paternal allele. Sites that are hyperreactive in vivo on the maternal chromosomes for DMS treatment, UV 6-4 photoproducts, and UV dimers are indicated by closed squares, circles, and triangles, respectively. Open symbols mark nucleotide positions that are hyporeactive in vivo. Each CpG site at which the formation of a 6-4 photoproduct is inhibited by CpG methylation on the paternal chromosomes is indicated by an M.
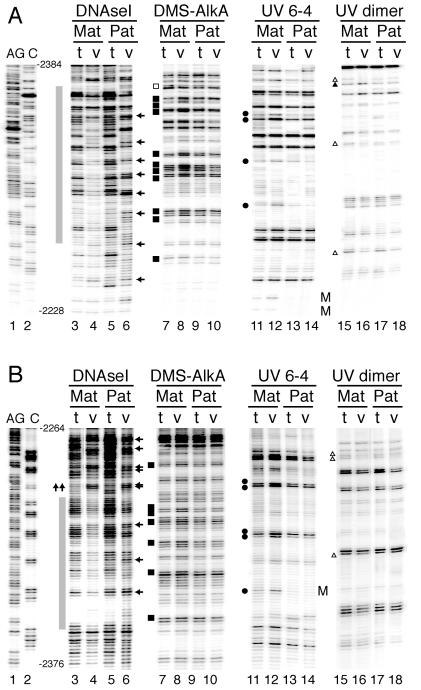
We detected an additional area of strong DNase I protection—FP4. Figure 2 shows the upper and lower stands of this area analyzed by DNase I, DMS, and UV photofootprinting. DNase I protection over an area of 110 bp was observed on both strands on the maternally derived chromosomes. By contrast, paternal chromosomes displayed periodically spaced DNase I hyperreactive sites. On the upper strand there was a strong periodicity of 10 to 11 bp, suggesting the presence of rotationally positioned nucleosomes. The DMS and UV footprinting data showed several hyperreactive and hyporeactive nucleotides within the DNase I-protected area on the maternal chromosome only, supporting the notion that the maternal sequences are associated with a sequence-specific DNA binding protein.
FIG. 2.
In vivo genomic footprinting of sequences in the H19-Igf2 ICR. (A) Upper strand; (B) lower strand. The footprinted area designated FP4 is adjacent to a degenerate CTCF site which does not bind CTCF. The gray boxes indicate the positions of the areas that show in vivo footprints on the maternal chromosomes. Arrows next to the DNase I lane derived from the paternal chromosome point to in vivo periodic DNase I hyperreactive sites, suggesting the existence of positioned nucleosomes. The symbols indicate in vitro versus in vivo reactivity differences on the maternally derived chromosomes and are the same as those described for Fig. 1.
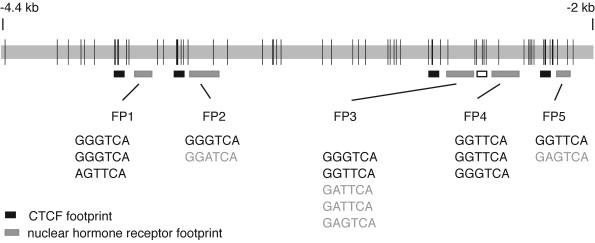
Using the technologies described for Fig. 1 and 2, we found three additional in vivo footprints in the 2.4-kb ICR. Strikingly, FP1, FP2, FP3, and FP5 were all localized 30 to 45 bp 3′ to each in vivo CTCF binding site. FP4 was located in the proximity of another, degenerated CTCF site that no longer can bind CTCF (8). The ICR therefore consists of a five-unit repeat structure. The arrangement of all identified in vivo footprints along the 2.4-kb ICR sequence is shown in Fig. 3.
FIG. 3.
Schematic representation of the location of in vivo footprints across the 2.4-kb H19-Igf2 ICR sequence. In vivo footprints identified by LMPCR analysis of the ICR sequence are indicated by boxes. Black boxes mark the location of the four in vivo CTCF footprints (34). Gray-shaded boxes indicate the positions of the newly identified NHR-like footprints (FP1, FP2, FP3, FP4, and FP5) found 30 to 45 bases 3′ to each CTCF footprint. The open box next to FP4 delineates the position of a degenerate CTCF site. NHR half sites found within the DNase I footprinted areas are shown below each respective footprint. PuGGTCA or PuGTTCA sites are shown with black characters; less-stringent half binding sites are shown in light-grey characters. Vertical lines mark the positions of CpG dinucleotides along the ICR.
