Abstract
AU-rich elements (AREs) control the expression of numerous genes by accelerating the decay of their mRNAs. Rapid decay and deadenylation of β-globin mRNA containing AU-rich 3′ untranslated regions of the chemoattractant cytokine interleukin-8 (IL-8) are strongly attenuated by activating the p38 mitogen-activated protein (MAP) kinase/MAP kinase-activated protein kinase 2 (MK2) pathway. Further evidence for a crucial role of the poly(A) tail is provided by the loss of destabilization and kinase-induced stabilization in ARE RNAs expressed as nonadenylated forms by introducing a histone stem-loop sequence. The minimal regulatory element in the IL-8 mRNA is located in a 60-nucleotide evolutionarily conserved sequence with a structurally and functionally bipartite character: a core domain with four AUUUA motifs and limited destabilizing function on its own and an auxiliary domain that markedly enhances destabilization exerted by the core domain and thus is essential for the rapid removal of RNA targets. A similar bipartite structure and function are observed for the granulocyte-macrophage colony-stimulating factor (GM-CSF) ARE. Stabilization in response to p38/MK2 activation is seen with the core domain alone and also after mutation of the AUUUA motifs in the complete IL-8 ARE. Stabilization by ARE binding protein HuR requires different sequence elements. Binding but no stabilization is observed with the IL-8 ARE. Responsiveness to HuR is gained by exchanging the auxiliary domain of the IL-8 ARE with that of GM-CSF or with a domain of the c-fos ARE, which results in even stronger responsiveness. These results show that distinct ARE domains differ in function with regard to destabilization, stabilization by p38/MK2 activation, and stabilization by HuR.
The rate of mRNA degradation is an important determinant of the extent and duration of gene expression, in particular for genes encoding proteins involved in acute responses, like many cytokines, growth factors, and proto-oncogenes. AU-rich sequences, originally identified in the 3′ untranslated region (UTR) of several cytokine genes (5, 37), function as potent destabilizing elements that cause rapid decay of the respective transcript. On the basis of structural and functional properties, AU-rich elements (AREs) have been grouped into three classes (9). Class I AREs (e.g., that of c-fos) contain one to three scattered AUUUA motifs within a U-rich region. Class II AREs, like that of the mRNA for granulocyte-macrophage colony-stimulating factor (GM-CSF), are characterized by multiple overlapping AUUUA motifs. Class III AREs (e.g., that of c-jun) lack the AUUUA pentamer but contain U-rich stretches. A recent bioinformatics approach identified several hundred genes containing putative AREs with at least one AUUUA motif (2).
ARE-dependent rapid decay is the target of several different signaling mechanisms. These include the Jun N-terminal kinase (7, 27), phosphatidylinositol-3-kinase (28), and p38 mitogen-activated protein (MAP) kinase pathways (12, 29, 34, 46). Activation of p38 MAP kinase, triggered by the inflammatory cytokine interleukin-1 (IL-1) or by bacterial lipopolysaccharide, induces stabilization of ARE-containing endogenous mRNAs and reporter mRNAs containing several different AREs, including those of IL-3, IL-6, IL-8, tumor necrosis factor alpha (TNF-α), cyclooxygenase 2, and c-fos. Experiments with cultured cells and with mice point to involvement of the p38 MAP kinase substrate MAP kinase-activated protein kinase 2 (MK2) in this effect of the p38 MAP kinase pathway (19, 22, 31, 46). Results obtained so far indicate that stabilization induced by this pathway is selective for certain AU-rich transcripts, whereas no stabilization is observed with certain other ARE-containing mRNAs (16, 22), as well as with short-lived mRNAs lacking AREs (4). The exact requirements for p38/MK2-induced stabilization in terms of both cis-acting sequences and trans-acting factors have not yet been defined.
mRNA decay begins with loss of the poly(A) tail (deadenylation), and the AREs were observed to enhance deadenylation (11, 38, 44). More recently, AREs have been suggested to mediate the recruitment of the exosome, a complex of RNases involved in 3′-to-5′ degradation of the mRNA body (8, 30). A number of proteins have been identified that can interact with AREs and/or affect the stability of ARE-containing transcripts. The most prominent among them include HuR, TTP, BRF1, KSRP, AUF1/hnRNP D, and other members of the hnRNP family of proteins. Except for HuR, all of the above-mentioned proteins can function as destabilizers of the ARE mRNA to which they bind. Since activation of the p38/MK2 pathway selectively stabilizes ARE mRNAs, the ARE binding proteins are likely targets of the pathway.
We previously observed in gel shift assays that HuR interacts with 3′ UTR sequences of IL-8 (4), a cytokine that is chemotactic for leukocytes, activates neutrophils and is involved in angiogenesis. HuR is a ubiquitously expressed member of the human ELAV family of RNA binding proteins (24) whose functional role in the stabilization of several ARE-containing mRNAs has been established (15, 23, 33, 35, 42, 43). It also plays a role in the nuclear export of mRNA (17) and in control of translation (20, 26).
Here we report that the IL-8 3′ UTR contains an ARE with two domains that differ in function. Destabilization by this ARE and mutant forms thereof is counteracted by p38/MK2 activation but not by HuR overexpression. Instead, HuR-induced stabilization can be imposed by sequences not required for responsiveness to p38/MK2. The results demonstrate that AREs can consist of different domains with different functions in destabilization and stabilization induced by different mechanisms.
MATERIALS AND METHODS
Plasmids.
The human cDNAs for full-length and 3′-truncated IL-8 [nucleotides (nt) 1 to 1652, 1 to 1310, and 1 to 1048; GenBank accession no. Y00787; numbering extended to the defined poly(A) addition site (49)] were amplified by RT-PCR with primer pairs containing XhoI sites and cloned into the pUHD10-3 plasmid downstream of the tTA-regulated promoter (18; kindly provided by Hermann Bujard). Plasmids ptet-BBB-IL8972-1310, ptet-BBB-GM-CSFARE, and ptet-BBB-c-fosARE encode the rabbit β-globin mRNA with AREs of the IL-8, GM-CSF, and c-fos transcripts, respectively, inserted into the BglII site of the β-globin 3′ UTR (46, 48). With BglII-flanked primers, IL-8 cDNA fragments were obtained by PCR amplification; short cDNAs corresponding to regions of IL-8 or GM-CSF mRNA, as well as those encoding chimeric AREs of GM-CSF, IL-8, and c-fos, were obtained by overlapping primer annealing as previously described (45). cDNA fragments were cloned into the BglII site of the β-globin 3′ UTR in ptet-BBB (48). To generate ptet-BBB-IL8972-1310Δ, in which nt 1017 to 1076 are deleted, the PCR-generated nt 1077-to-1310 fragment with a 5′ NheI site was inserted into the BglII site of ptet-BBB and then the nt 972-to-1016 fragment was inserted into the NheI site. cDNAs corresponding to the AREs of IL-8 and GM-CSF (accession no. M10663), as well as parts thereof, were also cloned into the BglII site of pBluescript for in vitro transcription (see below). Mutations of AUUUA motifs were performed by exchanging the central U for G by using the QuikChange kit (Stratagene). To express β-globin transcripts without a poly(A) tail, the polyadenylation signal was mutated to generate an AvaIII site and an RT-PCR-amplified, AvaIII-flanked DNA of the stem-loop of the human histone 1.3 gene and 3′ flanking sequences were inserted (ptet-BBB-HSL). ptet-BBB-IL-8ARE-HSL was generated by inserting nt 1017 to 1076 of IL-8 as described above. Expression plasmids for myc-tagged HuR and constitutively active MAP kinase kinase 6 (MKK62E) have been described previously (10, 46).
