Abstract
High levels of transcription are associated with increased mutation rates in Saccharomyces cerevisiae, a phenomenon termed transcription-associated mutation (TAM). To obtain insight into the mechanism of TAM, we obtained LYS2 forward mutation spectra under low- versus high-transcription conditions in which LYS2 was expressed from either the low-level pLYS2 promoter or the strong pGAL1-10 promoter, respectively. Because of the large size of the LYS2 locus, forward mutations first were mapped to specific LYS2 subregions, and then those mutations that occurred within a defined 736-bp target region were sequenced. In the low-transcription strain base substitutions comprised the majority (64%) of mutations, whereas short insertion-deletion mutations predominated (56%) in the high-transcription strain. Most notably, deletions of 2 nucleotides (nt) comprised 21% of the mutations in the high-transcription strain, and these events occurred predominantly at 5′-(G/C)AAA-3′ sites. No −2 events were present in the low-transcription spectrum, thus identifying 2-nt deletions as a unique mutational signature for TAM.
Transcription influences genomic stability in a complex manner by affecting DNA repair, recombination, mutagenesis, and chromatin structure (reviewed in references 1 and 31). In the subpathway of nucleotide excision repair known as transcription-coupled repair, for example, the encounter of RNA polymerase (Pol) with a transcription-blocking lesion on the transcribed strand specifically triggers repair of the damage, resulting in more efficient repair of lesions on the transcribed strand than on the nontranscribed strand (16). In addition to promoting strand-specific repair, high levels of transcription have been shown to elevate recombination rates in Saccharomyces cerevisiae (33, 36, 39, 41), Schizosaccharomyces pombe (15), and mammalian cells (34). Finally, increased transcription has been shown to stimulate spontaneous mutagenesis in Escherichia coli (3, 24, 44) and bacteriophage T7 (4) and to enhance deletions within an E. coli plasmid target (40). In S. cerevisiae, Datta and Jinks-Robertson demonstrated previously that an increased transcription level likewise stimulates spontaneous mutation rates, a phenomenon termed transcription-associated mutation, or TAM (8). Specifically, reversion of the lys2ΔBgl frameshift allele and forward mutation at the LYS2 locus increased 35- and 10-fold, respectively, in response to transcriptional induction from the pGAL1-10 promoter. Whereas the genetic requirements and underlying mechanisms of transcription-coupled repair (16, 38) and transcription-associated recombination are becoming clearer (11, 12, 21), the mechanism of TAM is still poorly understood.
The genetic requirements and mutation spectrum of TAM have been characterized in yeast strains using lys2ΔBgl, a +4 frameshift allele that reverts by acquisition of compensatory −1 frameshift mutations. In the high-transcription strain, 70% of the reversion mutations required Rev3p, a component of the translesion synthesis DNA Pol ζ, suggesting that TAM occurs at sites of DNA damage (8). In a subsequent report, it was demonstrated that strains deficient in nucleotide excision repair (rad1Δ, rad2Δ, or rad16Δ single mutants), recombination (rad52Δ), or base excision repair (ung1Δ or apn1Δ single mutants) exhibited synergistic increases in reversion rate in combination with high levels of transcription (32). Thus, bulky helix distortions and small DNA base damage both appear to contribute to TAM. In the same study, an analysis of the revertants in the high- and low-transcription strains showed that −1 frameshifts occurred more frequently at GC base pairs (74%) in the high-transcription strain but at AT base pairs (60%) in the low-transcription strain (32). Although the sequencing results demonstrated that high levels of transcription through a gene influence mutational specificity, they did not clarify the mechanism of TAM. In addition, the use of a reversion assay imposed an ascertainment bias, since only a specific subset of mutation types could be detected. Since it is not known if −1 frameshifts are a predominant class of mutations stimulated by high levels of transcription, previous studies may relate only to a subset of transcription-associated events. A forward mutation assay would be more appropriate for addressing these issues and exploring the mechanism of TAM.
In order to determine the full spectrum of TAMs, the present study examined the molecular nature of LYS2 forward mutations that arise under low- versus high-transcription conditions. We found that high levels of transcription through the LYS2 gene influence both the types and distributions of forward mutations. Specifically, we have identified a unique mutation signature, 2-nucleotide (nt) deletions, in the high-transcription strain.
MATERIALS AND METHODS
Media and growth conditions.