In vitro binding assays.
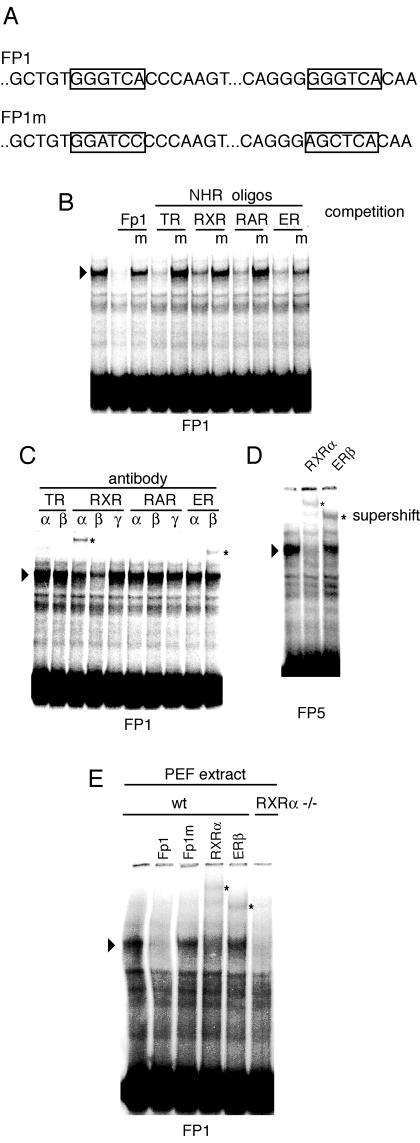
We next analyzed the sequences showing the novel in vivo footprints adjacent to CTCF binding sites to identify putative transcription factor binding elements. Each of the footprinted areas contained at least two NHR half sites (most commonly GGGTCA or GGTTCA). The identity of these sequences is shown in Fig. 3. To characterize the protein complexes interacting with these sequences, we carried out gel mobility shift experiments using the FP1 footprinted sequence as the labeled probe and nuclear extracts from wild-type PEFs. A single mobility shift band was specifically detected on the FP1 sequence (Fig. 4B). This complex was competed by an excess of wild-type oligonucleotide but not by an FP1 oligonucleotide in which the GGGTCA sequences had been mutated to either GGATCC or AGCTCA (Fig. 4A). The FP1 gel shift band was also competed by different oligonucleotides containing recognition sites for thyroid hormone receptor (TR), retinoic acid X receptor (RXR), retinoic acid receptor (RAR), or estrogen receptor (ER). Mutant forms of these NHR-binding sites did not compete or competed inefficiently (Fig. 4B). To clarify the identity of the gel shift complexes binding at the FP1 sequence, antibodies directed against TR alpha (TRα) and TRβ, RXR alpha (RXRα), RXRβ, and RXRγ, RAR alpha (RARα), RARβ, and RARγ, and ER alpha (ERα) and ERβ were used in gel mobility supershift assays. Antibodies directed against RXRα and ERβ produced clear supershifts. An antibody against RXRβ reduced the intensity of the FP1 gel shift band but did not produce a supershift. Antibodies directed against other NHR proteins did not produce significant supershifts and did not eliminate the FP1 band (Fig. 4C). We found that the same antibodies, RXRα and ERβ, supershifted the FP5 oligonucleotide (Fig. 4D).
FIG.4.
Gel mobility shift assays to identify protein binding at the in vivo footprinted sequences. (A) Schematic representation of NHR half sites within the FP1 sequence. The base changes in the FP1 mutant (FP1m) sequence are shown. (B) The FP1 footprint sequence was used as the labeled probe. As competitors, FP1, mutant (m) FP1, or oligonucleotides containing NHR consensus binding sites as well as their mutant forms were used. The FP1 gel shift band was competed by NHR oligonucleotides but not by their consensus site mutant forms. (C) Antibody supershift assays to identify components of the FP1 complex. Antibodies directed against TRα, TRβ, RXRα, RXRβ, RXRγ, RARα, RARβ, RARγ, ERα, and ERβ were used in gel mobility shift assays. Antibodies against RXRα and ERβ produced supershift bands. (D) Antibody supershift assays to identify components of the FP5 complex. The FP5 oligonucleotide was used as the labeled probe. Antibodies against RXRα and ERβ produced supershifted bands. (E) Rxrα−/− mice lack gel shift at the FP1 sequence. The FP1 oligonucleotide was used as probe and was incubated with either normal or Rxrα−/− PEF extracts. FP1 and FP1m competitors or antibodies against RXRα and ERβ were included with both extracts but are shown only for normal PEF. wt, wild type.