Transfection and determination of mRNA decay.
HeLa cells constitutively expressing the tetracycline-controlled transactivator protein (18; kindly provided by H. Bujard) were cultured in Dulbecco's modified Eagle medium complemented with 5% fetal calf serum. Transient transfections by the calcium phosphate method and RNA degradation kinetics were performed as previously described (46). Briefly, cells (4 × 106 seeded per 9-cm-diameter dish) were transfected with the indicated plasmids. Amounts of plasmid DNA within each experiment were kept constant by adding empty vector. For kinetics of mRNA degradation, cells from one dish were trypsinized following transfection and distributed into parallel cultures. The next day, transcription from the tetracycline-regulatable promoter was stopped by addition of doxycycline (3 μg/ml). At the indicated times thereafter, total RNA was isolated and Northern blot analysis was performed with digoxigenin-labeled antisense RNA probes. RNA half-lives were determined as described in reference 46, with a video imaging system and the Molecular Analyst program (Bio-Rad).
Analysis of poly(A) tail length.
RNA samples were mixed with an equal amount of formamide loading buffer (80% [vol/vol] formamide and 1 mg of bromophenol blue per ml in 1× TBE [89 mM Tris-borate, 2 mM EDTA, pH 8.3]), incubated at 65°C for 15 min, and loaded onto denaturing polyacrylamide gels (4% [wt/vol] acrylamide, 7 M urea). Running in 1× TBE was performed at about 10 V/cm for various times, depending on the size of the RNA monitored. Semidry electroblotting (Peqlabs) at 200 mA for 1 h with 1× TBE as buffer was used to transfer the RNAs to a nylon membrane for Northern blot analysis. Deadenylated transcripts were prepared in vitro by mixing total RNA from transfected cells with oligo(dT) (0.5 μg for every 10 μg of total RNA) in buffer containing 200 mM KCl and 1 mM EDTA (pH 8.0). The sample was incubated at 90°C for 2 min and then annealed at 25°C for 10 min. Two volumes of a solution containing 100 mM KCl, 0.5 mM EDTA (pH 8.0), 20 mM Tris-HCl (pH 8.0), 28 mM MgCl2, 20 U of RNase inhibitor, and 1 U of RNase H were then added. The sample was incubated at 37°C for 30 min and precipitated with ethanol.
Preparation of cytoplasmic extracts.
All steps were carried out in the cold. HeLa cells (106 per sample) were washed once with phosphate-buffered saline, harvested by scraping, pelleted by centrifugation, and resuspended in 200 μl of hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 0.5 mM phenylmethylsulfonyl fluoride). A 25-μl volume of the same buffer including 2.5% (vol/vol) Nonidet P-40 was then added. After centrifugation at 1,000 × g for 4 min, the supernatants were removed and cleared by centrifugation at 10,000 × g. Aliquots were frozen at −70°C.
In vitro transcription and electrophoretic mobility shift assays.
Labeled RNA (107 to 108 cpm/μg) was synthesized by incubating 1 to 3 μg of linearized plasmid DNA in a mixture of 50 μCi of [α-32P]UTP (400 Ci/mmol; Hartmann Analytic); 18 μM unlabeled UTP; 0.5 mM unlabeled ATP, CTP, and GTP; 2 U of T7 or T3 RNA polymerase per μl, as required (both enzymes were from Roche); and 2 U of RNase inhibitor (MBI) per μl for 1 h at 37°C. RNase-free DNase (Roche) was then added to a final concentration of 1 U/μl. After incubation for 15 min at 37°C, the RNA was passed through a NucTrap push column (Stratagene) to remove free nucleotides and stored at −80°C. Radiolabeled RNA probes (1.5 × 105 cpm) were incubated with cytoplasmic extracts (6 μg of protein per sample) in 20 μl of buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 2 mM MgCl2, 3% (vol/vol) glycerol, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg of pepstatin A per ml, and 200 μg of tRNA per ml for 10 min at 30°C. RNase T1 (30 U/sample) was then added, and incubation was continued for 20 min at 37°C. Where indicated, mouse monoclonal antibody 19F12 against HuR (0.75 μg, kindly donated by Henry Furneaux) or rabbit antiserum against hnRNP D/AUF1 (diluted 1:200; a gift of Gary Brewer) was included for the last 10 min. Samples were electrophoresed on a nondenaturing polyacrylamide gel (5% acrylamide in 0.25× TBE buffer). The gels were dried and autoradiographed.
RESULTS
Evidence for involvement of an ARE in controlling stability of the IL-8 mRNA.
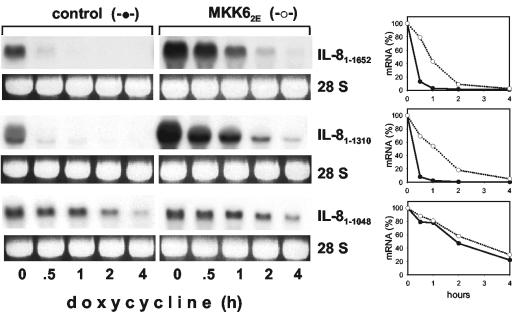
Evidence was provided previously that the stability of IL-8 mRNA is subject to modulation by the p38/MK2 pathway (46). Monitoring the stability of the full-length IL-8 mRNA in HeLa cells by using the tet-off system, we now confirm that the half-life in unstimulated cells is very short (<20 min) and is markedly increased upon selective activation of p38 MAP kinase by coexpression of a constitutively active mutant form of MKK6 (MKK62E) (Fig. 1). Regulation of stability was unaffected by deletion of about 300 nt from the 3′ end of the IL-8 mRNA. However, further deletion—including a region with four AUUUA motifs (nt 1048 to 1076, see Fig. 4A)—strongly impeded rapid basal turnover (construct IL-81-1048 in Fig. 1). Thus, the AUUUA-containing region appears to play an important role in destabilizing the IL-8 mRNA. As the IL-81-1048 transcript still is only moderately stable (half-life of about 2 h) other elements may contribute to a minor extent. Most importantly, no further stabilization was induced by MKK62E. This indicates that the AUUUA-containing region provides responsiveness to p38 MAP kinase-induced stabilization in a nonredundant manner and confirms earlier results that stabilization in response to p38 MAP kinase is selective (4, 16, 22).
FIG. 1.
Evidence for a crucial role of an ARE in the control of IL-8 mRNA decay. HeLa-tTA cells were transfected with a vector expressing constitutively active MKK6 (MKK62E) or empty vector (control) and with tet-off plasmids encoding the complete IL-8 mRNA (IL-81-1652) and 3′-shortened forms with (IL-81-1310) or without (IL-81-1048) a region with four AUUUA motifs. At the indicated times after transcription from the tet-off plasmids was stopped by addition of doxycycline (3 μg/ml), total RNA was isolated and analyzed by Northern blotting with an IL-8 antisense probe. Ethidium bromide staining of the 28S rRNA is shown to allow comparison of the RNA amounts loaded. Results were quantified by a video analyzer system (amount of mRNA at the time of doxycycline addition [0 min] = 100%).