Yeast strains were grown nonselectively in YEP medium (1% yeast extract, 2% peptone) supplemented with 1% galactose-1% raffinose (YEPGR) or 2% dextrose (YEPD). Minimal medium for selective growth contained 0.67% yeast nitrogen base without amino acids, 2% agar, and either 2% dextrose (SD medium) or 2% galactose-2% glycerol-2% ethanol (GGE medium). Lysine auxotrophs were selected on α-aminoadipate medium (α-AA; 2 g/liter) (7), containing 1% galactose-1% raffinose as carbon sources and supplemented with required nutrients. ura3 mutants were selected on medium containing 5-fluoroorotic acid (5) and required nutrients. Canavanine-resistant (Canr) mutants were recovered on SD medium supplemented with required nutrients and 60 μg of filter-sterilized l-canavanine sulfate/ml. All incubations were at 30°C.
Strain constructions.
A list of yeast strains is given in Table 1. The low-transcription (SJR282) and high-transcription (SJR371) strains were described previously (8). Strain SJR455 contains the lys2ΔRV::hisG allele, in which the internal EcoRV fragment of LYS2 (−19 to +3628 relative to the ATG start codon) was replaced with bacterial hisG sequences. To generate this allele, the internal EcoRV fragment of pDP6 (see below) was replaced with the hisG-URA3-hisG cassette (2), yielding plasmid pSR292. Following transformation of the parent strain of SJR455 with SalI-digested pSR292 and selection of Ura+ transformants, pop-outs that had lost the URA3 gene and one copy of hisG were identified on 5-fluoroorotic acid medium.
TABLE 1.
Yeast strains
| Strain | Genotypea | Reference or source |
|---|---|---|
| SJR282 | MATα ade2-101oc his3Δ200 ura3ΔNco suc2 gal80Δ::HIS3 | 8 |
| SJR371 | Same as SJR282, pGAL-LYS2 | 8 |
| SJR455 | MATaura3ΔNco his4-619 leu2-R lys2ΔRV::hisG | This study |
| SJR459 | SJR455 URA3::lys2ΔB-2 | This study |
| SJR460 | SJR455 URA3::lys2ΔB-22 | This study |
| SJR463 | SJR455 URA3::lys2ΔB-6 | This study |
| SJR462 | SJR455 URA3::lys2ΔA-22 | This study |
| SJR461 | SJR455 URA3::lys2Δ5-8 | This study |
| SJR1909 | SJR455 lys5Δ::kan | This study |
| SLY186 | MATa/MATα ura3/ura3 leu2/leu2 his3/his3 | D. Johnson |
| MJL4 | SJR282 kan-pGAL-lys2-G1382T | This study |
| MJL5 | SJR282 kan-pGAL-lys2-C1399T | This study |
| MJL6 | SJR282 kan-pGAL-lys2-C1400G | This study |
The lys2Δ3′ alleles are illustrated in Fig. 1.
Additional deletion mapping strains were constructed by transforming SJR455 with integrating plasmids containing nested lys2Δ3′ alleles and URA3. Each lys2Δ3′ plasmid was targeted to integrate at the URA3 locus by digestion with SmaI and, following selection of Ura+ transformants, integration of a single copy of the plasmid was confirmed by Southern blot analysis. The lys2Δ3′ plasmids were constructed as follows, starting with plasmid pDP6, a pUC9-based plasmid containing a 5-kb genomic XbaI/HindIII fragment that encompasses the entire LYS2 locus (10). pDP6 was digested with SstII and SmaI, which both cut in the LYS2-distal polylinker region. The linearized plasmid was then treated for variable times with exonuclease III, a 3′-to-5′ exonuclease that digests only from the blunt SmaI end (18). Following treatment with mung bean nuclease to remove remaining single-stranded tails, the plasmid was religated and deletion endpoints were determined by sequence analysis. Finally, a 5.5-kb EcoRI fragment containing the URA3 locus (from pSR91) (36) was inserted at a unique EcoRI site located in the polylinker region immediately adjacent to the deletion endpoint of each lys2Δ3′ plasmid.