We attempted to carry out chromatin immunoprecipitation analysis of the ICR sequences in embryo fibroblasts. This approach was not successful due to the limited affinity of the RXRα and ERβ antibodies available to us. To strengthen the connection of RXRα binding to the ICR hexad sites, we examined in vitro binding of FP1 oligonucleotide with PEF extract lacking RXRα. Rxrα−/− (33) PEF extract did not produce a gel shift at the FP1 sequence (Fig. 4E).
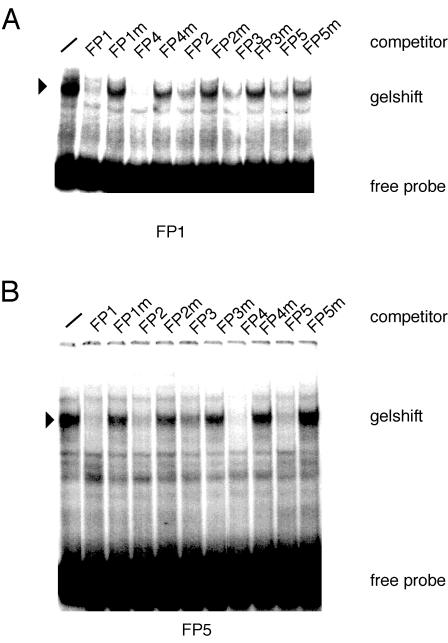
We then established the relationship between FP1-interacting proteins and the proteins binding at sites FP2 to FP5. Gel shift competition assays were conducted (Fig. 5). When the FP1 sequence was used as a probe, sequences from FP2, FP3, FP4, and FP5 all competed with the complexes forming on the FP1 oligonucleotide. Their respective mutants, in which the NHR half sites had been altered, did not compete or were much-less-efficient competitors. Similarly, when the FP5 sequence was used as a labeled probe sequences from FP1, FP2, FP3, and FP4 all competed efficiently with the complexes forming on the FP5 oligonucleotide. Mutants in which the NHR half sites had been altered did not compete or were much-less-efficient competitors (Fig. 5).
FIG. 5.
Gel mobility shift assays to characterize protein binding at the in vivo footprinted sequences. (A) Protein binding to the labeled FP1 sequence through the use of nuclear extracts from mouse embryo fibroblasts. Specificity was demonstrated by competition with the wild-type and mutant FP1 sequences. Excess unlabeled oligonucleotides representing the FP2, FP3, FP4, and FP5 sequences as well as their mutant forms were used in competition assays. The FP1 complexes were competed by all other FP oligonucleotides but less efficiently by their mutant forms. (B) The FP5 sequence was used as the labeled probe. The FP5-DNA protein complex was inhibited by unlabeled FP1, FP2, FP3, and FP4 sequences but not by their mutant forms. −, no competitor.
Binding of NHR complexes is insensitive to CpG methylation.
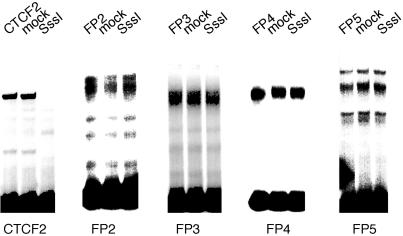
The binding of CTCF to its recognition sequences has been shown to be dependent on the unmethylated state of the target site (3, 8, 11, 34). We have used nuclear extracts from mouse embryo fibroblasts to verify the methylation dependence of CTCF binding (Fig. 6). It was of interest to determine whether the NHR complexes binding at the FP1 to FP5 sequences are also sensitive to CpG methylation. The FP2-to-FP5 oligonucleotide sequences were methylated at all CpG sites when SssI DNA methylase was used. FP1 did not contain a CpG sequence and was not used in these assays. The data (Fig. 6) clearly show that binding of the NHR complexes, unlike CTCF binding, is not sensitive to CpG methylation.
FIG. 6.
