Abstract
The structurally related exocyclic guanine adducts α-hydroxypropano-dG (α-OH-PdG), γ-hydroxypropano-dG (γ-OH-PdG), and M1dG are formed when DNA is exposed to the reactive aldehydes acrolein and malondialdehyde (MDA). These lesions are believed to form the basis for the observed cytotoxicity and mutagenicity of acrolein and MDA. In an effort to understand the enzymatic pathways and chemical mechanisms that are involved in the repair of acrolein- and MDA-induced DNA damage, we investigated the ability of the DNA repair enzyme AlkB, an α-ketoglutarate/Fe(II) dependent dioxygenase, to process α-OH-PdG, γ-OH-PdG, and M1dG in both single- and double-stranded DNA contexts. By monitoring the repair reactions using quadrupole time-of-flight (Q-TOF) mass spectrometry, it was established that AlkB can oxidatively dealkylate γ-OH-PdG most efficiently, followed by M1dG and α-OH-PdG. The AlkB repair mechanism involved multiple intermediates and complex, overlapping repair pathways. For example, the three exocyclic guanine adducts were shown to be in equilibrium with open-ring aldehydic forms, which were trapped using (pentafluorobenzyl)hydroxylamine (PFBHA) or NaBH4. AlkB repaired the trapped open-ring form of γ-OH-PdG but not the trapped open-ring of α-OH-PdG. Taken together, this study provides a detailed mechanism by which three-carbon bridge exocyclic guanine adducts can be processed by AlkB and suggests an important role for the AlkB family of dioxygenases in protecting against the deleterious biological consequences of acrolein and MDA.
Introduction
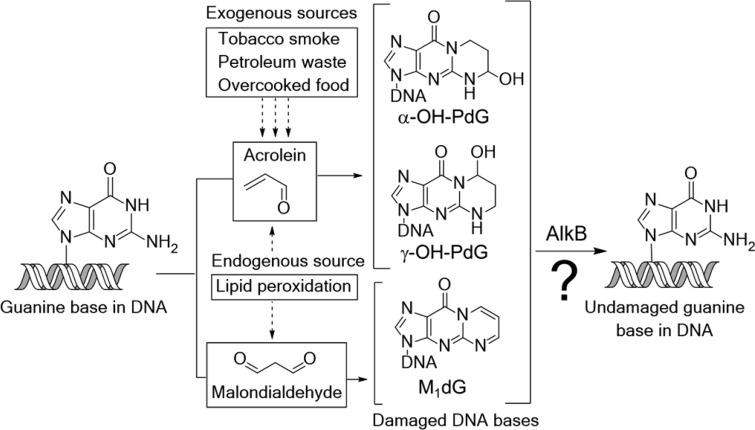
Reactive aldehydes, such as acrolein and malondialdehyde (MDA), react with DNA and form exocyclic adducts. Acrolein, an α,β-unsaturated aldehyde commonly found in tobacco smoke1 and other exogenous sources (petroleum industry waste,2 automobile exhaust,3,4 and overcooked food5) is a mutagenic agent6−10 that has been implicated in the etiology of lung cancer.11,12 Acrolein is also formed endogenously as a byproduct of lipid peroxidation,13,14 alongside structurally related molecules such as MDA. As a reactive aldehyde, acrolein condenses with deoxyguanosine (dG) through a Michael addition and subsequent cyclization15−17 to form two exocyclic adducts: α-OH-PdG (3-(2′-deoxy-β-d-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-6-hydroxypyrimido[1,2-α]purin-10(3H)-one) and γ-OH-PdG (3-(2′-deoxy-β-d-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2-α]purin-10(3H)-one) (Figure 1).
Figure 1.
Reactions of the environmental pollutant acrolein and of the lipid peroxidation byproduct malondialdehyde with DNA guanine bases generate the structurally similar exocyclic deoxyguanosine lesions α-OH-PdG, γ-OH-PdG, and M1dG.
The acrolein–DNA adducts have deleterious biological consequences and are believed to underlie the mutagenic effects of acrolein18−21 and its ability to promote carcinogenesis.22,23 α-OH-PdG can block DNA replication in human cells and can cause G to A and G to T mutations.24,25 By contrast, γ-OH-PdG is efficiently bypassed by certain polymerases in both bacterial and mammalian cells;26−29 however, γ-OH-PdG can also form inter- and intrastrand cross-links30−33 (via an open-ring aldehydic form), which are difficult to bypass and can cause mutations.34−36
MDA is an important biomarker of lipid peroxidation, a deleterious reaction between cellular lipids and reactive oxygen species produced by inflammatory processes.13,14,37 Similar to acrolein, MDA can also form an exocyclic dG adduct, M1dG (3-(2′-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one).38 This adduct is both a strong block to replication and mutagenic,39−41 properties that may contribute to inflammation-associated human malignancies, including aging42 and cancer.13,37
Given the toxicity and mutagenicity of the acrolein- and MDA-derived exocyclic deoxyguanosine adducts, it is important to investigate if these adducts are substrates for DNA repair pathways. While the nucleotide excision repair (NER) pathway can remove these lesions from DNA,23 acrolein, the agent responsible for generating the exocyclic dG lesions, inhibits NER.23 Little is known about the contribution of a direct reversal repair pathway for removal of acrolein- and MDA-derived exocyclic guanine DNA adducts.
AlkB, the Escherichia coli direct reversal DNA repair enzyme, can efficiently repair a wide range of DNA and RNA alkyl lesions.43−53 As an Fe(II)- and α-ketoglutarate-dependent dioxygenase, AlkB uses molecular oxygen to oxidize and remove simple alkyl DNA lesions (such as 3-methylcytosine,51 1-methyladenine,51 3-methylthymine,51 6-methyladenine,50 1- methylguanine (m1G),51 2-methylguanine(m2G),52 and 2-ethylguanine(e2G)52) and exocyclic bridged lesions (N1,N6-ethenoadenine (εA),46,47 N1,N6-ethanoadenine (EA),49,50 3,N4-ethenocytosine (εC),46,48 hydroxy-3,N4-ethanocytosine,48,53 and hydroxy-3,N4-propanocytosine).48,53 Given the ability of AlkB to remove alkyl groups from both N151 and N2 positions52 of guanine, we suspected that the exocyclic guanine adducts α-OH-PdG, γ-OH-PdG, and M1dG could also be potential substrates for AlkB.
This study investigated the ability of AlkB to repair the acrolein- and MDA-derived exocyclic dG lesions in single-straned (ssDNA) or double-stranded DNA (dsDNA). We established that all three exocyclic dG lesions can exist in open-ring forms in the sequence context studied, and we also investigated whether these open-ring forms are substrates for AlkB repair. Using high-resolution quadrupole time-of-flight (Q-TOF) MS, we found that AlkB can oxidize all three exocyclic dG adducts, in both the open- and closed-ring forms. The AlkB activity on the lesions was more efficient in ssDNA than in dsDNA, with the base opposite the lesion dictating the repair efficiency. Among the three lesions studied, γ-OH-PdG was most efficiently repaired, followed by M1dG and α-OH-PdG, respectively. By identifying and characterizing the AlkB reaction intermediates and products using MS, MS/MS, and biochemical trapping experiments, a model for the chemical mechanism of repair of exocyclic dG lesions by AlkB is proposed. The observation that these important lesions are oxidized by AlkB provides evidence that direct reversal enzymes, such as AlkB, may play important roles as modulators of the biological consequences of acrolein and MDA.
Experimental Procedures
Oligonucleotide Synthesis
Oligonucleotide 16-mers of sequence 5′-GAAGACCTXGGCGTCC-3′ (X = adduct) bearing exocyclic guanine adducts, α-OH-PdG,54,55 γ-OH-PdG,56,57 and M1dG58 (Figure 1) were prepared using the solid-phase methods59 and were deprotected, purified, and characterized as described previously. Table 1 shows the calculated MWs of all the oligonucleotides used in this study and their observed intermediates. To determine the oligonucleotides’ concentrations, the extinction coefficients (ε) of normal DNA bases at 260 nm were used; alkyl-modified bases were approximated as guanines.
Table 1. Calculated and Observed Monoisotopic Molecular Weights of Oligonucleotides Observed in the Present Study.
| lesion or base (structures) | MW (calcd) of neutral species | calcd monoisotopic m/z (−4 charge) | obsvd monoisotopic m/z (−4 charge) |
|---|---|---|---|
| (1a)/(1b) | 4960.88 | 1239.21 | 1239.16 |
| 1239.19 | |||
| 1239.20 | |||
| (2)/(18) | 4942.87 | 1234.71 | 1234.68 |
| 1234.70 | |||
| (3a)/(3b)/(21) | 4976.88 | 1243.21 | 1243.17 |
| 1243.18 | |||
| 1243.24 | |||
| (4)/(20) | 4958.87 | 1238.71 | 1238.67 |
| 1238.68 | |||
| 1238.70 | |||
| (5a)/(5b) | 4960.88 | 1239.21 | 1239.23 |
| (6) | 4904.86 | 1225.21 | 1225.17 |
| 1225.19 | |||
| (7) | 4940.86 | 1234.21 | 1234.17 |
| 1234.19 | |||
| (8) | 4956.85 | 1238.20 | 1238.19 |
| 1238.24 | |||
| (9)/(13)/(14) | 4974.86 | 1242.71 | 1242.73 |
| 1242.69 | |||
| (10) | 4992.87 | 1247.21 | 1247.19 |
| 1247.25 | |||
| (11a)/(11b)/ (12a)/(12b) | 4958.87 | 1238.71 | not observed |
| (15) | 4944.89 | 1235.21 | 1235.16 |
| (16) | 4962.90 | 1239.72 | 1239.71 |
| (17) | 4978.89 | 1243.72 | 1243.71 |
| (19) | 4958.68 | 1239.67 | 1239.67 |
AlkB Repair of Exocyclic dG Lesions (α-OH-PdG, γ-OH-PdG, and M1dG) in Single- and Double-Stranded DNA Oligonucleotides
AlkB incubations were performed with the AlkBΔN11 protein, a version of AlkB lacking the first 11 amino acids. This truncated protein has been previously shown to have activity similar to that of wild-type AlkB.49 The AlkBΔN11 protein was expressed in BL21(DE3) cells and purified as described previously.49 The AlkB incubations were performed at 37 °C for 2 h in a reaction buffer containing 45 mM HEPES (pH 8.0), 0.9 mM α-ketoglutarate, 67 μM Fe(NH4)2(SO4)2·6H2O, and 1.8 mM ascorbate, followed by HPLC/Q-TOF MS analysis. Typically, 100 pmol DNA was reacted with 5 μM AlkB (or enzyme diluent for control reactions) in the presence of all cofactors in a 20 μL reaction volume. The AlkB incubations with double-stranded DNA were performed under similar conditions except that prior to AlkB addition 1.2 equiv of complementary oligonucleotides were annealed by heating the mixture at 65 °C for 5 min. The mixtures were cooled at a rate of 0.1 °C/s to 4 °C and then used in AlkB reactions as described above.