SJR1909 is a lys5Δ (deletion of nt 61 to 759 of the 819-nt open reading frame) derivative of SJR455 constructed by transformation with a PCR-generated lys5Δ::kan fragment. Following selection of transformants on YEPD medium containing 200 mg of Geneticin (Sigma)/liter, replacement of LYS5 with lys5Δ::kan was confirmed by PCR. Strains MJL4, MJL5, and MJL6 are pGAL-lys2 derivatives of base substitution (BS) mutants isolated from the low-transcription strains containing the lys2-G1382T, lys2-C1399T, and lys2-C1400G alleles, respectively. A PCR-generated kanMX6-pGAL1 fragment was used to replace the pLYS2 promoter with kan-pGAL. Primers 5′-ATAAGTAACAAGCAGCCAATAGTATAAAAAAAAATCTGAGTTTATTACCTTTCCTGGAATGAATTCGAGCTCGTTTAAAC-3′ (forward) and 5′-ATGTGGTAACACTGAAAGAGTTGGATTATCCAACTTCTCTATCCAGACCTTTTCGTTAGTCATTTTGAGATCCGGGTTTT-3′ (reverse) were used to amplify a 2.1-kb fragment using plasmid pFA6a-kanMX6-PGAL1 (29). Following selection of trans-formants on Geneticin medium, replacement of pLYS2 with pGAL was confirmed by PCR.
Mutation rate measurements.
Independent 2-day-old colonies on YEPD agar (approximately 4 × 106 cells) were inoculated into 5 ml of YEPGR medium and grown nonselectively to a density of 1.2 × 108 cells/ml with shaking at 300 rpm for 3 to 4 days. Cells were washed once and resuspended in 5 ml of sterile distilled H2O. For LYS2 forward mutation rate experiments, strains SJR371 and SJR282 were plated at densities of 2 × 106 and 2 × 107 cells, respectively, per α-AA plate. Because previous experiments indicated that Lys− cells grew poorly when plated at low densities on α-AA medium (unpublished data), 8 × 107 SLY186 diploid cells were also plated on each α-AA plate. The SLY186 diploid strain was grown for 3 to 4 days in YEPD medium, washed twice, concentrated, and plated together with the experimental strains in a volume of less than 200 μl. Colonies began arising on day 8 after selective plating, and day 8 to 10 summed counts were used for rate calculations. For determination of Canr mutation rates, 2 × 107 cells were plated onto canavanine medium, and colonies were counted on day 4 after selective plating. Mutation rates were determined by the method of the median (25), and 95% confidence intervals (CIs) were calculated as described in reference 9. For all mutation rate calculations, data from 12 or more independent cultures were used.
Isolation and mapping of independent lys2 mutant alleles.
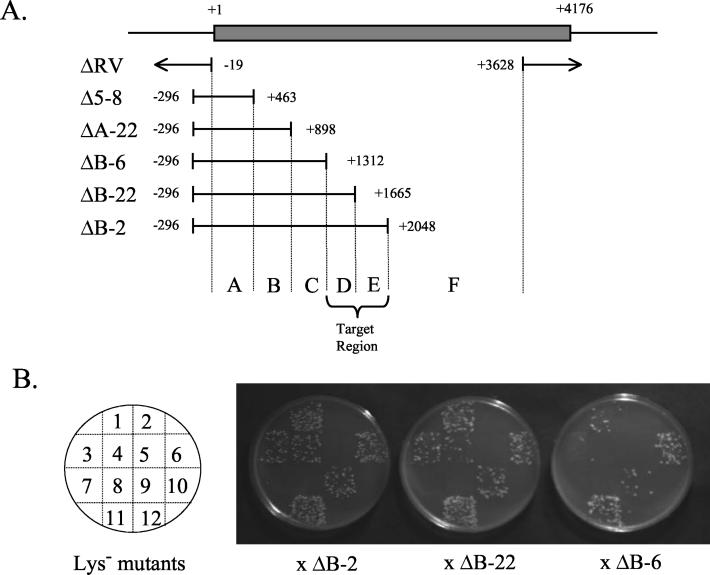
Independent cultures of high- and low-transcription strains were grown as described above for the mutation rate experiments. Aliquots containing approximately 3 × 106 or 3 × 107 cells of the high- and low-transcription strains, respectively, were combined with approximately 3 × 107 washed SLY186 cells and pelleted by centrifugation. Cell pellets were resuspended in 100 μl of distilled H2O, plated onto α-AA medium, and incubated for 8 to 10 days. To ensure independence of mutants, only a single α-AA-resistant (α-AAr) colony derived from each culture was subjected to further analysis. Following purification on YEPD agar, α-AAr colonies were patched onto SD medium lacking lysine to confirm a Lys− phenotype. Less than 5% of α-AAr colonies exhibited a strong Lys+ phenotype, and these were not analyzed further. The approximate position of the lys2 mutation in each Lys− mutant was determined by examining the production of prototrophic recombinants when the mutant was crossed to a series of tester strains containing defined lys2 deletion alleles (Fig. 1). Briefly, a plate containing patches of mutants and a plate containing a lawn of a given tester were each replica plated onto the same YEPD plate. The plate was incubated overnight in order to allow mating to occur and then was replica plated to selective medium on which only diploids would grow. Matings with SJR455 were replica plated to GGE minimal medium supplemented with uracil and lysine; matings with all other