Binding of NHR complexes at the ICR is independent of CpG methylation. The FP2, FP3, FP4, and FP5 oligonucleotides were methylated at all CpG sites through the use of SssI DNA methylase. FP1 does not contain a CpG site. The methylated and mock-methylated oligonucleotides were used in gel mobility shift assays with nuclear extracts from embryo fibroblasts. Binding was independent of CpG methylation. As a control, an oligonucleotide containing CTCF site 2 was methylated. Binding of CTCF was completely inhibited by CpG methylation.
Binding assays using germ cell extracts.
Genomic footprinting strongly suggested maternal allele-specific binding of both CTCF (34) and NHR complexes (Fig. 1 to 3) in somatic cells. However, genomic imprints are set in the germ line; thus, it becomes important to characterize protein binding at the ICR sequences in this lineage. We were particularly interested in determining whether there are differences between male and female germ cells in NHR complexes binding to ICR sequences. In the female germ line, the ICR sequences remain unmethylated; it is conceivable that de novo methylation of the ICR is prevented by bound protein complexes. Conversely, the ICR undergoes de novo methylation in the male germ line in germ cells from 14.5 to 18.5 dpc (6, 44), and this process could be facilitated by protein factors binding to the ICR in the male germ line. CTCF or other proteins that specifically bind to CTCF consensus sequences do not qualify for either of these activities, since the ICR still remains unmethylated in the maternal germ line and becomes methylated in the paternal germ line of mice in which all four CTCF sites have been dramatically mutated (39a).
Genomic footprint or chromatin immunoprecipitation analysis is not possible or is technically challenging in germ cells due to the limited material that can be obtained. However, in vitro binding assays with nuclear extracts are feasible. We have developed a transgenic mouse line expressing the EGFP reporter gene under control of the germ cell-specific Pou5f1 promoter which allows for purification of germ cells by cell sorting (35) (Fig. 7). Using flow cytometry, we isolated germ cells from 15.5- and 18.5-dpc embryos. Also, spermatogonia were purified from newborns and pachytene spermatocytes were purified from adult testes. The latter were sorted by flow cytometry on the basis of physical characteristics (19). We performed semiquantitative RT-PCR to assess the relative levels of abundance of Erβ and Rxrα transcripts in male and female germ cells. We found that both NHRs were expressed at much higher levels in male than in female germ cells of 15.5-dpc embryos (Fig. 7B). A protein complex binding to the FP4 sequence was present in male, but not female, germ cells at 15.5 dpc (Fig. 7C). A complex having a level of mobility identical to that seen in PEFs was detected at FP1 sequences in male germ cells, i.e., in 15.5- and 18.5-dpc germ cells and spermatogonia (Fig. 7D). This complex was supershifted with antibodies against ERβ and RXRα (Fig. 7D). The specific gel shift was not present in pachytene spermatocytes, which represent a stage past the time when methylation imprints are established for the ICR. More importantly, however, this complex was completely absent in 15.5- and 18.5-dpc female germ cells (Fig. 7D). These data are consistent with the possibility that the NHR complexes play a specific role in the male germ line. Their presence in male germ cells at the stage at which de novo methylation of the ICR occurs is consistent with the possibility that these complexes facilitate methylation of the ICR in male germ cells.
The NHR binding sites in the H19-Igf2 ICR are conserved.
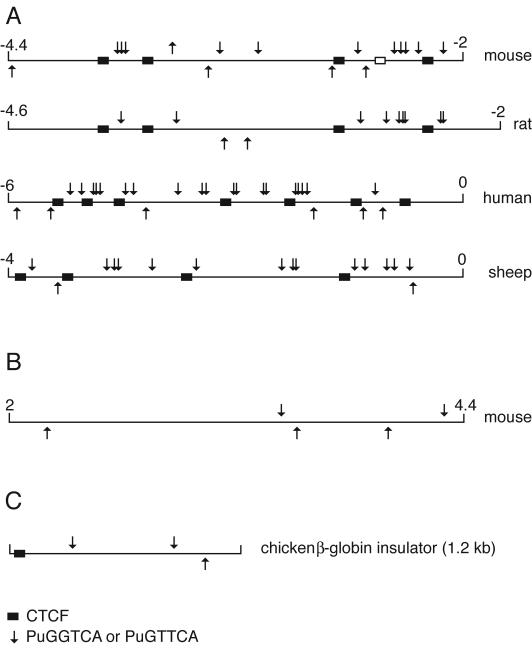
The biological significance of the hormone receptor binding sites in the ICR would be strengthened if these sites were conserved in different mammalian species. We have aligned the ICR sequences obtained from humans, mice, rats, and sheep. The alignments show the conservation of the CTCF sites (Fig. 8A) and also the NHR binding sites in all four species. NHR hexad sites are much less abundant in the body of the H19 gene, and their numbers are not biased towards the upper strand (Fig. 8B). NHR sites are less abundant on the chicken β-globin insulator and are located at a great distance from the CTCF site (Fig. 8C). Conservation of abundance of NHR sites in the ICR and their strand bias and proximity to CTCF binding sites in different species are consistent with these binding sites and their associated NHR complexes having functional significance.