Quantification of AlkB Repair Efficiency
The AlkB repair efficiencies were estimated from the fraction of starting material converted into intermediates and products (Table 2). These values were determined by integrating the extracted ion chromatogram (EIC) peaks corresponding to each of the species observed in the AlkB reactions (starting material, intermediates, and products) and calculating the percentage of each species formed after AlkB incubation relative to the total amount of material (Table 3). In each reaction, the total amount of material was calculated by summing the areas corresponding to the starting material and all intermediates and products derived from the starting material. All of the values were background-corrected for the amount of species (intermediate or product) that may have been present in the AlkB untreated control reactions.
Table 2. Percentage of Lesion-Containing Oligonucleotides Chemically Modified by AlkB in a 2 h Reaction.
| X = lesion | α-OH-PdG (%) | γ-OH-PdG (%) | M1dG (%) |
|---|---|---|---|
| ssDNA | 8.0 | 58.8 | 31.0 |
| dsDNA (T: X) | 1.6 | 17 | 45.5 |
| dsDNA (G: X) | 1.0 | 10.4 | 22.0 |
| dsDNA (C: X) | 0.6 | <0.5 | 18.1 |
| dsDNA (A: X) | 0.7 | 11.2 | 15.2 |
Table 3. Intermediates and Products from the 2 h Incubation of AlkB with α-OH-PdG (A), γ-OH-PdG (B), and M1dG (C).
| (A) | |||||
|---|---|---|---|---|---|
| α-OH-PdG (%) | intermediates
(%) |
product (%) | |||
| X = lesion (α-OH-PdG) | (1a)/(1b) 1239.20 | (2) 1234.70 | (3a)/(3b) 1243.18 | (4) 1238.68 | (6) 1225.20 |
| ssDNA | 74.0 | 18.0 | 5.5 | 2.5 | <0.1 |
| dsDNA (T: X) | 84.4 | 14.0 | 1.3 | 0.3 | <0.1 |
| dsDNA (G: X) | 84.0 | 14.9 | 0.9 | 0.1 | <0.1 |
| dsDNA (C: X) | 84.7 | 14.6 | 0.3 | 0.4 | <0.1 |
| dsDNA (A: X) | 84.2 | 15.1 | 0.6 | <0.1 | <0.1 |
| (B) | |||||
|---|---|---|---|---|---|
| γ-OH-PdG (%) | intermediates
(%) |
product (%) | |||
| X = lesion (γ-OH-PdG) | (5a)/(5b) 1239.17 | (3a)/(3b) 1243.17 | (7) 1234.17 | (4) 1238.67 | (6) 1225.17 |
| ssDNA | 41.2 | 45.0 | 3.5 | 1.3 | 9.0 |
| dsDNA (T: X) | 83.0 | 12.5 | 2.8 | 1.0 | 0.7 |
| dsDNA (G: X) | 89.5 | 9.0 | 0.7 | 0.4 | 0.4 |
| dsDNA (C: X) | 99.9 | <0.1 | <0.1 | <0.1 | <0.1 |
| dsDNA (A: X) | 88.7 | 8.3 | 1.9 | 0.7 | 0.4 |
| (C) | |||||
|---|---|---|---|---|---|
| M1dG (%) | intermediates
(%) |
product (%) | |||
| X = lesion (M1dG) | (7) 1234.19 | (8) 1238.19 | (9)/(13)/(14) 1242.69 | (10) 1247.19 | (6) 1225.19 |
| ssDNA | 69.0 | 8.2 | 6.7 | 7.9 | +8.1 |
| dsDNA (T: X) | 54.5 | 16.7 | 12.3 | 11.9 | 4.6 |
| dsDNA (G: X) | 78.0 | 6.4 | 5.7 | 6.1 | 3.8 |
| dsDNA (C: X) | 81.9 | 7.3 | 6.9 | 2.6 | 1.3 |
| dsDNA (A: X) | 84.8 | 4.4 | 4.1 | 3.8 | 2.9 |
Trapping Experiments of DNA Adducts with PFBHA
Trapping reactions of oligonucleotides with PFBHA were performed in water at room temperature for 1 h using 5 μM DNA and 500 μM PFBHA in a 10 μL incubation volume, followed by HPLC/Q-TOF MS analysis, as described previously.50 The calculated MWs of the trapped intermediates are included in Table S1.
Melting Temperature Analysis
Melting temperatures were measured on the dsDNA obtained by mixing the 16-mer oligonucleotides containing lesions with equimolar amounts of the complementary oligonucleotides containing each of the four possible natural bases (C, T, G, and A) opposite the lesion site. The oligonucleotides (4 μM in a total volume of 25 μL) were mixed in AlkB reaction buffer (45 mM HEPES, pH 8, 0.9 mM α-ketoglutarate, 1.8 mM ascorbate, 67 μM Fe(II)) supplemented with the high-resolution melting dye LCGreen Plus (Idaho Technologies Inc., Salt Lake City, UT) to a final concentration of 1×. Annealing was done by heating at 75 °C for 5 min, followed by a slow cooling (0.1 °C/s) to 4 °C. Melting temperature, Tm, was measured using a Roche LightCycler 480 spectrophotometer, by heating the samples from 37 to 95 °C at a rate of 0.1 °C/s and recording fluorescence five times every degree Celsius. Data analysis was performed using LightCycler480 software. For each oligonucleotide and complement pair, three independent measurements were performed and averaged.
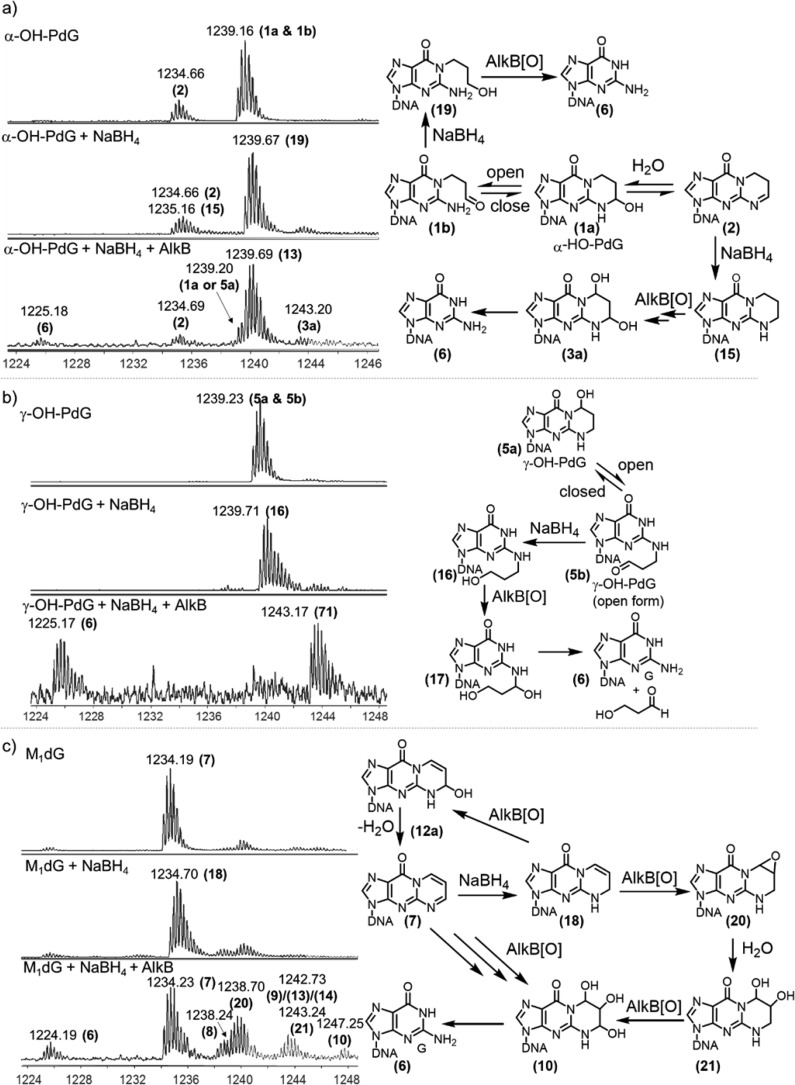
Reduction of Exocyclic dG Lesions by NaBH4 and Their Repair by AlkB
Reduction of exocylic dG lesions (α-OH-PdG, γ-OH-PdG, and M1dG) was performed by incubating ∼1 nmol of 16-mer oligonucleotide in 100 mM potassium phosphate, pH 8.0, with ∼1 M NaBH4 for 1 h at 37 °C in 10 μL reaction volume. To prevent excess pressure from hydrogen gas accumulation from the NaBH4 hydrolysis, the tubes were uncapped and continuously spun in a microfuge during the incubation. Subsequently, the excess NaBH4 and salts were removed by passing the reaction mixtures through homemade dry-spun Sephadex G50 Fine (Amersham Biosciences) columns. After concentration by centrifugation under vacuum, the purified reduced oligonucleotides were used in AlkB reactions using the conditions described previously. All samples (untreated, NaBH4 treated, and NaBH4 then AlkB treated) were analyzed using HPLC-ESI-TOF mass spectrometry.
HPLC/Q-TOF MS and MS/MS Analysis
Oligonucleotide analyses were performed using Agilent Q-TOF 6510 mass spectrometer (Palo Alto, CA) setup as follows: needle voltage, 3.5 kV; nitrogen drying gas, 8 L/min; heated capillary, 340 °C; and nebulizer, 30 psig. The reaction mixtures were separated by HPLC on a Zorbax SB-Aq column (2.1 × 150 mm; 3.5 μm; Agilent Technologies, Palo Alto, CA) at a 0.2 mL/min flow rate at room temperature. Solvents were 10 mM ammonium acetate in water (A) and 100% acetonitrile (B). A linear gradient of 1–18% B over 21 min was used. Data analysis was performed using the Agilent MassHunter workstation software.