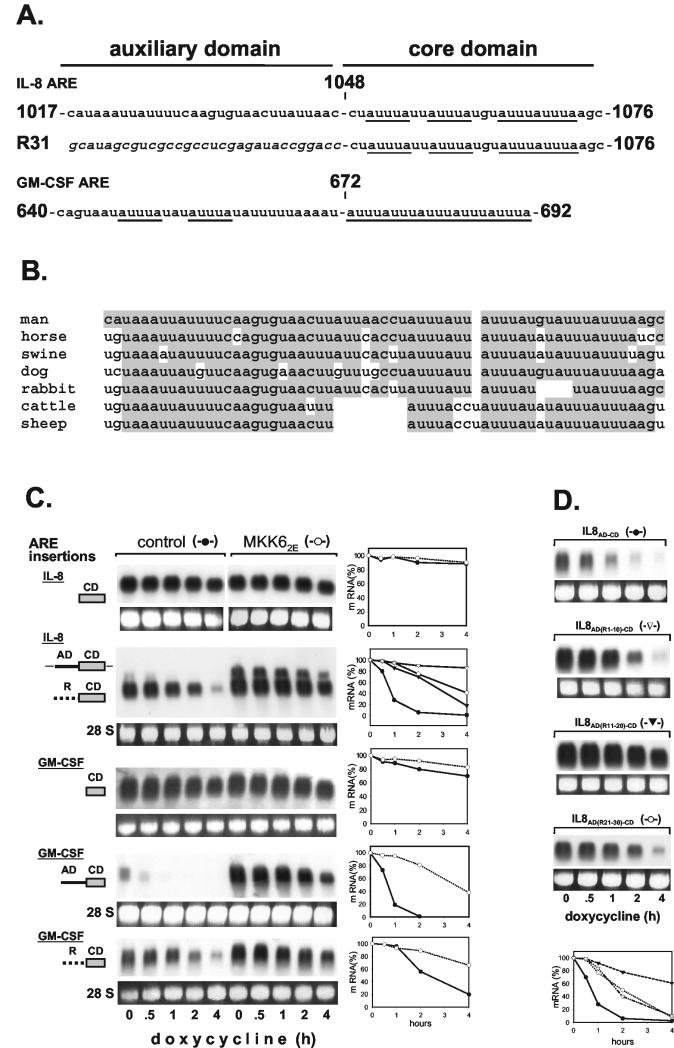
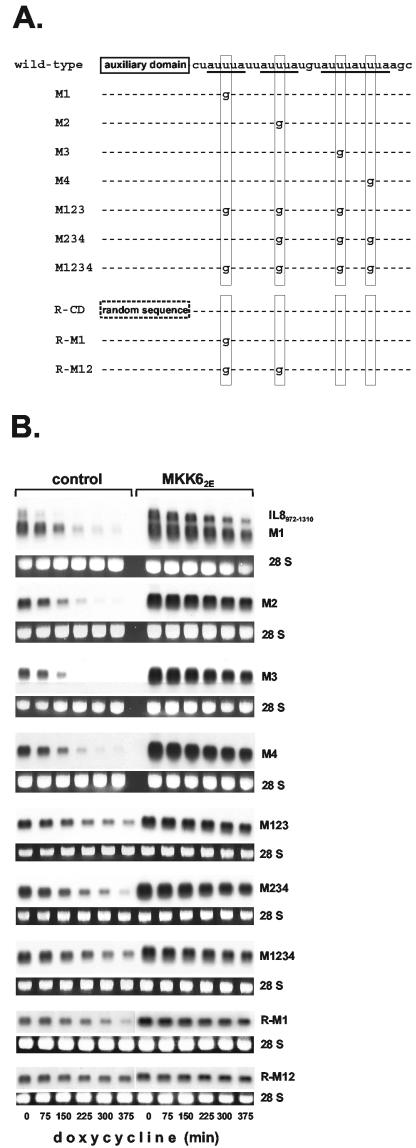
FIG.4.
Two-domain structure of IL-8 and GM-CSF AREs. (A) Sequences of the AREs are shown with AUUUA motifs underlined. A random sequence used to replace the auxiliary domain (R31) is in italics. (B) Comparison of the human IL-8 ARE with corresponding regions of IL-8 transcripts from different species. Nucleotides identical to those in the human sequence are shaded. (C) Basal decay and MKK62E-induced stabilization were analyzed for β-globin mRNAs with domains of IL-8 and GM-CSF AREs inserted as indicated (CD, core domain; AD, auxiliary domain; R, random sequence; ▾, control; ▿, MKK62E for IL-8 R-CD). (D) Destabilizing activity of the IL-8 ARE (IL-8AD-CD) and derivatives in which the auxiliary domain was mutated by exchanging 10 nt of its 5′ part (IL-8AD(R1-10)-CD), its central part (IL-8AD(R11-20)-CD), or its 3′ part (IL-8AD(R21-30)-CD) with a random sequence (GCCGCCUCGA). mRNA stability was assayed and quantified as described in the legend to Fig. 1.
Identification of a 60-nt regulatory ARE in the IL-8 3′ UTR.
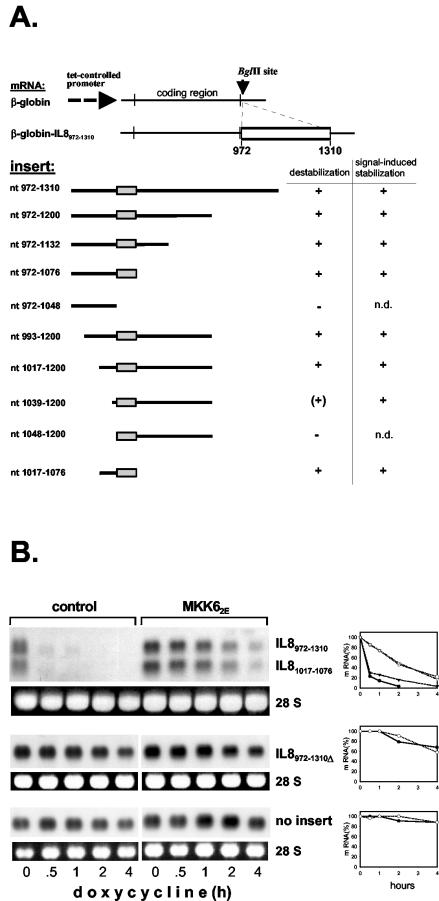
As reported previously (46), a region of the IL-8 3′ UTR that encompasses the four AUUUA motifs (nt 972 to 1310) renders the stable β-globin mRNA unstable and responsive to MKK62E-induced stabilization. Further localization of the regulatory elements was performed by inserting fragments shortened from both ends into the stable β-globin RNA and assaying destabilization and MKK62E-induced stabilization (Fig. 2A). Destabilization was lost when shortening from the 3′ end deleted the region containing the four AUUUA motifs (insertion of nt 972 to 1048). Shortening from the 5′ end resulted in loss of regulation even before this region was reached. Expressing β-globin RNA with an insertion of nt 972 to 1310 or 1017 to 1076 shows equal destabilization and MKK62E-induced stabilization for both mRNAs (Fig. 2B). Furthermore, deleting nt 1017 to 1076 from the nt 972-to-1310 fragment results in strongly increased basal stability and loss of responsiveness to MKK62E, characteristics resembling those of the β-globin mRNA with no insertion (Fig. 2B). These data identify nt 1017 to 1076 as the minimal regulatory element within this region of the IL-8 mRNA.