FIG. 8.
H19-Igf2 ICR hexad nuclear receptor binding sites in different species. (A) Mouse, rat, human, and sheep ICR sequences were analyzed for the presence of G/AGG/TTCA sites (arrows) on top and bottom strands. Note the higher frequency of sites in the upper strand. CTCF sites are indicated with black boxes. (B) As a control sequence, the mH19 gene was analyzed from kb +2 to +4.4. (C) Occurrence of G/AGG/TTCA sequences in the 1.2-kb chicken β-globin insulator. Numbers indicate locations of the analyzed sequences relative to the transcriptional start site in kilobases.
Functional analysis of the NHR footprints by gene targeting.
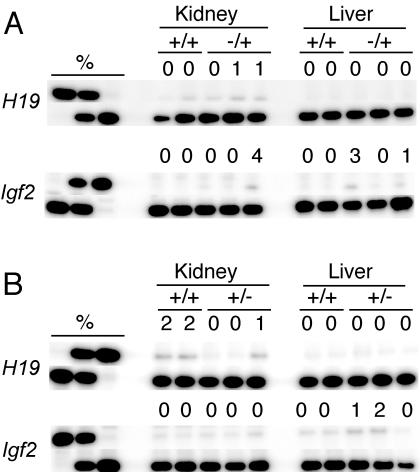
We deleted FP1 and FP4 by homologous recombination in mice. Gene targeting was done as described before (39). On paternal or maternal inheritance, the mutant ICR had no effect on the normal allele-specific expression pattern of Igf2 and H19. Both genes were expressed monoallelically in the liver and kidney of 17.5-dpc +/−(P) and −(M)/+ fetuses (Fig. 9). Thus, the mutant ICR, lacking protein binding at FP1 and FP4, successfully substituted for the insulator and imprinting functions of the normal ICR. This result is consistent with the possibility that the nuclear hormone binding sites in the ICR act redundantly, and inactivation of all the five footprints may be necessary to reveal their function in somatic cells and in the germ line.
FIG. 9.
Phenotype of the 17.5-dpc fetuses inheriting FP1 and FP2 deletions in the H19-Igf2 ICR. The results of RT-PCR single-nucleotide primer extension assays are shown. (A) Maternal transmission of the deletions. (B) Paternal transmission of the deletions. The value above each band represents the amount of RNA contributed by the presumptive inactive allele (top row) as a percentage of the total. Each lane represents an individual embryo.
DISCUSSION
The mechanistic basis for the phenomenon of parental imprinting has remained unclear. Many imprinted gene clusters contain one or more differentially methylated regions. In some cases, these DMRs are directly associated with a CpG island spanning the promoter of an imprinted gene. However, the DMRs are often located at a distance from the promoter. For Igf2 and H19, the best-studied pair of imprinted genes, the DMR is 80 kb distant from the Igf2 promoter and yet methylation of the ICR determines whether Igf2 is expressed or not. For the H19-Igf2 ICR, it has been demonstrated that the Igf2 promoter is regulated by an insulator function residing in the ICR. This chromatin insulation function, which shields the Igf2 promoter from an enhancer located 3′ to the H19 gene, is carried out by the zinc finger protein CTCF (3, 4, 8, 11, 32, 34). Despite the clear role that CTCF plays in chromatin insulation at the H19-Igf2 ICR and its role in protecting the ICR from methylation in somatic cells (27), it has remained unclear what function of the ICR is responsible for the differential methylation set up in the germ line. The ICR still remains unmethylated in the female germ line in which all four CTCF sites have been drastically mutated, and this mutant ICR becomes methylated de novo in the male germ line (39a). In addition, when the ICR sequence was replaced with the chicken β-globin insulator (39), these sequences did not undergo de novo methylation in the male germ line. Again, this indicates that CTCF is not involved in the de novo methylation process and that the chicken insulator does not carry sequences signaling de novo methylation in the germ line.