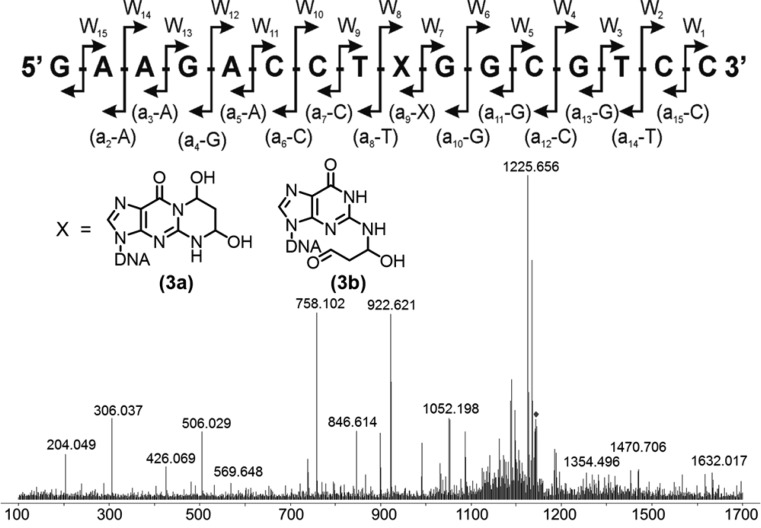
LC–MS/MS analyses were performed on the Agilent 6510 Q-TOF instrument, operated in the negative ion mode with the following parameters: gas temperature, 340 °C; ESI capillary voltage, 3.5 kV; nebulizer pressure, 30 psi; drying nitrogen gas, 8 L/min; and fragmentation energy, 30–35 V. The theoretical fragmentation pattern of a 16-mer oligonucleotide is shown in Figure 2A; the expected monoisotopic masses of the fragments were calculated using Mongo Oligo Mass Calculator, version 2.06 (http://mods.rna.albany.edu/Masspec-Toolbox). A typical fragmentation spectrum (e.g., intermediate 3a/3b) is shown in Figure 2B. The comparison between the calculated and observed m/z values for the MS/MS spectrum of 3a/3b is shown in Figure 2C. MS/MS analyses of the observable reactants, intermediates, and products for all of the AlkB reactions with 16-mer oligonucleotides containing α-OH-PdG, γ-OH-PdG, and M1dG are included in the Supporting Information (Figures S10–S17 and Tables S2–S9).
Figure 2.
MS/MS analysis of prototypic 16-mer oligonucleotide containing the AlkB-oxidized form of γ-OH-PdG. (Top) Predicted collision-induced dissociation (CID) fragmentation pattern of the 16-mer oligonucleotide. X denotes the lesion or repair reaction intermediate or product. (Bottom) MS/MS fragmentation spectrum of 16-mer containing the AlkB-oxidized form of γ-OH-PdG. A comparison of predicted and observed m/z values for the MS/MS fragmentation pattern shown at the bottom of the figure is included in the Supporting Information (Table S6).
Results
Exocyclic Guanine DNA Adducts Are Substrates for AlkB in ssDNA
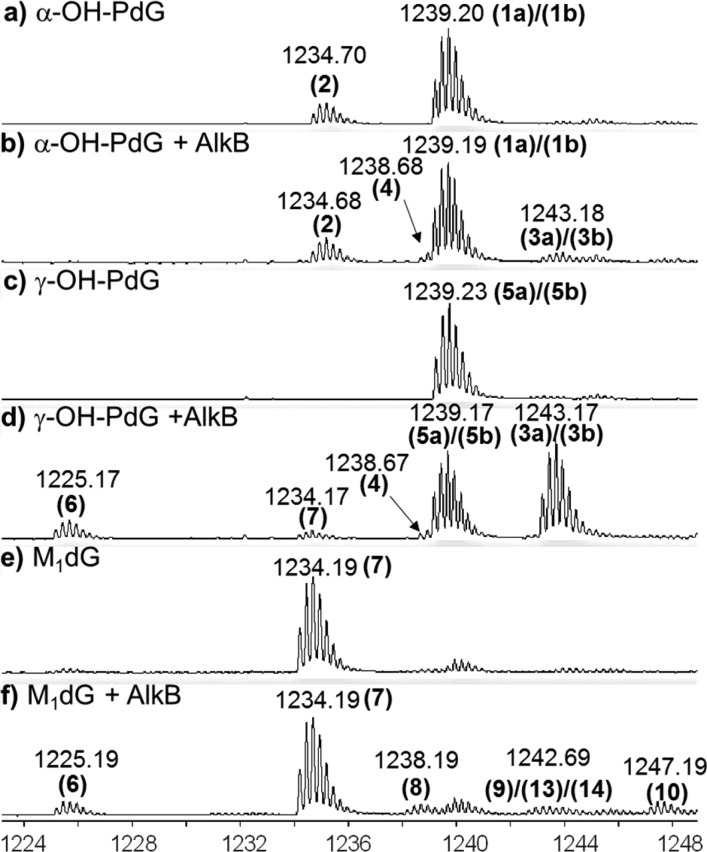
The ability of AlkB to repair acrolein- and MDA-derived exocyclic dG adducts was measured by incubating site-specifically modified 16-mer oligonucleotides with purified AlkB protein. Following a 2 h incubation at 37 °C, the reaction mixtures were analyzed using high-resolution MS.50,52 For each lesion, experiments were conducted in both the presence and absence of the AlkB protein, with all of the necessary cofactors. Figure 3 shows representative MS spectra corresponding to each oligonucleotide containing an exocyclic dG lesion before and after AlkB treatment. The molecular weight (MW) of each of the 16-mer oligonucleotides employed was calculated, from which the −4 charge monoisotopic mass (all 12C, 14N, etc.) was determined (Table 1). For example, the MW of the 16-mer containing the α-OH-PdG lesion is 4960.88 (Da); therefore, its monoisotopic −4 charge state has a theoretical m/z of 1239.21. In this case, an m/z of 1239.20 was observed experimentally (Figure 3a), which correlated well with the theoretical value (Table 1). Because the MS conditions used throughout this study produced robust −4 charge states for all the oligonucleotides analyzed, all of the m/z numbers discussed below refer to −4 charge states, unless otherwise specified. The chemical structures corresponding to the peaks labeled in Figure 3 as well as their proposed AlkB-catalyzed transformations are shown in Figure 4.
Figure 3.
Q-TOF mass spectrometry analysis of reactants and products of the oligonucleotides containing exocyclic guanine lesions incubated with AlkB for 2 h. Data represent the −4 charge envelopes; multiple ion mass peaks associated with each envelope reflect mostly the number of 13C atoms in each −4 charge packet. The monoisotopic peak (all 12C, 14N, etc.) value is labeled above each peak envelope. (a) α-OH-PdG; (b) α-OH-PdG + AlkB; (c) γ-OH-PdG; (d) γ-OH-PdG + AlkB; (e) M1dG; and (f) M1dG + AlkB.
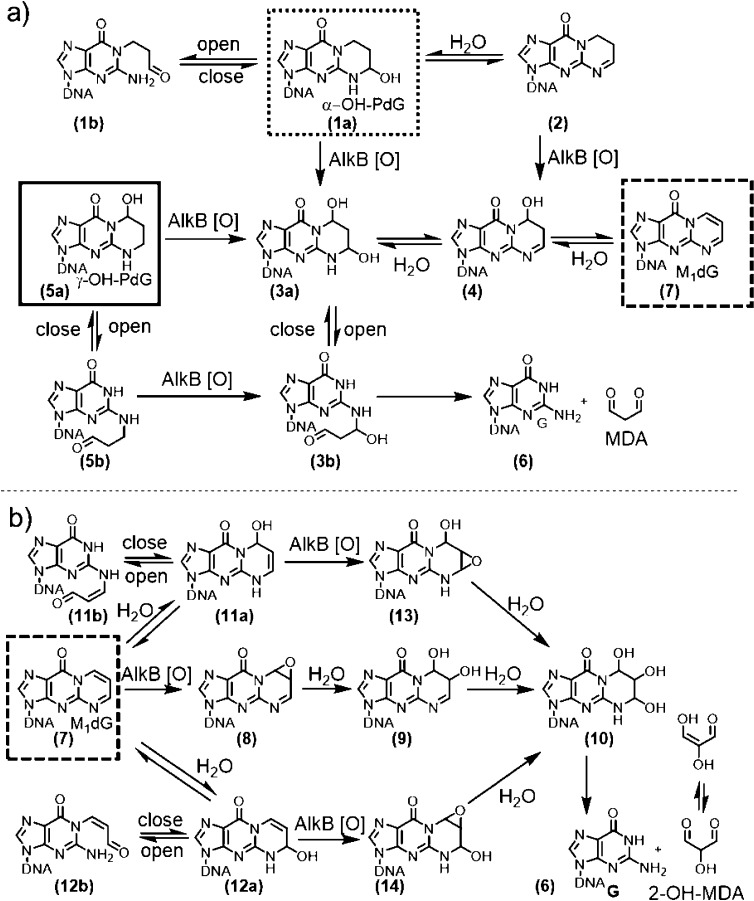
Figure 4.
Chemical structures and proposed pathways for AlkB-mediated exocyclic guanine lesions transformation: (a) α-OH-PdG (dotted border) and γ-OH-PdG (solid border); (b) M1dG (dashed border).
Interestingly, the mass spectrum of α-OH-PdG in the absence of AlkB showed two peaks: a major peak at m/z 1239.20, which corresponds to the 16-mer containing the α-OH-PdG lesion (structure 1a, calculated m/z 1239.21), and a minor peak at m/z 1234.70, which corresponds to a dehydrated form of α-OH-PdG (structure 2, calculated m/z 1234.71) (Figures 3a and 4a). The chemical structure of 2 was confirmed by MS/MS fragmentation analysis (Figures S10–S11 and Tables S2–S3). Upon addition of AlkB protein, two new products were observed with m/z 1238.68 and 1243.18 (Figures 3b, S1a, and S2). The product with mass 1243.18 was assigned to the oxidized form of the parent α-OH-PdG lesion (structure 3a, calculated m/z 1243.21), consistent with the 4 m/z units increase, which in the −4 charge state corresponds to 16 Da, the mass of an oxygen atom. The product with mass 1238.68 is similarly 4 m/z units higher than the dehydrated form at 1234.70, suggesting that it could be an oxidized form of 2 (structure 4, calculated m/z 1238.71) (Figures S1 and S2). However, structure 4 could also arise from dehydration of the oxidized intermediate (structure 3a), being 4.5 m/z units lower than the 1243.18 species (which, in the −4 charge state, corresponds to the loss of a water molecule, 18 Da). These data suggest that, under these reaction conditions, AlkB can oxidize the α-OH-PdG lesion (and its dehydrated form), transforming it into the diol 3a that could spontaneously lose the exocyclic bridge to restore the parental deoxyguanosine (Figure 4a). However, the fully repaired deoxyguanosine product was not observed, perhaps because the amount of oxidized intermediate 3a formed was relatively small, with only 5.5% conversion (Table 3A).