FIG. 2.
Localization of a regulatory ARE in the IL-8 mRNA. (A) Summary of results for basal decay and MKK62E-induced stabilization of β-globin-IL-8 hybrid mRNAs (n.d., not determined owing to a long basal half-life). RNAs were expressed by plasmid ptet-BBB containing the indicated IL-8 sequences inserted into the BglII site. The AUUUA-containing region is boxed. (B) Comparison of basal decay and MKK62E-induced stabilization was performed for β-globin mRNA without insertions or with IL-8 nt 972 to 1310, 1017 to 1076, or 972 to 1310 with nt 1017 to 1076 deleted (IL-8972-1310Δ). Decay kinetics were analyzed by using a β-globin antisense probe and quantified as described in the legend to Fig. 1 (triangles, IL-8972-1310; closed symbols, control; open symbols, MKK62E).
p38 MAP kinase-induced stabilization via the IL-8 ARE involves inhibition of deadenylation.
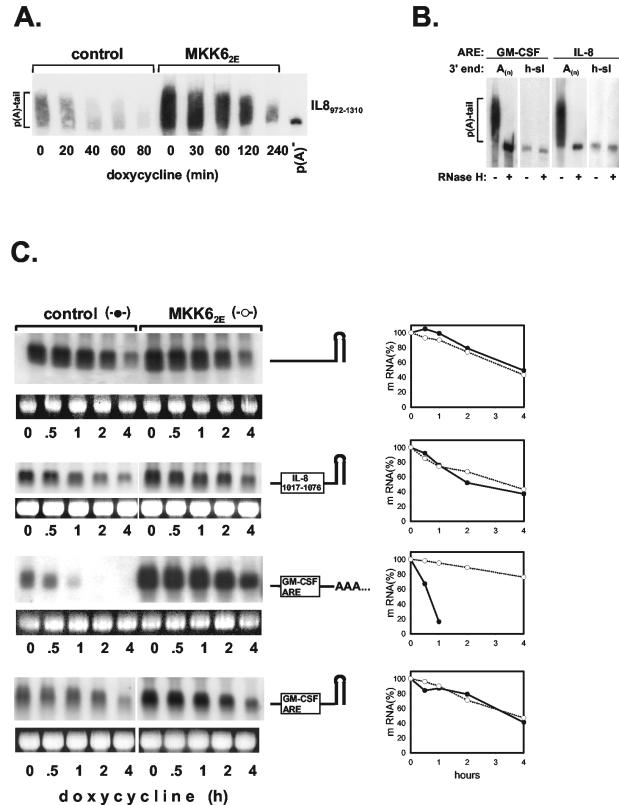
Deadenylation is considered the first step in the degradation of ARE-containing mRNAs (11, 38, 44). To find out if activation of the p38/MK2 pathway imposes changes in this process, the poly(A) tail of β-globin mRNA containing the IL-8 ARE, isolated from cells cotransfected with MKK62E or empty vector, was analyzed by polyacrylamide gel electrophoresis. While under unstimulated conditions, the poly(A) tail length decreased rapidly along with degradation of the RNA; stabilization by coexpression of MKK62E was accompanied by inhibition of deadenylation (Fig. 3A). Further evidence of involvement of the poly(A) tail in ARE-based regulation of mRNA decay was obtained by expressing β-globin transcripts without or with the IL-8 ARE (nt 1017 to 1076) as nonadenylated forms. The polyadenylation signal in the β-globin gene was replaced with the stem-loop and 3′ flanking sequences of the human histone 1.3 gene (32). This resulted in expression of nonadenylated β-globin and β-globin-IL-8 chimeric RNAs (Fig. 3B). Comparison of their half-lives revealed that ARE-based control of decay was lost: the nonadenylated β-globin RNAs without and with the ARE had comparable basal decay rates, which were not affected by MKK62E coexpression (Fig. 3C, compare with IL-81017-1076 in Fig. 2B). Similarly, a β-globin mRNA containing the ARE of GM-CSF also lost rapid decay and MKK62E-induced stabilization when expressed without a poly(A) tail (Fig. 3C).
FIG. 3.
Evidence for a role of the poly(A) tail in ARE-mediated control of mRNA stability. (A) HeLa-tTA cells were transfected with empty vector (control) or a vector expressing MKK62E and plasmid ptet-BBB-IL-8972-1310. At the indicated times after transcription from ptet-BBB-IL-8972-1310 was stopped with doxycycline (3 μg/ml), total RNA was isolated, separated on a polyacrylamide gel, and analyzed by Northern blotting with a β-globin antisense probe. An aliquot of the RNA was subjected to deadenylation in vitro [p(A)−]. (B) Absence of a poly(A) tail in mRNAs containing a histone mRNA-derived stem-loop was confirmed by polyacrylamide gel electrophoresis of total RNA from cells expressing β-globin RNAs with the indicated ARE without or with the histone stem-loop (h-sl) before and after digestion with RNase H plus oligo(dT). (C) Decay kinetics were determined for polyadenylated and nonadenylated, histone stem-loop-containing β-globin mRNAs without or with the indicated AREs. Basal decay and MKK62E-induced stabilization were assayed as described in the legend to Fig. 1.
The IL-8 ARE consists of two functionally different domains, and the AUUUA-containing domain is sufficient for p38 MAP kinase-induced stabilization.
In its 3′ part, the nt 1017-to-1076 fragment contains a core domain (nt 1048 to 1076) with four AUUUA motifs, two of which overlap (Fig. 4A). The 5′ part (nt 1017 to 1047, termed the auxiliary domain hereafter) lacks AUUUA motifs. Comparison of the corresponding sequences in different species reveals marked homology of the auxiliary domain, as well as the core domain (Fig. 4B). Note that the organization of two separate and two overlapping AUUUA motifs is preserved.
Further investigation showed that these two domains differ in function. An RNA that contained only the auxiliary domain (Fig. 2A, nt 972 to 1048) was stable, indicating that this domain cannot destabilize on its own. Lack of destabilization was also observed upon insertion of a fragment that contains only the AUUUA-containing core domain and lacks the auxiliary domain (Fig. 4C, construct IL-8 CD). However, owing to the site of insertion in the β-globin construct, the fragments are located directly after the stop codon. To account for a possible effect of close proximity to the stop codon, the half-life of the construct containing just the core domain was compared to that of a construct in which spacing from the stop codon was preserved by inserting a random sequence identical in length to the 5′ domain. That construct (IL-8 R-CD)—in contrast to the one without spacing—clearly exhibits destabilization (Fig. 4C). Yet destabilization is much weaker compared to that of an RNA with the complete ARE (compare IL-8 R-CD and AD-CD in Fig. 4C). Thus, spacing between the stop codon and a destabilizing AU-rich sequence might be critical for its functioning. Lack of destabilization by the auxiliary domain alone is not related to that, since in the nt 972-to-1048 construct (Fig. 2A) there is a distance of about 50 nt between the auxiliary domain and the stop codon. Taken together, the results show that the 5′ part of the IL-8 ARE functions as an auxiliary domain that does not destabilize on its own but strongly enhances the moderate destabilization exerted by the core domain. Replacement of 10 nt in the 5′, central, or 3′ part of the auxiliary domain with a 10-nt random sequence resulted in loss of destabilizing activity, with the strongest impairment noted when the central part was replaced (Fig. 4D). This indicates that sequences most crucial for enhancing destabilization are located in the central part of the auxiliary domain.