In this study, we identified five NHR binding sites at 30 to 50 bp adjacent to each CTCF site and one such site adjacent to a degenerate CTCF site. These binding sites do not conform to standard NHR binding sites in which two half sites are separated by one to eight nucleotides. Rather, the GGGTCA and GGTTCA hexad sites are scattered through larger areas. Nonetheless, these sites are functional in protein binding in vivo and interact specifically with complexes containing RXRα and ERβ in vitro. It has been shown that widely spaced half-palindromic ER sites can function synergistically (13), and widely spaced, directly repeated PuGGTCA elements were shown to act as promiscuous response elements for different NHRs (12). Both hormone receptors are able to form heterodimers with other receptors. Heterodimer formation between RXRα and ERβ is not unprecedented. A ligand-independent direct interaction between RXRα and ERβ has been demonstrated by the yeast two-hybrid test and glutathione S-transferase pulldown assays (31).
An association between CTCF and NHR sites has been observed before. CTCF binding sites are often flanked at a distance of 10 to 13 bp by thyroid hormone response elements (TRE), for example, at the chicken lysozyme upstream silencer and the human c-myc gene, and these CTCF-hormone response element composite elements are regulated by thyroid hormone (1, 18). The spacing of hexad binding sites in the H19-Igf2 ICR is very different from the spacing of the typical TR binding sites found in the lysozyme and c-myc genes, and we did not identify TR as a component of the complexes binding to these sites in PEFs or germ cells. Instead, EMSAs suggest that RXRα and ERβ may be components of the ICR complexes. These NHR footprints are in close proximity to CTCF sites; therefore, these complexes might modulate the efficiency of the insulator in the maternally transmitted chromosome in somatic cells in a tissue-specific manner.
We found in vivo protein binding to these sequences in PEFs exclusively in the maternal chromosome (Fig. 1 and 2). NHRs can have activating or repressing transcriptional activity depending on association with their ligand and with coactivators or corepressors (47). Maternal allele-specific binding of RXRα and ERβ and their coactivators might support CTCF in keeping the maternal chromosome unmethylated in somatic cells. In vitro binding to these sequences does not depend on methylation (Fig. 6); therefore, the paternal chromosome most likely has a higher-order chromatin structure that is not accessible to NHR binding. Alternatively, an unidentified protein complex much less abundant in cell extracts than the NHRs may bind to the ICR sequences in vivo in a methylation-sensitive fashion.
Intriguingly, Rxrα and Erβ transcripts were much more abundant in male than in female germ cells and their protein complexes were present in male, but not female, germ cells at a stage at which de novo methylation of the male-germ-line ICR sequences occurs (Fig. 7). This temporal and parent-of-origin-specific correlation is consistent with the possibility that the NHR complexes are involved in de novo methylation. Due to the absence of the NHR complexes in female germ cells at the same stage of development, this de novo methylation would not occur on maternal chromosomes. Since some corepressor complexes are associated with histone deacetylase or histone methylase activities (5, 17, 47), it is possible that histone modification at the ICR induces de novo DNA methylation, although more direct connections between NHRs and the DNA methylation machinery could be imagined (7). Interestingly, ERβ was shown to bind N-CoR in the presence of estrogen agonists in vivo and in vitro (45).
Prior to the identification of all five in vivo FP sequences, we generated a mouse line from which the entire FP1 and FP4 sequences, including six NHR hexad binding sites, were deleted by gene targeting. In these mice, imprinting of Igf2 and H19 loci was correctly maintained (Fig. 9). Due to the existence of a multitude of such binding sites (no fewer than 15), however, this initial result could be explained by functional redundancy of the hexad binding sites. We are therefore currently producing a mouse line in which all 15 hexad sites in FP1 to FP5 are mutated. In addition, to shed light on a general role of NHR binding in imprinting we are examining the methylation status of germ cells in Rxrα−/−fetuses, which die at approximately 15.5 dpc (33).