The oligonucleotide containing γ-OH-PdG had an experimental m/z of 1239.23 (structure 5a, calculated m/z 1239.21). Incubation with AlkB resulted in the conversion of about 60% of the γ-OH-PdG starting material into four new species (Figure 3d and Table 2) with m/z values of 1225.17 (dG, structure 6), 1243.17 (diol intermediate 3a), 1238.67 (single-dehydration product 4), and 1234.17 (double-dehydration product 7, M1dG). Relative mass differences between these peaks (Figure S1b) were used to propose chemical structures for all of the intermediates (Figure 4a), which were further confirmed by an MS/MS fragmentation analysis (Figures S12–S14 and Tables S4–S6). The appearance of these intermediates and products suggests the following reaction course for direct reversal of γ-OH-PdG damage: AlkB oxidizes the methylene group adjacent to the N2-position of γ-OH-PdG to generate α,γ-dihydroxy-PdG (3a), which can release MDA to form undamaged guanine (6). The final step of MDA release from 3a is analogous to the release of glyoxal from the εA glycol intermediate that forms when AlkB repairs the εA lesion.46 However, intermediate 3a can also dehydrate to form 4 and, upon loss of another water molecule, intermediate 7, which is the MDA adduct M1dG (Figure 4a). As will be shown later, M1dG is itself a substrate for AlkB oxidation. However, given the small amounts M1dG formed in the reaction of γ-OH-PdG with AlkB, the intermediates of the M1dG reaction with AlkB likely were below the limit of detection and therefore were not observed.
When the oligonucleotide containing M1dG (starting material m/z of 1234.19) was incubated with AlkB under similar conditions, four new mass envelopes were observed in the mass spectrum (Figures 3f and S1c), besides the unreacted starting material. These peaks suggested a reaction course for direct reversal of M1dG damage by AlkB (Figure 4b). Specifically, the mass envelope at m/z 1238.19 was consistent with the AlkB-catalyzed formation of an epoxide for M1dG (structure 8 in Figure 4b, calculated m/z 1238.20, Table 1). Hydrolysis of the epoxide would generate glycol 9, observed at m/z 1242.69, consistent with the calculated m/z 1242.71. The additional species observed at m/z 1247.19 indicated the presence of the trihydroxylated intermediate 10 (calculated m/z 1247.21), which would be the hydration product of glycol 9. The spontaneous loss of 2-hydroxy-MDA from the trihydroxylated intermediate 10 would generate the undamaged guanine base, observed at m/z 1225.19 (calculated m/z 1225.21) (Figures 3f and 4b). The assignment of intermediates was confirmed by MS/MS fragmentation analysis (Figures S15–S17 and Tables S7–S9).
Exocyclic Guanine DNA Adducts Are Substrates for AlkB in dsDNA
AlkB can repair alkylated nucleic acid bases in both ssDNA and dsDNA. While repair in the single-stranded context is typically more efficient,61 there are examples of lesions that are repaired by AlkB equally well in ssDNA and dsDNA.62 Therefore, it was of interest to investigate whether the acrolein- and MDA-derived exocyclic dG lesions were also substrates for AlkB in dsDNA.
AlkB repair reactions were carried out with each of the three exocyclic dG adducts opposite each of the four canonical bases (T, C, G, and A) (Figures S3–S5). The amount of the starting material converted by AlkB (Table 2) was estimated by integrating the extracted ion chromatograms (EIC) of each peak in the mass spectra (see Experimental Procedures for details). Likewise, the relative amounts of the intermediates and repair products formed in each case were also estimated (Table 3A–C). The data show that the efficiency of AlkB-mediated oxidation of exocyclic dG adducts depended not only on the type of lesion (as seen in ssDNA) but also on the nature of the base placed opposite the lesion. In general, the oxidation was less efficient in dsDNA compared with that in ssDNA, with M1dG being a notable exception, for which the maximum conversion of starting material was observed in dsDNA when placed opposite T (Table 2). As a general trend, lesions were more efficiently converted when placed opposite T than opposite any other base. The smallest conversion was observed when lesions were placed opposite C (Table 2).
The quantification of intermediates and fully repaired product for each of the AlkB reactions above provided some additional insights. For γ-OH-PdG, the AlkB conversion was at least 3-fold lower in dsDNA compared with that in ssDNA, and the amount of diol intermediate (structure 3a) formed in the dsDNA reaction was reduced by a similar amount. However, the amount of fully repaired product, dG, was disproportionally reduced (by at least 10-fold with respect to ssDNA, Table 3B), indicating that the final step of MDA release from 3a is impeded when AlkB repair occurs in dsDNA. For M1dG opposite T, the AlkB conversion was higher in dsDNA than that in ssDNA. However, the amount of fully repaired product was maximal in ssDNA (8.1%) compared with that in dsDNA (1.3–4.6%) (Table 3C), suggesting, as before, that the final step of the release of the oxidized exocyclic bridge from intermediate 10 was impeded in dsDNA. For α-OH-PdG in dsDNA, AlkB oxidation was even less efficient than that in ssDNA, with the oxidized intermediates detected at correspondingly lower levels (Table 3A). Just as in ssDNA, no fully repaired dG product was observed in the AlkB reaction of α-OH-PdG in dsDNA. However, it appears that the equilibrium between the hydrated (structure 1a) and dehydrated (structure 2) forms of α-OH-PdG shifted toward the structure 1a in dsDNA.
Cytosine Opposite Exocyclic Guanine Adducts Stabilizes Duplex DNA
The reactions of AlkB with dsDNA consistently showed that when the acrolein- and MDA-derived exocyclic dG adducts were placed opposite C the efficiency of repair was significantly diminished (Table 2). We hypothesize that this effect was due to the ability of exocyclic dG lesions to exist in open-ring structures.63−65 In the case of γ-OH-PdG64 and M1dG,65 the presence of the cognate base pairing partner (dC) in the opposite strand has been shown to induce the opening of the exocyclic ring to expose the Watson–Crick base-pairing side of the damaged guanine and allow it to form hydrogen bonds. To test this hypothesis in our sequence context, the thermal melting temperatures (Tm) of the oligonucleotides containing exocyclic dG adducts opposite all four canonical bases were measured (Table 4). Consistent with this hypothesis and previous observations,64,65 the melting temperatures containing γ-OH-PdG and M1dG opposite C were the highest in each case (as compared to other complementary bases) and approached the Tm of a duplex containing a canonical G:C base pair at the lesion site. When compared with a normal guanine, the presence of M1dG increased the melting temperature of the duplex when opposite T, G, or A. This effect was likely due to the extended aromatic ring system of M1dG, which allowed for additional favorable stacking interactions. By contrast with the other two lesions, the oligonucleotide containing α-OH-PdG was not stabilized opposite any of the four canonical bases. This observation was consistent with the fact that the open-ring structure of α-OH-PdG would allow only the N2 position of guanine to interact with the opposite base, while the N1 position would be still blocked.
Table 4. Melting Temperature of Oligonucleotides Containing α-HOPG, γ-HOPG, M1dG, or G Annealed to a Complementary Strand Containing C, T, G, or A Opposite the Lesion Site.
| oligonucleotides | C | T | G | A |
|---|---|---|---|---|
| G | 63.6 ± 0.3 | 57.1 ± 0.2 | 54.1 ± 0.3 | 56.5 ± 0.3 |
| α-HOPG | 53.0 ± 0.5 | 49.8 ± 0.2 | 53.4 ± 0.2 | 52.8 ± 0.7 |
| γ-HOPG | 59.0 ± 0.3 | 54.5 ± 0.3 | 55.0 ± 0.3 | 54.0 ± 0.3 |
| M1dG | 62.2 ± 0.3 | 60.1 ± 0.3 | 56.6 ± 0.2 | 56.6 ± 0.2 |
PFBHA Trapping Reveals Open-Ring Forms of Exocyclic Guanine DNA Lesions
The γ-OH-PdG63,64,66 and M1dG67−70 lesions have been reported to equilibrate between ring-closed and ring-open forms. In dsDNA, γ-OH-PdG predominantly exists in open-ring form when placed opposite dC,64 whereas in ssDNA, it exists mostly in the closed-ring form. In both cases, γ-OH-PdG could be trapped by peptides, with the conjugation being more effective in dsDNA than in ssDNA.66 The reversible, hydrolytic ring-opening of M1dG has also been well-studied,67−69,65 with the ring-open form of M1dG predominating in dsDNA when M1dG is placed opposite dC.65,67
The presence of open-ring forms of the three exocyclic dG lesions was studied in our sequence context by incubating the site-specifically modified oligonucleotides used in the AlkB repair studies with O-(2,3,4,5,6-pentafluorobenzyl)hydroxylamine (PFBHA), a nucleophilic alkoxyamine that forms stable oxime linkages with free aldehydes.50 After 1 h at room temperature, the reaction mixtures were analyzed using high-resolution MS (Figure 5). The calculated and experimentally observed m/z values for the oligonucleotides trapped with PFBHA are listed in Table S1.
Figure 5.