testers were replica plated to GGE complete medium deficient in uracil and leucine. After 3 days of growth, the diploid patches were replica plated to GGE complete medium deficient in lysine. These plates were irradiated with UV in order to induce recombination, immediately wrapped in aluminum foil, and then incubated for 3 days before scoring prototroph (papillae) production. Production of large numbers of papillae from the SJR455-derived diploids (SJR455 contains only the lys2ΔRV::hisG allele, which removes from −19 to +3628) could represent lys2 alleles with high reversion rates, or lys2 alleles mapping upstream of the LYS2 coding sequence or at the 3′ end of the LYS2 coding sequence. These were not pursued further. The pattern of papillae production from diploids derived from matings with the other five lys2Δ3′ tester strains indicated the interval (A to F) within which a given lys2 allele was located (Fig. 1). Some diploids exhibited confluent growth on lysine-deficient medium (i.e., complementation) and were assumed to represent lys5 mutations, as α-AA selects for mutations at either LYS2 or LYS5 (7). This was confirmed by observing noncomplementation when the haploid was mated to a lys5Δ strain (SJR1909).
FIG. 1.
LYS2 locus and deletion mapping. (A) The 4.2-kb LYS2 locus is shown as a shaded rectangle, below which the lys2ΔRV deletion allele (ΔRV) and five lys2Δ3′ alleles (Δ5-8 through ΔB-2) are shown. Deletion end points define six LYS2 subregions (A to F), to which forward mutations were mapped. (B) Typical mapping results are shown. Twelve Lys− mutants were mated to each of three deletion tester strains, SJR459, SJR460, and SJR463, containing alleles ΔB-2, ΔB-22, or ΔB-6 (Table 1), which possess different lys2Δ3′ alleles as indicated in panel A. Lys+ colonies (“papping”) on lysine-deficient medium result from homologous recombination between the uncharacterized lys2 forward mutation and nonoverlapping deletion alleles. The specific testers that resulted in Lys+ recombinants when mated to mutants allowed determination of the position of the uncharacterized forward mutation in each lys2 allele. For example, mutant 2 did not form recombinants with any of the three testers (−, −, −), indicating that its mutation maps distal to nt 2048; mutant 3 produced a (+, +, −) pattern, indicating its mutation is in region D.
DNA sequence analysis.
We sequenced mutations mapping within a 736-bp LYS2 region spanning nt 1312 to 2048, henceforth referred to as the LYS2 target region (subregions D and E in Fig. 1). Total genomic DNA was isolated from 5-ml YEPD cultures by glass bead lysis (19) and resuspended in 100 μl of Tris-EDTA, pH 7.6. The LYS2 target region was amplified from 1-μl genomic DNA samples in 50-μl reaction volumes using 200 nM concentrations of forward primer (5′-AGGTGTTGTAGTTGGACCAGATT-3′; LYS2F +1218) and reverse primer (5′-TACCGCAACATTCACAGTCA-3′; RLYS2 +2089). PCR mixtures (3.6 μl) were purified by treatment with ExoSAP-IT (U.S. Biochemicals) and sequenced using the BigDye Terminator cycle sequencing ready reaction kit (ABI Prism). Mutations in regions D and E were sequenced using primers 5′-AAGGGTGTTCTTGGTAGA-3′ (forward, LYS2F +1285) and 5′-AAGCTACTACACCATTCC-3′ (forward, LYS2F +1561), respectively. Sequencing reaction mixtures were sent to the Iowa State DNA sequencing facility for analysis. DNA sequence data were submitted to the Saccharomyces Genome Database website for BLASTn analysis to obtain nucleotide alignments to wild-type LYS2 sequence, and mutations were identified as nonaligned sequence.
Statistical analysis.
Mutation distributions and occurrences were compared using either standard chi-square or contingency chi-square analysis as appropriate. These analyses were done either manually or using the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html). Mutation rates were compared using 95% CIs (9).
RESULTS
Mapping forward mutation positions to LYS2 subregions.