Acknowledgments
We are grateful to Shih-Huey E. Tang for technical assistance, Lucy Brown, Claudio Spalla, and Jim Bolen for flow cytometry, Francisco Silva and Walter Tsark for blastocyst injection, Timothy O'Connor for his gift of AlkA protein, Steven Lloyd for endonuclease V, and Henry Sucov for the RXRα+/− mice. We also thank Frederic Chedin for his comments on the manuscript.
This work was supported by National Institutes of Health grant GM064378 to J.R.M. and National Cancer Institute grant CA33572-21 for the flow cytometry core facility.
REFERENCES
- 1.Arnold, R., M. Burcin, B. Kaiser, M. Muller, and R. Renkawitz. 1996. DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic Acids Res. 24:2640-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 5.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 6.Davis, T. L., J. M. Trasler, S. B. Moss, G. J. Yang, and M. S. Bartolomei. 1999. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58:18-28. [DOI] [PubMed] [Google Scholar]
- 7.Di Croce, L., V. A. Raker, M. Corsaro, F. Fazi, M. Fanelli, M. Faretta, F. Fuks, F. Lo Coco, T. Kouzarides, C. Nervi, S. Minucci, and P. G. Pelicci. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295:1079-1082. [DOI] [PubMed] [Google Scholar]
- 8.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 9.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Kaffer, C. R., M. Srivastava, K. Y. Park, E. Ives, S. Hsieh, J. Batlle, A. Grinberg, S. P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14:1908-1919. [PMC free article] [PubMed] [Google Scholar]
- 11.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 12.Kato, S., H. Sasaki, M. Suzawa, S. Masushige, L. Tora, P. Chambon, and H. Gronemeyer. 1995. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 15:5858-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, S., L. Tora, J. Yamauchi, S. Masushige, M. Bellard, and P. Chambon. 1992. A far upstream estrogen response element of the ovalbumin gene contains several half-palindromic 5′-TGACC-3′ motifs acting synergistically. Cell 68:731-742. [DOI] [PubMed] [Google Scholar]
- 14.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 15.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9:2079-2089. [DOI] [PubMed] [Google Scholar]
- 16.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 17.Li, J., Q. Lin, H. G. Yoon, Z. Q. Huang, B. D. Strahl, C. D. Allis, and J. Wong. 2002. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol. Cell. Biol. 22:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz, M., L. J. Burke, P. LeFevre, F. A. Myers, A. W. Thorne, C. Crane-Robinson, C. Bonifer, G. N. Filippova, V. Lobanenkov, and R. Renkawitz. 2003. Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J. 22:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malkov, M., Y. Fisher, and J. Don. 1998. Developmental schedule of the postnatal rat testis determined by flow cytometry. Biol. Reprod. 59:84-92. [DOI] [PubMed] [Google Scholar]
- 20.Mann, J. R., P. E. Szabó, M. R. Reed, and J. Singer-Sam. 2000. Methylated DNA sequences in genomic imprinting. Crit. Rev. Eukaryot. Gene Expr. 10:241-257. [DOI] [PubMed] [Google Scholar]
- 21.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaughlin, K. J., P. Szabó, H. Haegel, and J. R. Mann. 1996. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development 122:265-270. [DOI] [PubMed] [Google Scholar]
- 23.Nabetani, A., I. Hatada, H. Morisaki, M. Oshimura, and T. Mukai. 1997. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol. Cell. Biol. 17:789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pant, V., P. Mariano, C. Kanduri, A. Mattsson, V. Lobanenkov, R. Heuchel, and R. Ohlsson. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeifer, G. P., and A. D. Riggs. 1991. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 5:1102-1113. [DOI] [PubMed] [Google Scholar]
- 26.Ripoche, M. A., C. Kress, F. Poirier, and L. Dandolo. 1997. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 11:1596-1604. [DOI] [PubMed] [Google Scholar]
- 27.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 28.Searle, A. G., and C. V. Beechey. 1990. Genome imprinting phenomena on mouse chromosome 7. Genet. Res. 56:237-244. [DOI] [PubMed] [Google Scholar]