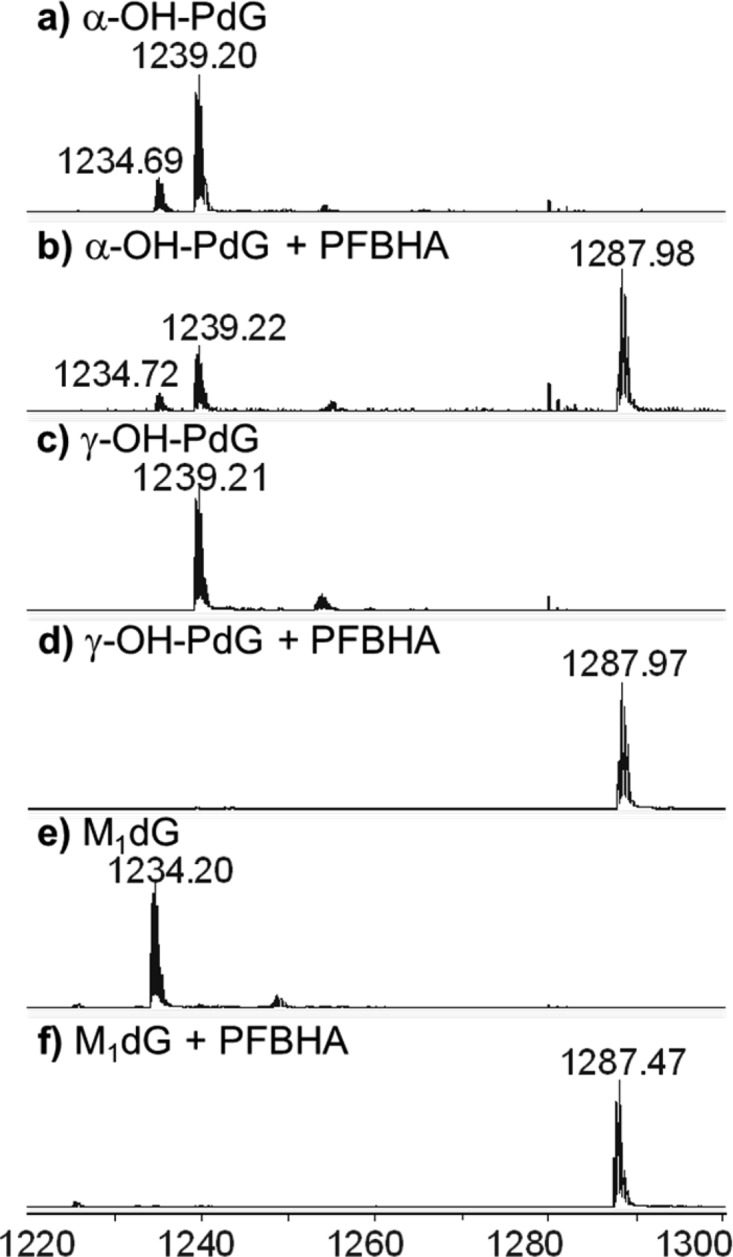
Q-TOF mass spectrometry analysis of the PFBHA trapping reactions of oligonucleotides containing exocyclic guanine lesions. Data represent the −4 charge envelopes, as described in Figure 3. (a) α-OH-PdG; (b) α-OH-PdG + PFBHA; (c) γ-OH-PdG; (d) γ-OH-PdG + PFBHA; (e) M1dG; and (f) M1dG + PFBHA.
All three oligonucleotides containing the exocyclic dG lesions were trapped by PFBHA, indicating that they can potentially exist in open-ring forms containing free aldehyde groups (Figure 5). In the case of γ-OH-PdG, PFBHA trapping provides strong evidence that the exocyclic adduct (5a) exists in equilibrium with an open-ring aldehyde form (5b). For M1dG (7), however, the complete trapping with PFBHA does not unequivocally demonstrate the existence of the ring-open aldehyde forms (11b or 12b), because PFBHA can react directly with the closed-ring form of M1dG and generate the oxime product without the formation of a free aldehyde. This alternative course of reaction is based on the observation that the rate of ring opening of M1dG at neutral pH in water is slower than the rate of trapping of M1dG with hydroxylamine.71
The presence of PFBHA completely trapped the oligonucleotide containing γ-OH-PdG and M1dG. The parent peak of γ-OH-PdG at m/z 1239.21 (calculated m/z 1239.21) shifted quantitatively to a PFBHA-trapped species at m/z 1287.97 (calculated m/z 1287.97). Similarly, the parent peak of M1dG at m/z 1234.20 (calculated m/z 1234.21) was completely converted to a PFBHA-trapped species at m/z 1287.47 (calculated m/z 1287.46). In the case of α-OH-PdG, 1a (observed m/z 1239.20, calculated m/z 1239.21) can form an open-ring structure (1b) that can be trapped with PFBHA. However, as described previously, 1a is also in equilibrium with the imine 2, which could also react with PFBHA to give a trapped product. Only a partial trapping of the α-OH-PdG was observed (PFBHA-trapped species at m/z 1287.98, calculated m/z 1287.97), with the relative amounts of untrapped oligonucleotides suggesting that PFBHA reacted primarily with 1b and not with 2. Assuming PFBHA trapping of 1b is fast, these data also suggest that the formation of the open-ring form of α-OH-PdG is slower than that for γ-OH-PdG.
The Exocyclic dG Adducts Reduced with NaBH4 Are Also Substrates for AlkB
Given the presence of the open-ring forms of the three exocyclic dG lesions, it was of interest to investigate whether AlkB could also act on the open-ring forms. Sodium borohydride (NaBH4) buffered with sodium phosphate was used to lock the open-ring forms by reducing the open-ring aldehydes to alcohols,72 which were detected using high-resolution MS (Figure 6). For γ-OH-PdG, upon treatment with NaBH4, the parent peak at m/z 1239.23 was quantitatively converted to a new peak at m/z 1239.71, consistent with the addition of two hydrogens. Because there are no functional groups reducible by NaBH4 in the closed-ring structure of γ-OH-PdG, or on any other normal base, the peak at m/z 1239.71 must correspond to the open-ring alcohol of γ-OH-PdG, structure 16 (calculated m/z 1239.72) (Figures 6b and S7). Similarly, when the oligonucleotide containing α-OH-PdG was treated with NaBH4, the parent peak at m/z 1239.16 shifted to m/z 1239.67 (Figures 6a and S6). Similar to the argument made for γ-OH-PdG, the only functional group that could be reduced by NaBH4 is the aldehyde of the open-ring form of α-OH-PdG (structure 1b, Figure 4a) to generate an open-ring alcohol (structure 19, calculated m/z 1239.67). The dehydrated form of α-OH-PdG (structure 2), a cyclic imine, was also reduced by NaBH4, as suggested by the change in the isotopic distribution pattern and the expansion of the peak envelope toward higher masses (Figure 6a). These changes are consistent with the appearance of a peak at m/z 1235.16 (Figures 6a and S6), which presumably corresponded to a fully saturated 1,N2-propano-dG (structure 15, calculated m/z 1235.21) (Figure 6a). Both reduction products (19 and 15) are consistent with the reported NaBH4 reduction of the α-OH-PdG free nucleoside.54 Additional evidence for the formation of 15 was obtained by running the NaBH4 reaction at a lower pH (5.8 instead of 7.0), which improved the yield of reduction of 2 (Figure S9). However, even at low pH, reduction of 2 was not quantitative, perhaps due to the lesser reactivity of the aromatically stabilized cyclic imine toward NaBH4 compared to that of an aldehyde.
Figure 6.
Q-TOF mass spectrometry analysis of the reactions of oligonucleotides containing exocyclic guanine lesions with NaBH4 and the subsequent incubation of the reduced forms with AlkB for 2 h. Each peak is labeled with its monoisotopic m/z value, in the −4 charge state. The corresponding chemical structures and proposed transformations are included next to each set of mass spectrometry ion traces. (a) (Top) α-OH-PdG, (Middle) α-OH-PdG + NaBH4, (Bottom) α-OH-PdG + NaBH4 + AlkB. (b) (Top) γ-OH-PdG, (Middle) γ-OH-PdG + NaBH4, (Bottom) γ-OH-PdG + NaBH4 + AlkB. (c) (Top) M1dG, (Middle) M1dG + NaBH4, (Bottom) M1dG + NaBH4 + AlkB.
When exposed to NaBH4, the M1dG-containing oligonucleotide (observed m/z 1234.19, calculated m/z 1234.21) was converted to a reduced species detected at m/z 1234.70 (Figures 6c and S8). This product has been previously reported to be structure 18 (Figure 6c).73,74 The reduced ring-open form of M1dG requires the initial addition of a water molecule to produce 11a or 12a (calculated m/z 1238.71) (Figure 4b), which could subsequently be reduced with NaBH4. However, the open-ring form of M1dG is known to enolize readily and thus is not very reactive toward NaBH4.75 Consistent with this fact, a product with an m/z of 1239.21 corresponding to a reduced version of 11a or 12a could not be detected (Figures 6c and S8); therefore, under the reaction conditions, M1dG could not be trapped as an open-ring species by NaBH4. Nevertheless, the reaction of AlkB with the reduced form of M1dG provided additional insights into the regioselectivity of the AlkB repair reactions on exocyclic dG structures.
After NaBH4 treatment, the oligonucleotides were first purified using Sephadex G-50 columns to remove excess NaBH4 and other salts and were then allowed to react with AlkB under the same conditions as those previously described. AlkB produced more fully repaired product (dG) when incubated with NaBH4-treated lesions as compared to the incubations with untreated lesions. For α-OH-PdG, these reaction conditions produced detectable levels of fully repaired dG (6). The amount of the reduced open-ring alcohol 19 did not change upon treatment with AlkB, whereas the amount of 15 was diminished in the AlkB reaction (Figure 6a). These observations indicated that the fully repaired dG was not generated from the reduced open-ring form 19, but rather from the 1,N2-propano-dG reduced intermediate (15) via two successive oxidations by AlkB, which would generate 3a (Figure 6a).
For the reduced ring-open form of γ-OH-PdG (16), the AlkB reaction intermediates were consistent with oxidation at the carbon attached to the N2 position of guanine with subsequent release of a three-carbon aldehyde to generate fully repaired dG (Figure 6b). This reaction was very efficient, with most of the starting material being consumed along with a high yield (∼50%) of dG. For NaBH4-treated M1dG, the AlkB reaction generated several intermediates (Figure 6c), consistent with oxidation of 18 at the carbon attached to the N2 position of guanine to generate 12a, which upon dehydration yielded the original M1dG lesion (structure 7, observed m/z 1234.23). Subsequently, M1dG (7) reacted with AlkB, as shown before (Figures 3f and 4b), to generate epoxide 8, triol 10, and, upon spontaneous loss of 2-hydroxy-MDA, the fully repaired product, dG. In an alternative reaction course, 18 could react with AlkB to generate epoxide 20 and upon hydration, diol 21 (Figure 6c). Subsequently, 21 can be further oxidized by AlkB to generate triol 10, leading to additional repair product dG. These additional reaction pathways could explain the improved yield of fully repaired product, dG, when NaBH4-treated M1dG was incubated with AlkB.