Importantly, activation of the p38 MAP kinase cascade by MKK62E induced further stabilization of the RNA containing the random sequence plus the core domain (IL-8 R-CD, Fig. 4C). Thus, the AUUUA motif-containing core domain is sufficient to confer sensitivity to p38 MAP kinase-induced stabilization.
Evidence for a bipartite structure has been presented for the GM-CSF ARE (47) (Fig. 4A), which mediates extents of destabilization and MKK62E-induced stabilization very similarly to the IL-8 ARE (46; Fig. 4C). As observed for the IL-8 ARE, replacement of the auxiliary domain with a random sequence resulted in diminished destabilization. Furthermore, this mRNA was also stabilized by coexpression of MKK62E, confirming the role of the AUUUA-containing domain in p38/MK2-induced stabilization (Fig. 4C).
Role of AUUUA motifs in destabilization and p38 MAP kinase-induced stabilization.
While the GM-CSF ARE contains five overlapping AUUUA motifs, the IL-8 ARE contains four, with only two of them overlapping. It contains no UUAUUUA(U/A)(U/A) nonamer, which has been described as a key destabilizing sequence (21, 50). To investigate the contributions of the individual AUUUA motifs, they were destroyed singly and in combinations by introducing U→G point mutations of the central U (Fig. 5A contains a scheme of the mutant sequences analyzed). Elimination of the first motif resulted in significantly decreased destabilizing efficiency of the ARE, compared to that of the intact ARE without (data not shown) or with (Fig. 5B, compare transcript M1 to IL-8972-1310) adjacent sequences. Destroying each of the other motifs separately (Fig. 5B, control for mutants M2 to M4) also induced marked loss of destabilization. Progressively mutating the remaining AUUUA motifs in the complete ARE, yielding RNAs with two intact motifs (data not shown), only one intact motif (mutants M123 and M234), or no intact motif left (M1234) resulted in progressive loss of the destabilizing effect. Unexpectedly, destroying the motif overlap by mutating motif 3 or 4 or both did not result in a more pronounced loss of destabilizing activity. Thus, no evidence was obtained for a superior function of overlapping as opposed to dispersed motifs. Rather, Fig. 5 and additional experiments show a tendency to more strongly compromised function owing to loss of motif 1. Importantly, in all of the mutants, MKK62E expression still induced stabilization of the RNA (Fig. 5B). It is noteworthy that while the half-life of the rapidly degraded reporters containing the AREs of GM-CSF and IL-8 is extended to about 2 to 3 h (Fig. 4C), the half-life of the IL-8 ARE transcripts containing single mutations in their AUUUA motifs, which is about 2 h in unstimulated cells, is extended to more than 6 h (Fig. 5). Even for mutant M1234—which has only very weak residual destabilizing capacity—stabilization in response to activation of the p38/MK2 pathway was clearly discernible (half-life of 8 h versus 4.2 h without MKK62E). This demonstrates that RNA elements without an intact AUUUA motif can mediate p38 MAP kinase-regulated mRNA stability.
FIG. 5.
Role of AUUUA motifs in the regulatory function of the IL-8 ARE. (A) Scheme of mutants analyzed. (B) Decay kinetics in the absence and presence of MKK62E expression were compared for β-globin mRNAs containing the IL-8 ARE and mutations thereof as outlined in panel A. (C) Quantification of the results from panel B for basal decay of the indicated mRNAs was performed as for Fig. 1.
Mutation of the first AUUUA motif in a construct lacking the auxiliary domain caused almost complete loss of destabilization. The half-life of the mRNA containing that insert (R-M1) is comparable to that of the mRNA containing the complete ARE with all of the AUUUA motifs mutated (Fig. 5B and C, compare transcript R-M1 to M1234). Also, with R-M1, some stabilization in response to MKK62E is still detectable. Since mutating one AUUUA motif and replacing the auxiliary domain with a random sequence causes the same extent of loss of destabilizing activity as mutating four AUUUA motifs, the auxiliary domain can compensate for the loss of three AUUUA motifs in the core domain. Furthermore, an mRNA without the auxiliary domain and two AUUUA motifs mutated (R-M12) no longer showed destabilization compared to the β-globin mRNA with no insertion (half-life of >10 h) (Fig. 5B). These results confirmed the importance of the auxiliary domain for destabilization.
Interaction of the ARE with cytoplasmic proteins involves both domains and is not affected by p38 activation.
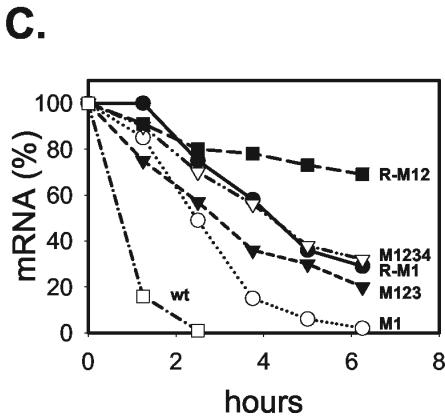
Gel shift assays with radiolabeled RNA representing the complete IL-8 ARE show two prominent and additional minor complexes formed between the RNA and HeLa cytoplasmic proteins (Fig. 6A). Mutating one AUUUA motif (mutant M3) resulted in a slight loss of the intensity of the complexes, whereas mutating all four motifs (M1234) caused a dramatic loss of the major complexes. Thus, intensity of complex formation correlates with the extent of destabilization. No change in complex formation was observed when activating the p38/MK2 pathway by MKK62E (Fig. 6A). Complex formation was impaired, resembling the result obtained with the M1234 mutant, when assaying RNA corresponding to the auxiliary domain alone or the core domain alone (Fig. 6B). Thus, both domains contribute to formation of the two major protein complexes. Furthermore, comparing the complete GM-CSF ARE and its two domains separately also shows a different and weaker interaction with proteins for both domains. To identify the proteins in the complexes, antibodies against the ARE-binding proteins hnRNP D/AUF1 and HuR were included in the assay (Fig. 6B). Anti-hnRNP D/AUF1 antibodies did not affect complex formation with the IL-8 ARE, whereas they induced a shift with the GM-CSF ARE, serving as a positive control. Antibodies against HuR induced a supershift with both AREs (Fig. 6B), indicating that they both can interact with HuR.
FIG. 6.
Interaction of the IL-8 and GM-CSF AREs with cytoplasmic proteins. Radiolabeled in vitro-transcribed RNA fragments corresponding to the complete AREs (AD+CD) or their mutants (M3 and M1234) or domains (AD, auxiliary domain; CD, core domain), as indicated, were incubated with cytoplasmic (cytopl.) extract from HeLa cells. Normal immunoglobulin G (IgG) or antibodies against hnRNP D/AUF1 or HuR were included where indicated. Protein-RNA complexes were analyzed by nondenaturing gel electrophoresis. Complexes are marked by black arrowheads, and complexes supershifted by antibodies are marked by open arrowheads.