- 29.Shibata, H., T. Ueda, M. Kamiya, A. Yoshiki, M. Kusakabe, C. Plass, W. A. Held, S. Sunahara, M. Katsuki, M. Muramatsu, and Y. Hayashizaki. 1997. An oocyte-specific methylation imprint center in the mouse U2afbp-rs/U2af1-rs1 gene marks the establishment of allele-specific methylation during preimplantation development. Genomics 44:171-178. [DOI] [PubMed] [Google Scholar]
- 30.Shibata, H., Y. Yoda, R. Kato, T. Ueda, M. Kamiya, N. Hiraiwa, A. Yoshiki, C. Plass, R. S. Pearsall, W. A. Held, M. Muramatsu, H. Sasaki, M. Kusakabe, and Y. Hayashizaki. 1998. A methylation imprint mark in the mouse imprinted gene Grf1/Cdc25Mm locus shares a common feature with the U2afbp-rs gene: an association with a short tandem repeat and a hypermethylated region. Genomics 49:30-37. [DOI] [PubMed] [Google Scholar]
- 31.Song, M. R., S. K. Lee, Y. W. Seo, H. S. Choi, J. W. Lee, and M. O. Lee. 1998. Differential modulation of transcriptional activity of oestrogen receptors by direct protein-protein interactions with retinoid receptors. Biochem. J. 336(Pt. 3):711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava, M., S. Hsieh, A. Grinberg, L. Williams-Simons, S. P. Huang, and K. Pfeifer. 2000. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 14:1186-1195. [PMC free article] [PubMed] [Google Scholar]
- 33.Sucov, H. M., E. Dyson, C. L. Gumeringer, J. Price, K. R. Chien, and R. M. Evans. 1994. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 8:1007-1018. [DOI] [PubMed] [Google Scholar]
- 34.Szabó, P., S. H. Tang, A. Rentsendorj, G. P. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607-610. [DOI] [PubMed] [Google Scholar]
- 35.Szabó, P. E., K. Hubner, H. Scholer, and J. R. Mann. 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115:157-160. [DOI] [PubMed] [Google Scholar]
- 36.Szabó, P. E., and J. R. Mann. 1995. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 9:1857-1868. [DOI] [PubMed] [Google Scholar]
- 37.Szabó, P. E., G. P. Pfeifer, and J. R. Mann. 1998. Characterization of novel parent-specific epigenetic modifications upstream of the imprinted mouse H19 gene. Mol. Cell. Biol. 18:6767-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabó, P. E., G. P. Pfeifer, F. Miao, T. R. O'Connor, and J. R. Mann. 2000. Improved in vivo dimethyl sulfate footprinting using AlkA protein: DNA-protein interactions at the mouse H19 gene promoter in primary embryo fibroblasts. Anal. Biochem. 283:112-116. [DOI] [PubMed] [Google Scholar]
- 39.Szabó, P. E., S. H. Tang, M. R. Reed, F. J. Silva, W. M. Tsark, and J. R. Mann. 2002. The chicken beta-globin insulator element conveys chromatin boundary activity but not imprinting at the mouse Igf2/H19 domain. Development 129:897-904. [DOI] [PubMed] [Google Scholar]
- 39a.Szabó, P. E., S.-H. E. Tang, F. J. Silva, W. M. K. Tsark, and J. R. Mann. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 24:4791-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, S. H., F. J. Silva, W. M. Tsark, and J. R. Mann. 2002. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32:199-202. [DOI] [PubMed] [Google Scholar]
- 41.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tornaletti, S., and G. P. Pfeifer. 1995. UV light as a footprinting agent: modulation of UV-induced DNA damage by transcription factors bound at the promoters of three human genes. J. Mol. Biol. 249:714-728. [DOI] [PubMed] [Google Scholar]
- 43.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda, T., K. Abe, A. Miura, M. Yuzuriha, M. Zubair, M. Noguchi, K. Niwa, Y. Kawase, T. Kono, Y. Matsuda, H. Fujimoto, H. Shibata, Y. Hayashizaki, and H. Sasaki. 2000. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5:649-659. [DOI] [PubMed] [Google Scholar]
- 45.Webb, P., C. Valentine, P. Nguyen, R. H. Price, Jr., A. Marimuthu, B. L. West, J. D. Baxter, and P. J. Kushner. 2003. ERbeta binds N-CoR in the presence of estrogens via an LXXLL-like motif in the N-CoR C-terminus. Nucl. Recept. 1:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyszomierski, S. L., J. Yeh, and J. M. Rosen. 1999. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13:330-343. [DOI] [PubMed] [Google Scholar]
- 47.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 48.Zemel, S., M. S. Bartolomei, and S. M. Tilghman. 1992. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 2:61-65. [DOI] [PubMed] [Google Scholar]