Discussion
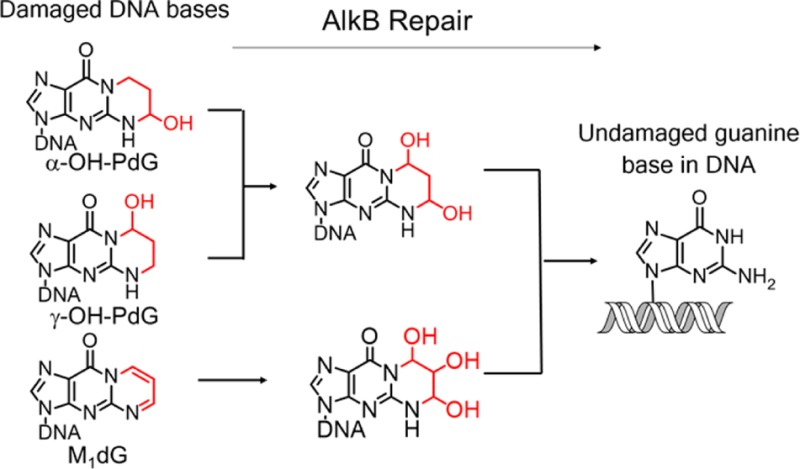
The three exocyclic dG lesions investigated (i.e., α-OH-PdG, γ-OH-PdG, and M1dG) are important biomarkers of exposure to ubiquitous environmental chemicals5 and inflammation-induced lipid peroxidation byproducts,13,14,37 acrolein and MDA. The present study demonstrates that the exocyclic dG lesions are processed by AlkB by a complex but chemically understandable biochemical pathway. Among the three lesions studied in ssDNA, γ-OH-PdG was repaired most efficiently, followed by M1dG and α-OH-PdG (Table 2 and Table 3A–C). The overall AlkB efficiency at repairing the lesions was generally lower in dsDNA than that in ssDNA, as evidenced by the lower amounts of fully repaired product formed in each case (Table 3A–C). However, in the one case, when M1dG was opposite T, the total amount of starting material consumed was higher than in the incubation of AlkB with M1dG in ssDNA. The base placed opposite the lesion also influenced the repair reactions. Generally, when the lesions were placed opposite T, the repair was most efficient, with C being the opposite base that gave the lowest conversions (Tables 2 and 3A–C). These observations can be rationalized by the base-flipping mechanism proposed for AlkB,76 in which a mispaired base (i.e., lesion opposite T) is more likely to splay out into the enzyme active site to be repaired. By contrast, when the exocyclic dG lesions are placed opposite C, a “correct base pair” is formed (in the case of γ-OH-PdG and M1dG, via their open-ring forms), which hinders base-flipping, effectively hiding the lesion from AlkB and reducing the repair efficiency. This mechanism is consistent with our previous study of AlkB activity on N2-dG alkyl lesions,52 such as m2G, e2G, and the bulkier N2-furfuryl-dG and N2-tetrahydrofurfuryl-dG, which are good AlkB substrates in ssDNA but are poorly repaired when placed in dsDNA opposite dC.52 Furthermore, the preference of AlkB for repairing exocyclic dG lesions in ssDNA suggests that transcriptionally active regions of the genome are likely to be most protected by AlkB activity, which complements other repair pathways that target exocyclic dG lesions only in dsDNA contexts (i.e., NER).23
All three exocyclic dG lesions studied have been proposed to exist in equilibrium with open-ring forms.33,63,64 The trapping of the α-OH-PdG and γ-OH-PdG with PFBHA (Figure 5) suggests that there is a low but kinetically useful concentration of the ring-opened aldehyde form in equilibrium with the closed-ring form. Subsequently, by using NaBH4 to reduce the aldehydes and trap the open-ring forms of α-OH-PdG and γ-OH-PdG, the activity of AlkB on the open-ring forms was also investigated. N2-(3-Hydroxypropyl)-G, the NaBH4-reduced open-ring form of γ-OH-PdG, was shown to be a good substrate for AlkB repair, even better than the closed-ring γ-OH-PdG substrate (Figure 6b). The reduced open-ring form of α-OH-PdG, N1-(3-hydroxypropyl)-G, was not significantly oxidized by AlkB; instead, the reduced version of the dehydrated α-OH-PdG, 1,N2-propano-dG (structure 15), reacted with AlkB (Figure 6a).
For M1dG, however, the PFBHA trapping does not necessarily support the existence of the ring-opened aldehyde form, because the trapped product can also be formed by a direct reaction of PFBHA amine at the carbon adjacent to N1 of M1dG.65,75 Consistent with this argument, M1dG incubation with NaBH4 did not generate a measurable amount of trapped open-ring forms; the reduction yielded the closed-ring product 18, which was also a good substrates for AlkB, on par with the parent species M1dG. Taken together, the trapping experiments demonstrated that AlkB can process both open-ring species (such as the open-ring of γ-OH-PdG) and closed-ring species (such as the ones derived from M1dG). However, due to equilibria between open- and closed-ring forms, the AlkB repair reactions of exocyclic dG lesions are complex, involving overlapping pathways with multiple convergence points (Figure 4).
Given the complexity of the repair pathways (Figure 4), it is challenging to establish unambiguously which reaction pathway (i.e., oxidation of open- or closed-ring intermediates) is predominant. Specifically, is AlkB processing the exocyclic dG lesions by directly oxidizing the closed-ring forms or is AlkB reacting primarily with the open-ring forms, essentially simpler N1- and N2-alkyl guanines that are known to be AlkB substrates? For α-OH-PdG, the data were more consistent with the AlkB oxidation on the closed-ring form. When this substrate was reduced to the open-ring form with NaBH4, no corresponding AlkB oxidation intermediate was detected; the AlkB reaction presumably occurred only with the reduced dehydrated version of α-OH-PdG (structure 2), which is a closed-ring form (Figure 6a). A possible explanation for this preference may be that the open-ring form of α-OH-PdG generated a bulky alkyl group on the N1 position of guanine, which did not fit in the active site of AlkB, unlike a smaller N1-methyl-dG substituent, which is repaired by AlkB.51
For γ-OH-PdG, the data suggest that both the closed- and open-ring forms could be substrates for AlkB. We demonstrated that AlkB more efficiently repaired the reduced, open-ring form of γ-OH-PdG than the unreduced, closed-ring γ-OH-PdG lesion. This observation is consistent with previous work, which demonstrated that AlkB can repair guanines substituted at N2 positions with alkyl groups of varying sizes, including bulky alkyl substitutents.52 It is noted, however, that the observed repair of the open-ring form does not rule out the possibility that AlkB can additionally oxidize the closed-ring form of γ-OH-PdG (i.e., the 5a to 3a step of Figure 4a).
The data on the AlkB repair of M1dG lesion are most consistent with the formation of an epoxide intermediate (8) of the closed-ring form, which subsequently gets hydrated twice to form triol 10. Intermediate 10 can also be obtained from the oxidation and hydration of intermediates 11a or 12a, which are hydrated forms of M1dG. However, the presence of these hydrated forms was not detectable in the starting material, and their reduced forms were not observed in the NaBH4 reaction (Figure 6c). Therefore, the alternative pathways involving 11a and 12a are expected to play a minor role, if any, in the AlkB reaction (Figure 4b). The data on M1dG repair by AlkB suggest that the enzyme, similar to its mechanism on εA, primarily acts on the closed-ring form of the substrate.
The AlkB repair data on exocyclic dG lesions also provided some insight on the AlkB selectivity between oxidation at N1- or N2-attached carbons, in the context of an exocyclic propano adduct. By placing the three-carbon exocycle into a published AlkB structure,76 it was determined that the exocyclic carbon atoms adjacent to the N1 and N2 positions are situated at similar distances relative to the iron center of AlkB (Figure S19). This suggests that AlkB, in principle, should oxidize an exocyclic propano adduct equally well at either the N1 or N2 position. However, in the present study, AlkB repaired the two OH-PdG lesions with very different efficiencies (Table 2), with γ-OH-PdG being repaired more efficiently than α-OH-PdG. One explanation for the different AlkB reactivity toward these isomeric lesions may be the propensity to form open-ring species. The open-ring form of α-OH-PdG shifts the entire alkyl substituent to the N1 position, which likely cannot be accommodated in the AlkB active site; by contrast, the open-ring form of γ-OH-PdG contains the alkyl substituent at N2, which can swivel away from the protein to potentially reduce steric clashes. On the basis of these considerations, we speculate that the most likely reaction course of the fully reduced exocyclic propano-dG (16) with AlkB involves oxidations first at the N1 carbon (γ-position) and subsequently at the N2 carbon (α-position) to generate the fully repaired dG.
Compared with other known exocyclic AlkB substrates, such as εA and EA, the acrolein- and MDA-derived dG lesions are repaired less efficiently under similar conditions.46,49,50 One possible explanation is that three-carbon bridges are bulkier than two-carbon bridges and some of them (i.e., α-OH-PdG, γ-OH-PdG) can form puckered structures that are harder to accommodate in the active site of the enzyme. Furthermore, the position of a three-carbon exocycle between N1 and N2 of guanine could be suboptimal in the AlkB active site compared with the two-carbon bridge between N6 and N1 of adenine. Finally, AlkB has been demonstrated to prefer alkylated adenines compared to alkylated guanines; specifically, N1-methyl-adenine is repaired significantly more efficiently than N1-methyl-guanine.51 This difference in reactivity is believed to be due to the fact that N1-methyl-adenine has a positive charge on N1, whereas N1-methyl-guanine is a neutral species. Maciejewska et al. proposed that, in general, the preference of AlkB toward positively charged lesions is likely due to the presence of negatively charged aspartate residue (Asp-135) in the active site.53 Therefore, it is not surprising that this trend extends to exocyclic alkyl lesions, where the exocyclic guanines in the present study are repaired less efficiently by AlkB than exocyclic adenines, in part because they are neutral species. Furthermore, the higher repair efficiency of AlkB when acting on positively charged lesions could also be explained by the propensity of the AlkB oxidation intermediates of such lesions to decompose and generate the fully repaired base. Consequently, it is anticipated that AlkB oxidation intermediates with pKa’s of the base nitrogens altered in a direction that favors protonation would convert more efficiently to the fully repaired product. However, in the case of the exocyclic dG lesions, the AlkB oxidation intermediates are generally more abundant than fully repaired dG, suggesting that the pKa’s of the N1 or N2 nitrogen of the intermediates are not altered significantly during the AlkB reactions.