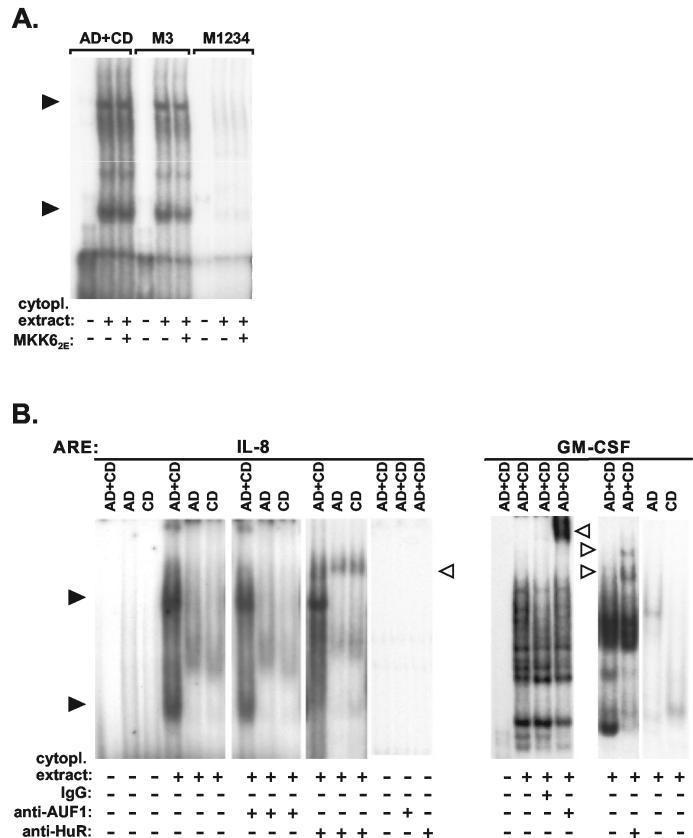
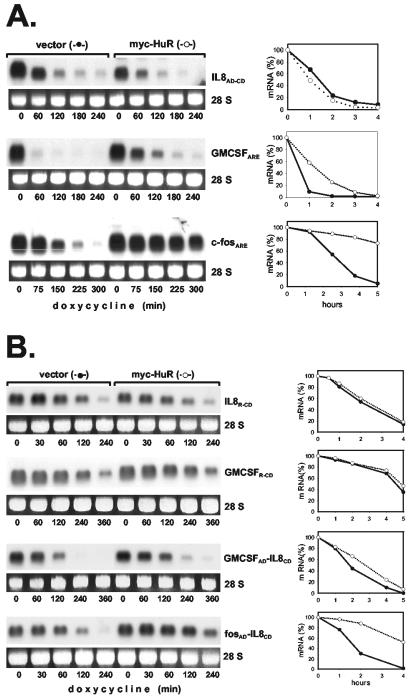
mRNA stabilization by HuR is selective and can be imposed by auxiliary domains.
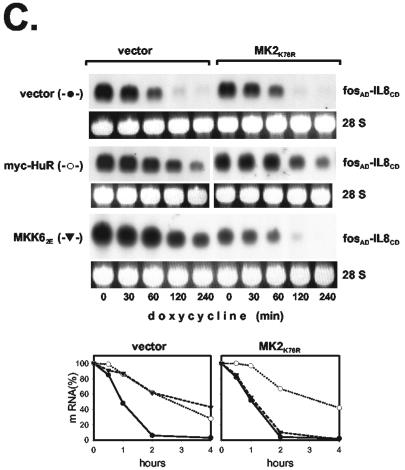
HuR interacts with both the IL-8 and GM-CSF AREs in gel shift assays (Fig. 6) and has been shown to stabilize several ARE-containing RNAs (15, 23, 33). To compare that mode of stabilization to the effect of the p38/MK2 pathway, we assayed how HuR affects the stability of ARE-containing β-globin mRNAs. Degradation of the IL-8 ARE-containing RNA was not affected by overexpression of myc-tagged HuR (Fig. 7A), whereas clear stabilization was observed for the RNA containing the ARE of GM-CSF, and—in line with results of others (10)—even much stronger stabilization occurred for an RNA containing the c-fos ARE (Fig. 7A). Investigating further the reason for different responsiveness to HuR, we observed that—like the IL-8 ARE-containing transcript—a transcript in which the auxiliary domain of the IL-8 ARE was replaced with a random sequence (IL-8R-CD) was not stabilized by HuR (Fig. 7B). This ruled out an inhibitory effect of the auxiliary domain. The construct containing the GM-CSF core domain behind a random sequence (GMCSFR-CD) also showed no significant stabilization, although the complete GM-CSF ARE did, indicating that the effect of HuR might depend on properties of the auxiliary region. To test this, the auxiliary domain of the IL-8 ARE was replaced with the auxiliary domain of the other AREs. The RNA containing a chimeric ARE with the auxiliary domain of GM-CSF and the core domain of IL-8 was stabilized significantly by HuR (GMCSFAD-IL-8CD in Fig. 7B). Even stronger stabilization was observed for the RNA containing the IL-8 core domain fused to the auxiliary domain of the c-fos ARE (fosAD-IL-8CD in Fig. 7B). This domain lacks AUUUA motifs and was recently identified by Shyu and coworkers (10) as the major domain responsible for HuR-induced stabilization in the c-fos ARE. Thus, unresponsiveness of the IL-8 ARE to HuR-induced stabilization is not a property of the core domain but results from the lack of an HuR-responsive auxiliary domain. The results suggest different sequence requirements for stabilization by HuR as opposed to p38/MK2 activation. As shown in Fig. 7C, the RNA containing the fosAD-IL-8CD sequence is also stabilized by MKK62E. While that effect—expectedly—was inhibited by coexpression of a dominant-negative form of MK2 (MK2K76R), stabilization by HuR was not. This rules out the possibility that MK2 is involved in the effect of HuR. This and the involvement of different ARE domains in each of the two effects indicate that stabilization of ARE-containing mRNAs by HuR overexpression and that by p38 MAP kinase activation occur via different mechanisms.
FIG. 7.
Unresponsiveness of the IL-8 ARE to HuR-mediated stabilization and its reversal by auxiliary domains of the GM-CSF ARE or the c-fos ARE. β-Globin mRNAs containing the ARE of IL-8, GM-CSF, or c-fos (A) or the core domain of the IL-8 or GM-CSF ARE 3′ adjacent to a random sequence (R-IL-8CD or R-GMCSFCD, respectively) or chimeric AREs consisting of the core domain of the IL-8 ARE and the auxiliary domain of the GM-CSF or c-fos ARE (GMCSFAD-IL-8CD or fosAD-IL-8CD, respectively) (B) were coexpressed with empty vector or myc-HuR as indicated. (C) Cells were transfected with plasmids encoding the β-globin-fosAD-IL-8CD mRNA and myc-HuR or MKK62E together with empty vector or an expression vector for MK2K76R. mRNA stability was determined and quantified as described in the legend to Fig. 1.
DISCUSSION
In this report, we define an ARE in the 3′ UTR of IL-8 and present evidence for functionally different domains involved in destabilization and stabilization induced by the p38/MK2 pathway or by the ARE-binding protein HuR. Determining the half-life of the stable β-globin mRNA with insertions of IL-8 3′ UTR regions, expressed from tet-off vectors in HeLa cells, a minimal region of 60 nt was found to exert potent destabilization of the mRNA in unstimulated cells. Activation of the p38/MK2 pathway by coexpressing constitutively active MKK6 resulted in marked stabilization (Fig. 2). This was accompanied by inhibition of deadenylation (Fig. 3), as reported recently for the cyclooxygenase 2 ARE (13). This, together with our previous finding that PABP1 associates with the ARE and is a substrate of MK2 in vitro (3) and the loss of ARE-mediated control of mRNA stability in transcripts lacking a poly(A) tail (Fig. 3), indicates a crucial role of the poly(A) tail and proteins interacting with it in ARE-mediated decay and its control.