The present work established the chemical competence of the E. coli dioxygenase AlkB to oxidize and repair acrolein- and MDA-derived exocyclic guanine lesions, suggesting that one role of AlkB may be to alleviate the mutagenic and toxic consequences of reactive aldehydes derived from these primary toxicants. Additionally, the ability of AlkB to repair the open-ring forms of exocyclic guanine lesions suggests that AlkB may also modulate the toxicity and mutagenicity associated with the formation of inter- and intrastrand cross-links30−33 generated by the open-ring forms of the acrolein- and MDA-derived exocyclic guanine lesions. Given that AlkB has nine mammalian homologues77,78 (ABH1–8, FTO), it is anticipated that these repair pathways may also be operating in mammalian cells; however, the extent to which these pathways play a role in modulating the mutagenic and carcinogenic risk associated with exposure to acrolein and inflammation-derived MDA is not known and remains to be established.
Acknowledgments
We thank Professor Catherine Drennan for providing the AlkB expression plasmid, Dr. Koli Taghizadeh for helpful mass spectrometry discussions, Dr. John Wishnok for insightful comments on the manuscript, and Dr. Agnes Rinaldo-Matthis at Karolinska Institute, Stockholm, for help with modeling M1dG in the active site of AlkB. We also thank the MIT Center for Environmental Health Sciences Bioanalytical Core Facility and the Wishnok/Tannenbaum Mass Spectrometry Laboratory for providing access to mass spectrometers.
Glossary
Abbreviations
- Q-TOF
quadrupole time-of-flight
- MS/MS
tandem MS
- dG
deoxyguanosine
- EIC
extracted ion chromatogram
- PFBHA
(pentafluorobenzyl)hydroxylamine
- α-OH-PdG
3-(2′-deoxy-β-d-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-6-hydroxypyrimido[1,2-α]purin-10(3H)-one
- γ-OH-PdG
3-(2′-deoxy-β-d-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2-α]purin-10(3H)-one)
- M1dG
3-(2′-deoxy-β-d-erythro-pentofuranosyl) pyrimido[1,2-α]purin-10(3H)-one
- MDA
malondialdehyde
- NaBH4
sodium borohydride
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
Supporting Information Available
Eighteen figures and nine tables, including MS/MS analyses for oligonucleotides containing α-OH-PdG, γ-OH-PdG, and M1dG and their AlkB reaction intermediates and products; the effect of pH on NaBH4 reduction reactions; and a model of M1dG in the active site of AlkB. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# (J.C.D.) Visterra Inc., Cambridge, MA 02139
Author Contributions
⊥ V.S., B.I.F., and D.L. contributed equally to this work.
This work was supported by National Institutes of Health grant nos. R01 CA080024 (J.M.E), R01 CA26731 (J.M.E.), R01 CA87819 (L.J.M.), P01 ES05355 (C.J.R.), P01 CA160032 (C.J.R.), and Center grant nos. P30 ES02109 (J.M.E.), P30 ES00267 (C.J.R. and L.J.M.), and P30 CA068485 (C.J.R. and L.J.M.). V.S. was supported by NIH Training Grant T32 ES007020.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bein K.; Leikauf G. D. (2011) Acrolein—a pulmonary hazard. Mol. Nutr. Food Res. 55, 1342–1360. [DOI] [PubMed] [Google Scholar]
- Hesterberg T. W.; Lapin C. A.; Bunn W. B. (2008) A comparison of emissions from vehicles fueled with diesel or compressed natural gas. Environ. Sci. Technol. 42, 6437–6445. [DOI] [PubMed] [Google Scholar]
- Jaganjac M.; Prah I. O.; Cipak A.; Cindric M.; Mrakovcic L.; Tatzber F.; Ilincic P.; Rukavina V.; Spehar B.; Vukovic J. P.; Telen S.; Uchida K.; Lulic Z.; Zarkovic N. (2012) Effects of bioreactive acrolein from automotive exhaust gases on human cells in vitro. Environ. Toxicol. 27, 644–652. [DOI] [PubMed] [Google Scholar]
- Smith D.; Cheng P.; Spanel P. (2002) Analysis of petrol and diesel vapour and vehicle engine exhaust gases using selected ion flow tube mass spectrometry. Rapid Commun. Mass Spectrom. 16, 1124–1134. [DOI] [PubMed] [Google Scholar]
- Stevens J. F.; Maier C. S. (2008) Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 52, 7–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett L. J.; Hurd H. K.; Hollstein M. C.; Levin D. E.; Esterbauer H.; Ames B. N. (1985) Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat. Res. 148, 25–34. [DOI] [PubMed] [Google Scholar]
- Curren R. D.; Yang L. L.; Conklin P. M.; Grafstrom R. C.; Harris C. C. (1988) Mutagenesis of xeroderma pigmentosum fibroblasts by acrolein. Mutat. Res. 209, 17–22. [DOI] [PubMed] [Google Scholar]
- Smith R. A.; Cohen S. M.; Lawson T. A. (1990) Acrolein mutagenicity in the V79 assay. Carcinogenesis 11, 497–498. [DOI] [PubMed] [Google Scholar]
- Kawanishi M.; Matsuda T.; Nakayama A.; Takebe H.; Matsui S.; Yagi T. (1998) Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 417, 65–73. [DOI] [PubMed] [Google Scholar]
- Wang H.-T.; Zhang S.; Hu Y.; Tang M.-S. (2009) Mutagenicity and sequence specificity of acrolein–DNA adducts. Chem. Res. Toxicol. 22, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.; Hu W.; Hu Y.; Tang M. (2006) Acrolein is a major cigarette-related lung cancer agent: preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U.S.A. 103, 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.; Wang H.; Hu Y.; Chen W.-S.; Akao M.; Feng Z.; Hu W. (2011) Acrolein induced DNA damage, mutagenicity and effect on DNA repair. Mol. Nutr. Food Res. 55, 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U.; Bartsch H.; Nair J. (2007) Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radical Biol. Med. 43, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Burcham P. C. (1998) Genotoxic lipid peroxidation products: their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis 13, 287–305. [DOI] [PubMed] [Google Scholar]
- Galliani G.; Pantarotto C. (1983) The reaction of guanosine and 2′-deoxyguanosine with acrolein. Tetrahedron Lett. 24, 4491–4492. [Google Scholar]
- Chung F. L.; Young R.; Hecht S. S. (1984) Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 44, 990–995. [PubMed] [Google Scholar]
- Zhang S.; Villalta P. W.; Wang M.; Hecht S. S. (2007) Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 20, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I. Y.; Hossain M.; Miller H.; Khullar S.; Johnson F.; Grollman A.; Moriya M. (2001) Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 276, 9071–9076. [DOI] [PubMed] [Google Scholar]
- Yang I.-Y.; Johnson F.; Grollman A. P.; Moriya M. (2002) Genotoxic mechanism for the major acrolein-derived deoxyguanosine adduct in human cells. Chem. Res. Toxicol. 15, 160–164. [DOI] [PubMed] [Google Scholar]
- VanderVeen L. A.; Hashim M. F.; Shyr Y.; Marnett L. J. (2003) Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 100, 14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Zhu M.; Xie J. (2010) Mutagenicity of acrolein and acrolein-induced DNA adducts. Toxicol. Mech. Methods 20, 36–44. [DOI] [PubMed] [Google Scholar]
- Cohen S. M.; Garland E. M.; St. John M.; Okamura T.; Smith R. A. (1992) Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 52, 3577–3581. [PubMed] [Google Scholar]
- Wang H.-T.; Hu Y.; Tong D.; Huang J.; Gu L.; Wu X.-R.; Chung F.-L.; Li G.-M.; Tang M. (2012) Effect of carcinogenic acrolein on DNA repair and mutagenic susceptibility. J. Biol. Chem. 287, 12379–12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I.-Y.; Chan G.; Miller H.; Huang Y.; Torres M. C.; Johnson F.; Moriya M. (2002) Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry 41, 13826–13832. [DOI] [PubMed] [Google Scholar]
- Sanchez A. M.; Minko I. G.; Kurtz A. J.; Kanuri M.; Moriya M.; Lloyd R. S. (2003) Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived alpha-HOPdG and gamma-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 16, 1019–1028. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Washington M. T.; Kanuri M.; Prakash L.; Prakash S.; Lloyd R. S. (2003) Translesion synthesis past acrolein-derived DNA adduct, gamma-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J. Biol. Chem. 278, 784–790. [DOI] [PubMed] [Google Scholar]
- Washington M. T.; Minko I. G.; Johnson R. E.; Wolfle W. T.; Harris T. M.; Lloyd R. S.; Prakash S.; Prakash L. (2004) Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol. Cell. Biol. 24, 5687–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Yamanaka K.; Kozekov I. D.; Kozekova A.; Indiani C.; O’Donnell M. E.; Jiang Q.; Goodman M. F.; Rizzo C. J.; Lloyd R. S. (2008) Replication bypass of the acrolein-mediated deoxyguanine DNA–peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 21, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuri M.; Minko I. G.; Nechev L. V.; Harris T. M.; Harris C. M.; Lloyd R. S. (2002) Error prone translesion synthesis past gamma-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 277, 18257–18265. [DOI] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Sanchez A.; Harris C. M.; Lloyd R. S.; Harris T. M. (2001) Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem. Res. Toxicol. 14, 1482–1485. [DOI] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Moseley M. S.; Harris C. M.; Rizzo C. J.; Stone M. P.; Harris T. M. (2003) DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 125, 50–61. [DOI] [PubMed] [Google Scholar]
- Sanchez A. M.; Kozekov I. D.; Harris T. M.; Lloyd R. S. (2005) Formation of inter- and intrastrand imine type DNA–DNA cross-links through secondary reactions of aldehydic adducts. Chem. Res. Toxicol. 18, 1683–1690. [DOI] [PubMed] [Google Scholar]
- Stone M. P.; Cho Y.-J.; Huang H.; Kim H.-Y.; Kozekov I. D.; Kozekova A.; Wang H.; Minko I. G.; Lloyd R. S.; Harris T. M.; Rizzo C. J. (2008) Interstrand DNA cross-links induced by alpha,beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 41, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Lao Y.; Yang I.-Y.; Hecht S. S.; Moriya M. (2006) Replication-coupled repair of crotonaldehyde/acetaldehyde-induced guanine-guanine interstrand cross-links and their mutagenicity. Biochemistry 45, 12898–12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A.; Minko I. G.; Harbut M. B.; Finkel S. E.; Goodman M. F.; Lloyd R. S. (2008) Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J. Biol. Chem. 283, 27433–27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Harbut M. B.; Kozekov I. D.; Kozekova A.; Jakobs P. M.; Olson S. B.; Moses R. E.; Harris T. M.; Rizzo C. J.; Lloyd R. S. (2008) Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 283, 17075–17082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch H.; Nair J. (2005) Accumulation of lipid peroxidation-derived DNA lesions: potential lead markers for chemoprevention of inflammation-driven malignancies. Mutat. Res. 591, 34–44. [DOI] [PubMed] [Google Scholar]