Results in Fig. 4 show a two-domain structure of the 60-nt region. A core domain containing four AUUUA motifs apparently plays an important role in the rapid decay of the IL-8 mRNA, since a truncated transcript that lacks that domain has a much longer half-life (about 2 h compared to <20 min, Fig. 1). It is possible that the 2-h half-life reflects the function of additional destabilizing elements, as has been observed for the TNF-α mRNA (39). Importantly, stabilization in response to p38/MK2 signaling is completely lost in the mRNA lacking the core domain. This indicates that the core domain is essential for regulation by this pathway.
Our own previous studies (46) and a report on the effect of 3′ UTR regions of IL-8 on chloramphenicol acetyltransferase expression (49) indicated, however, that the AUUUA motif-containing core region alone has low destabilizing activity. We now identify a 30-nt region immediately 5′ to the core as a potent codestabilizing domain. A two-domain structure has been reported for the ARE of c-fos (6) that consists of a 5′ domain containing three AUUUA motifs and a U-rich 3′ domain. The IL-8 ARE is organized more similarly to the GM-CSF and IL-3 AREs, which contain clusters of overlapping AUUUA motifs in the 3′ part and AU-rich sequences 5′ to the AUUUA motifs that contribute to destabilization (47). Evidence for the importance of upstream sequences has also been presented for the TNF-α ARE (40). Interestingly—unlike the upstream region of the IL-8 ARE—that of the GM-CSF ARE contains two AUUUA motifs which, however, are dispensable for its destabilizing activity (47). Both upstream domains are A and U rich (100% for GM-CSF, 81% for IL-8). Further characteristics of auxiliary domains have to be defined in future studies. The importance of such domains could be to provide buffer capacity against mutations of AUUUA motifs (Fig. 5B and reference 6).
While the auxiliary domains of IL-8 and GM-CSF strongly contribute to destabilization, they are dispensable for the stabilizing effect of the p38/MK2 pathway. Furthermore, severely compromising ARE function by mutating the AUUUA motifs—which results in reduced interaction with proteins (Fig. 6A), presumably including those involved in destabilization—did not yield RNAs that lose responsiveness to p38/MK2. A database for ARE-containing transcripts has been established on the basis of the sequence WWWUAUUUAUWWW and allowing one mismatch on either side of the UAUUUAU motif (2). In the IL-8 ARE, the second and third AUUUA motifs form the centers of sequences fulfilling these criteria. Interestingly, mutants in which both of these AUUUA motifs are destroyed still mediate destabilization and MKK6-induced stabilization. Thus, the sequence requirements for this type of regulation are even more flexible than initially assumed and can be met by transcripts not included in the ARED database. The results presented in Fig. 4 and 5 show that an optimal ARE function requires the context of several AUUUA motifs plus an upstream domain. Mutating any of these components results in impaired destabilization. Importantly, in all cases in which significant destabilization occurred, it was inhibited by activation of p38/MK2. This was even true for ARE sequences without an AUUUA motif. In unstimulated cells, the respective mRNA, containing the M1234 mutant ARE, was only moderately destabilized compared to the β-globin mRNA with no insertion and thus lost the physiologically important feature of allowing a sharp peak of gene expression in response to stimulation. However, in other mRNAs, ARE-like elements may exist in which the lack of AUUUA motifs is compensated for by even more effective auxiliary domains.
The overall picture that emerges from these results is that as long as there is measurable destabilization through an ARE, activation of p38/MK2 induces stabilization. On the other hand, short-lived mRNAs that lack AREs are not affected by p38/MK2 activation (4). These observations are compatible with a model in which this signaling pathway—rather than activating an independent stabilizing mechanism—impairs a distinct mechanism of destabilization that is dependent on the recognition of an ARE by a certain protein(s).
A likely possibility is that MK2 phosphorylates and thereby compromises the function of a protein directly involved in destabilization. Our observation that expressing MKK62E does not affect RNA-protein complex formation (Fig. 6A) suggests a functional change other than binding to the ARE. Candidate proteins for which evidence has been presented that they are phosphorylated by MK2 and selectively interact with AREs include hnRNP A0 (36), PABP1 (3), and TTP (25).
It has to be noted, however, that not all AREs mediate stabilization through this pathway. A number of mRNAs fulfilling the criteria of the ARED database are not stabilized in a p38 MAP kinase-dependent manner in response to lipopolysaccharide as an activator of p38/MK2 signaling (16). There could be several not mutually exclusive reasons for this observation, one of them being that certain AREs lack features that we have not yet identified. Another possible reason is that additional instability determinants not responsive to p38/MK2-induced stabilization override the effect of that signaling mechanism. This appears quite likely in view of our results showing rather flexible requirements for this type of stabilization, and indeed such a situation has been shown recently for the TNF-α mRNA (39), which contains a constitutive decay element. Such a mechanism can impose selectivity on stabilization within a broad range of transcripts whose AREs may fulfill the requirements for p38/MK2-induced stabilization.
In this paper, we demonstrate a fundamental difference between sequence requirements for stabilization through the p38/MK2 pathway as opposed to stabilization due to increased cytoplasmic HuR. HuR has been found to interact with several AREs in vitro and in cells, and its overexpression induced stabilization of mRNAs for vascular endothelial growth factor (23), nitric oxide synthase II (35), eotaxin (1), and different ARE-containing reporter RNAs (14, 15, 33). Furthermore, suppression of HuR amounts by antisense RNA or small interfering RNA approaches inhibited stabilization induced by hypoxia (23), cytokines (35), or UV light (43) or during cell cycle progression (42). Most recently, constitutively active MK2 (41), as well as stimuli known to activate the p38/MK2 pathway (H2O2 [41] and TNF-α [1]), was found to induce cytoplasmic accumulation of HuR. Thus, HuR could play a role in p38/MK2-induced stabilization. The lack of stabilization of an IL-8 ARE reporter RNA (Fig. 7), however, argues against this possibility. Furthermore, an AUUUA motif-containing core domain clearly allows stabilization of the mRNA by the p38/MK2 pathway, as shown for the core domains of GM-CSF and IL-8 (Fig. 4), whereas it does not suffice to allow stabilization by HuR. Rather, a separate domain can impose stabilization through HuR, as shown for the c-fosAD-IL-8CD chimeric ARE (Fig. 7). Different mechanisms of stabilization are also supported by the characteristics of the stabilized transcripts. HuR has been shown to prevent decay of the RNA body, rather than deadenylation (33).
The observation that HuR binds to the IL-8 ARE in vitro yet does not stabilize an IL-8 ARE-containing reporter RNA when overexpressed can be explained in different ways. For example, the affinity of HuR for the IL-8 ARE may be too low, or HuR may have to bind in a distinct way and/or in combination with certain other proteins to stabilize an RNA. Alternatively, binding in cells might be prevented by proteins that do not bind to the GM-CSF ARE or the c-fos ARE. Irrespective of the reason, the absence of a stabilizing effect of HuR on IL-8 ARE-containing mRNA argues against increased levels of HuR in the cytoplasm as the sole mechanistic basis for p38/MK2-induced stabilization. In additional experiments (data not shown), activation of p38/MK2 in cells overexpressing HuR did not result in increased stabilization of an IL-8 ARE reporter RNA compared to that in cells without ectopically expressed HuR. Taken together, these results suggest that HuR and p38/MK2 activation can induce mRNA stabilization by two independent mechanisms. This does not exclude the possibility that—as an additional mode of stabilization—p38/MK2 activation might also stabilize indirectly through HuR by inducing its cytoplasmic translocation (41).