- Basu A. K.; O’Hara S. M.; Valladier P.; Stone K.; Mols O.; Marnett L. J. (1988) Identification of adducts formed by reaction of guanine nucleosides with malondialdehyde and structurally related aldehydes. Chem. Res. Toxicol. 1, 53–59. [DOI] [PubMed] [Google Scholar]
- Otteneder M. B.; Knutson C. G.; Daniels J. S.; Hashim M.; Crews B. C.; Remmel R. P.; Wang H.; Rizzo C.; Marnett L. J. (2006) In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc. Natl. Acad. Sci. U.S.A. 103, 6665–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson C. G.; Akingbade D.; Crews B. C.; Voehler M.; Stec D. F.; Marnett L. J. (2007) Metabolism in vitro and in vivo of the DNA base adduct, M1G. Chem. Res. Toxicol. 20, 550–557. [DOI] [PubMed] [Google Scholar]
- Fink S. P.; Reddy G. R.; Marnett L. J. (1997) Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 94, 8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T.; Holbrook N. J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. [DOI] [PubMed] [Google Scholar]
- Sedgwick B.; Bates P. A.; Paik J.; Jacobs S. C.; Lindahl T. (2007) Repair of alkylated DNA: recent advances. DNA Repair 6, 429–442. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. (2004) Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157. [DOI] [PubMed] [Google Scholar]
- Aas P. A.; Otterlei M.; Falnes P. O.; Vågbø C. B.; Skorpen F.; Akbari M.; Sundheim O.; Bjørås M.; Slupphaug G.; Seeberg E.; Krokan H. E. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421, 859–863. [DOI] [PubMed] [Google Scholar]
- Delaney J. C.; Smeester L.; Wong C.; Frick L. E.; Taghizadeh K.; Wishnok J. S.; Drennan C. L.; Samson L. D.; Essigmann J. M. (2005) AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 12, 855–860. [DOI] [PubMed] [Google Scholar]
- Mishina Y.; Yang C.-G.; He C. (2005) Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 127, 14594–14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska A. M.; Ruszel K. P.; Nieminuszczy J.; Lewicka J.; Sokołowska B.; Grzesiuk E.; Kuśmierek J. T. (2010) Chloroacetaldehyde-induced mutagenesis in Escherichia coli: the role of AlkB protein in repair of 3,N4-ethenocytosine and 3,N4-alpha-hydroxyethanocytosine. Mutat. Res. 684, 24–34. [DOI] [PubMed] [Google Scholar]
- Frick L. E.; Delaney J. C.; Wong C.; Drennan C. L.; Essigmann J. M. (2007) Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl. Acad. Sci. U.S.A. 104, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Delaney J. C.; Page C. M.; Yang X.; Chen A. S.; Wong C.; Drennan C. L.; Essigmann J. M. (2012) Exocyclic Carbons Adjacent to the N6 of Adenine are Targets for Oxidation by the Escherichia coli Adaptive Response Protein AlkB. J. Am. Chem. Soc. 134, 8896–8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (2004) Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 14051–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Fedeles B. I.; Shrivastav N.; Delaney J. C.; Yang X.; Wong C.; Drennan C. L.; Essigmann J. M. (2013) Removal of N-alkyl modifications from N2-alkylguanine and N4-alkylcytosine in DNA by the adaptive response protein AlkB. Chem. Res. Toxicol. 26, 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska A. M.; Poznanski J.; Kaczmarska Z.; Krowisz B.; Nieminuszczy J.; Polkowska-Nowakowska A.; Grzesiuk E.; Kusmierek J. T. (2013) AlkB dioxygenase preferentially repairs protonated substrates: specificity against exocyclic adducts and molecular mechanism of action. J. Biol. Chem. 288, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechev L. V.; Kozekov I. D.; Brock A. K.; Rizzo C. J.; Harris T. M. (2002) DNA adducts of acrolein: site-specific synthesis of an oligodeoxynucleotide containing 6-hydroxy-5,6,7,8-tetrahydropyrimido[1,2-a]purin-10(3H)-one, an acrolein adduct of guanine. Chem. Res. Toxicol. 15, 607–613. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Cecilia Torres M.; Iden C. R.; Johnson F. (2003) Synthesis of the minor acrolein adducts of 2′-deoxyguanosine and their generation in oligomeric DNA. Bioorg. Chem. 31, 136–148. [DOI] [PubMed] [Google Scholar]
- Khullar S.; Varaprasad C. V.; Johnson F. (1999) Postsynthetic generation of a major acrolein adduct of 2′-deoxyguanosine in oligomeric DNA. J. Med. Chem. 42, 947–950. [DOI] [PubMed] [Google Scholar]
- Nechev L. V.; Harris C. M.; Harris T. M. (2000) Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem. Res. Toxicol. 13, 421–429. [DOI] [PubMed] [Google Scholar]
- Reddy G. R.; Marnett L. J. (1995) Synthesis of an oligodeoxyribonucleotide containing the alkaline labile malondialdehyde–deoxyguanosine adduct pyrimido[1,2-a]purin-10(3H)-one. J. Am. Chem. Soc. 117, 5007–5008. [Google Scholar]
- Caruthers M. H.; Barone A. D.; Beaucage S. L.; Dodds D. R.; Fisher E. F.; McBride L. J.; Matteucci M.; Stabinsky Z.; Tang J. Y. (1987) Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol. 154, 287–313. [DOI] [PubMed] [Google Scholar]
- Yang C.-G.; Garcia K.; He C. (2009) Damage detection and base flipping in direct DNA alkylation repair. ChemBioChem 10, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav N.; Li D.; Essigmann J. M. (2010) Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis 31, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Santos C.; Zaliznyak T.; Johnson F. (2001) NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct gamma-OH-1-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 276, 9077–9082. [DOI] [PubMed] [Google Scholar]
- Cho Y.-J.; Kim H.-Y.; Huang H.; Slutsky A.; Minko I. G.; Wang H.; Nechev L. V.; Kozekov I. D.; Kozekova A.; Tamura P.; Jacob J.; Voehler M.; Harris T. M.; Lloyd R. S.; Rizzo C. J.; Stone M. P. (2005) Spectroscopic characterization of interstrand carbinolamine cross-links formed in the 5′-CpG-3′ sequence by the acrolein-derived gamma-OH-1,N2-propano-2′-deoxyguanosine DNA adduct. J. Am. Chem. Soc. 127, 17686–17696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim M. F.; Riggins J. N.; Schnetz-Boutaud N.; Voehler M.; Stone M. P.; Marnett L. J. (2004) In vitro bypass of malondialdehyde–deoxyguanosine adducts: differential base selection during extension by the Klenow fragment of DNA polymerase I is the critical determinant of replication outcome. Biochemistry (Moscow) 43, 11828–11835. [DOI] [PubMed] [Google Scholar]
- Kurtz A. J.; Lloyd R. S. (2003) 1,N2-Deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 278, 5970–5976. [DOI] [PubMed] [Google Scholar]
- Mao H.; Schnetz-Boutaud N. C.; Weisenseel J. P.; Marnett L. J.; Stone M. P. (1999) Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl. Acad. Sci. U.S.A. 96, 6615–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins J. N.; Daniels J. S.; Rouzer C. A.; Marnett L. J. (2004) Kinetic and thermodynamic analysis of the hydrolytic ring-opening of the malondialdehyde-deoxyguanosine adduct, 3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purin-10(3H)-one. J. Am. Chem. Soc. 126, 8237–8243. [DOI] [PubMed] [Google Scholar]
- Riggins J. N.; Pratt D. A.; Voehler M.; Daniels J. S.; Marnett L. J. (2004) Kinetics and mechanism of the general-acid-catalyzed ring-closure of the malondialdehyde–DNA adduct, N2-(3-oxo-1-propenyl)deoxyguanosine (N2OPdG–), to 3-(2′-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG). J. Am. Chem. Soc. 126, 10571–10581. [DOI] [PubMed] [Google Scholar]
- Jeong Y.-C.; Sangaiah R.; Nakamura J.; Pachkowski B. F.; Ranasinghe A.; Gold A.; Ball L. M.; Swenberg J. A. (2005) Analysis of M1G-dR in DNA by aldehyde reactive probe labeling and liquid chromatography tandem mass spectrometry. Chem. Res. Toxicol. 18, 51–60. [DOI] [PubMed] [Google Scholar]
- Schnetz-Boutaud N.; Daniels J. S.; Hashim M. F.; Scholl P.; Burrus T.; Marnett L. J. (2000) Pyrimido[1,2-α]purin-10(3H)-one: a reactive electrophile in the genome. Chem. Res. Toxicol. 13, 967–970. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Kozekova A.; Harris T. M.; Rizzo C. J.; Lloyd R. S. (2008) Mutagenic potential of DNA–peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 637, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVeen L. A.; Druckova A.; Riggins J. N.; Sorrells J. L.; Guengerich F. P.; Marnett L. J. (2005) Differential DNA recognition and cleavage by EcoRI dependent on the dynamic equilibrium between the two forms of the malondialdehyde–deoxyguanosine adduct. Biochemistry 44, 5024–5033. [DOI] [PubMed] [Google Scholar]
- Knutson C. G.; Rubinson E. H.; Akingbade D.; Anderson C. S.; Stec D. F.; Petrova K. V.; Kozekov I. D.; Guengerich F. P.; Rizzo C. J.; Marnett L. J. (2009) Oxidation and glycolytic cleavage of etheno and propano DNA base adducts. Biochemistry 48, 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely J.; Rizzo C. J.; Marnett L. J. (2008) Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J. Am. Chem. Soc. 130, 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C.; Jia G.; Hou G.; Dai Q.; Zhang W.; Zheng G.; Jian X.; Yang C.-G.; Cui Q.; He C. (2010) Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature 468, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski M. A.; Bhagwat A. S.; Papaj G.; Bujnicki J. M. (2003) Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T.; Girard C. A.; Tung Y.-C. L.; Webby C. J.; Saudek V.; Hewitson K. S.; Yeo G. S. H.; McDonough M. A.; Cunliffe S.; McNeill L. A.; Galvanovskis J.; Rorsman P.; Robins P.; Prieur X.; Coll A. P.; Ma M.; Jovanovic Z.; Farooqi I. S.; Sedgwick B.; Barroso I.; Lindahl T.; Ponting C. P.; Ashcroft F. M.; O’Rahilly S.; Schofield C. J. (2007) The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.