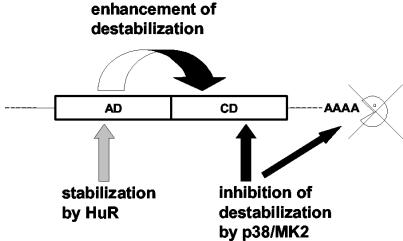
In summary, our data show that AREs can consist of functionally distinct domains (Fig. 8). The IL-8 and GM-CSF AREs contain a core domain with several AUUUA motifs and moderate destabilizing activity by a mechanism sensitive to inhibition by p38/MK2 activation. The 5′ auxiliary domain strongly enhances the moderate destabilizing activity of the core domain and thus is mandatory for most rapid mRNA degradation, the hallmark of ARE-mediated regulation. Stabilization by HuR requires features absent in the IL-8 ARE but present in auxiliary domains of GM-CSF and c-fos. The fact that mRNA stabilization by the p38/MK2 kinase cascade and that by HuR require differing sequence characteristics and consequently have different transcript selectivities suggests that the two modes of stabilization can operate in different physiological situations.
FIG. 8.
Scheme of structure-function relationships in bipartite AREs (see Discussion).
Acknowledgments
We thank H. Bujard for providing HeLa tTA cells and plasmid pUHD10-3, A.-B. Shyu for the plasmid for myc-HuR, and H. Furneaux and G. Brewer for antibodies against HuR and hnRNP D/AUF1, respectively. We thank Birgit Ritter and Thomas Wollert for technical assistance.
REFERENCES
- 1.Atasoy, U., S. L. Curry, I. Lopez De Silanes, A. B. Shyu, V. Casolaro, M. Gorospe, and C. Stellato. 2003. Regulation of eotaxin gene expression by TNF-α and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 171:4369-4378. [DOI] [PubMed] [Google Scholar]
- 2.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollig, F., R. Winzen, M. Gaestel, S. Kostka, K. Resch, and H. Holtmann. 2003. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 301:665-670. [DOI] [PubMed] [Google Scholar]
- 4.Bollig, F., R. Winzen, M. Kracht, B. Ghebremedhin, B. Ritter, A. Wilhelm, K. Resch, and H. Holtmann. 2002. Evidence for general stabilization of mRNAs in response to UV light. Eur. J. Biochem. 269:5830-5839. [DOI] [PubMed] [Google Scholar]
- 5.Caput, D., B. Beutler, K. Hartog, R. Thayer, S. Brown-Shimer, and A. Cerami. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA 83:1670-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C.-Y., T.-M. Chen, and A.-B. Shyu. 1994. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol. Cell. Biol. 14:416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. Y., N. Xu, and A. B. Shyu. 2002. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 22:7268-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couttet, P., M. Fromont-Racine, D. Steel, R. Pictet, and T. Grange. 1997. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94:5628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, J. L., M. Brook, A. R. Clark, and J. Saklatvala. 1999. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J. Biol. Chem. 274:264-269. [DOI] [PubMed] [Google Scholar]
- 13.Dean, J. L., S. J. Sarsfield, E. Tsounakou, and J. Saklatvala. 2003. p38 mitogen-activated protein kinase stabilizes mRNAs that contain cyclooxygenase-2 and tumor necrosis factor AU-rich elements by inhibiting deadenylation. J. Biol. Chem. 278:39470-39476. [DOI] [PubMed] [Google Scholar]
- 14.Dean, J. L., R. Wait, K. R. Mahtani, G. Sully, A. R. Clark, and J. Saklatvala. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 18.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. D. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat. Cell Biol. 1:94-97. [DOI] [PubMed] [Google Scholar]
- 20.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′ UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagnado, C. A., C. Y. Brown, and G. J. Goodall. 1994. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A). Mol. Cell. Biol. 14:7984-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasa, M., K. R. Mahtani, A. Finch, G. Brewer, J. Saklatvala, and A. R. Clark. 2000. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 20:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, N. S., S. Chung, H. Furneaux, and A. P. Levy. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273:6417-6423. [DOI] [PubMed] [Google Scholar]
- 24.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 25.Mahtani, K. R., M. Brook, J. L. E. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazan-Mamczarz, K., S. Galban, I. Lopez de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ming, X. F., G. Stoecklin, M. Lu, R. Looser, and C. Moroni. 2001. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 21:5778-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazawa, K., A. Mori, H. Miyata, M. Akahane, Y. Ajisawa, and H. Okudaira. 1998. Regulation of interleukin-1β-induced interleukin-6 gene expression in human fibroblast-like synoviocytes by p38 mitogen-activated protein kinase. J. Biol. Chem. 273:24832-24838. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 32.Neu-Yilik, G., N. H. Gehring, R. Thermann, U. Frede, M. W. Hentze, and A. E. Kulozik. 2001. Splicing and 3′ end formation in the definition of nonsense-mediated decay-competent human β-globin mRNPs. EMBO J. 20:532-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridley, S. H., J. L. Dean, S. J. Sarsfield, M. Brook, A. R. Clark, and J. Saklatvala. 1998. A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett. 439:75-80. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Pascual, F., M. Hausding, I. Ihrig-Biedert, H. Furneaux, A. P. Levy, U. Forstermann, and H. Kleinert. 2000. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J. Biol. Chem. 275:26040-26049. [DOI] [PubMed] [Google Scholar]
- 36.Rousseau, S., N. Morrice, M. Peggie, D. G. Campbell, M. Gaestel, and P. Cohen. 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 21:6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 38.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 39.Stoecklin, G., M. Lu, B. Rattenbacher, and C. Moroni. 2003. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol. Cell. Biol. 23:3506-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoecklin, G., P. Stoeckle, M. Lu, O. Muehlemann, and C. Moroni. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7:1578-1588. [PMC free article] [PubMed] [Google Scholar]
- 41.Tran, H., F. Maurer, and Y. Nagamine. 2003. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol. 23:7177-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, W., M. C. Caldwell, S. Lin, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, T., and R. Treisman. 1988. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336:396-399. [DOI] [PubMed] [Google Scholar]
- 45.Winzen, R., S. Kafert, B. Preiss, H. A. Mylius-Spencker, K. Resch, and H. Holtmann. 1996. Interaction between the mRNA of the 55-kDa tumor necrosis factor receptor and cellular proteins. Possible involvement in post-transcriptional regulation of receptor expression. J. Biol. Chem. 271:13461-13467. [DOI] [PubMed] [Google Scholar]
- 46.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, N., C. Y. Chen, and A. B. Shyu. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, N., P. Loflin, C. Y. Chen, and A. B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, Y., and K. Chadee. 2001. The 3′-untranslated region of human interleukin-8 mRNA suppresses IL-8 gene expression. Immunology 102:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zubiaga, A. M., J. G. Belasco, and M. E. Greenberg. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15:2219-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]