The low-transcription and high-transcription strains (SJR282 and SJR371, respectively) (Table 1) differ only with respect to the promoter that drives expression of the chromosomal LYS2 locus. In SJR282 expression is regulated by the low-level pLYS2 promoter, while in SJR371 pLYS2 was replaced by the highly inducible pGAL1-10 promoter (8). Under the growth conditions used here, there was an approximately 20-fold increase in steady-state LYS2 mRNA levels in the high- relative to the low-transcription strain (data not shown). Consistent with a previous report (8), the LYS2 forward mutation rate was elevated significantly in the high- relative to the low-transcription strain (17.5 × 10−7 versus 4.8 × 10−7, respectively) (Table 2). In contrast, the forward mutation rate at the CAN1 locus, which experienced the same level of transcription in both strains, did not differ (data not shown). The demonstration of TAM in a forward mutation assay suggested that transcription might stimulate a broader range of mutation types than those detected by reversion of the lys2ΔBgl frameshift allele. In order to determine the full spectrum of TAMs, we isolated and sequenced forward mutations at the LYS2 locus from both high- and low-transcription strains. Due to the large size of the LYS2 open reading frame (4.2 kb), mutations were first assigned to specific subregions of LYS2 by deletion mapping, and those mutations mapping to a specific region were sequenced. Briefly, individual alleles were mapped by crossing each Lys− mutant to haploid tester strains containing a nested series of lys2Δ3′ alleles. The resulting diploids were assessed for their ability to produce Lys+ recombinants, thus allowing individual mutations to be mapped to one of six defined LYS2 subregions (Fig. 1). Of the 931 Lys− mutants analyzed, 195 (21%) produced recombinants with all testers and, therefore, could not be classified (see Materials and Methods). In addition, 40 mutants (4.3%) complemented the lys2Δ3′ alleles in all testers and were confirmed to contain mutations at the LYS5 locus.
TABLE 2.
LYS2 forward mutation rates and spectra of target region mutations in low- and high-transcription strains
| Mutation type | Low-transcription strain
|
High-transcription strain
|
Ratio of rates (high/low) | ||
|---|---|---|---|---|---|
| No. (%) | Estimated rate (10−7)a | No. (%) | Estimated rate (10−7)a | ||
| Total (all types) | 73 (100) | 4.8b | 82 (100) | 17.5b | 3.6 |
| Transitions | 20 (27) | 1.32 | 8 (9.8) | 1.71 | 1.3 |
| AT→GC | 1 (1.4) | 0.066 | 1 (1.2) | 0.21 | 3.2 |
| GC→AT | 19 (26) | 1.25 | 7 (8.5) | 1.49 | 1.2 |
| Transversions | 27 (37) | 1.78 | 18 (22) | 3.84 | 2.2 |
| AT→CG | 3 (4.1) | 0.20 | 1 (1.2) | 0.21 | 1.1 |
| TA→AT | 1 (1.4) | 0.066 | 1 (1.2) | 0.21 | 3.2 |
| GC→TA | 14 (19) | 0.91 | 8 (9.8) | 1.72 | 1.9 |
| GC→CG | 9 (12) | 0.58 | 8 (9.8) | 1.72 | 3.0 |
| Total BS | 47 (64) | 3.09 | 26 (32) | 5.55 | 1.8 |
| Short Ins-Del | |||||
| +1 | 3 (4.1) | 0.20 | 6 (7.3) | 1.28 | 6.4 |
| +2 | 0 | <0.066 | 1 (1.2) | 0.21 | >3.2 |
| −1 | 17 (23) | 1.12 | 17 (21) | 3.63 | 3.2 |
| −2 | 0 | <0.066 | 17 (21) | 3.63 | >55 |
| −3 | 0 | <0.066 | 2 (2.4) | 0.43 | >6.5 |
| −4 | 0 | <0.066 | 3 (3.6) | 0.64 | >9.7 |
| Total Ins-Del | 20 (27) | 1.32 | 46 (56) | 9.82 | 7.4 |
| Deletions (size) | 1 (33 nt) | 0.066 | 2 (18 or 26 nt) | 0.43 | 6.5 |
| Duplication (size) | 0 | <0.066 | 1 (115 nt) | 0.21 | >3.2 |
| Complex | 1 | 0.066 | 6 (7.3) | 1.28 | 19.4 |
| Multiple BS | 4 (5.5) | 0.263 | 1 (1.2) | 0.213 | 0.81 |
Estimated rates were determined by multiplying the proportion occurrence of specific mutation types by the total mutation rate for that strain. When no events were observed, the rate was estimated assuming the occurrence of one event.
95% confidence intervals for the low- and high-transcription strains were (4.4 to 5.6) × 10−7 and (15.1 to 21.4) × 10−7, respectively.
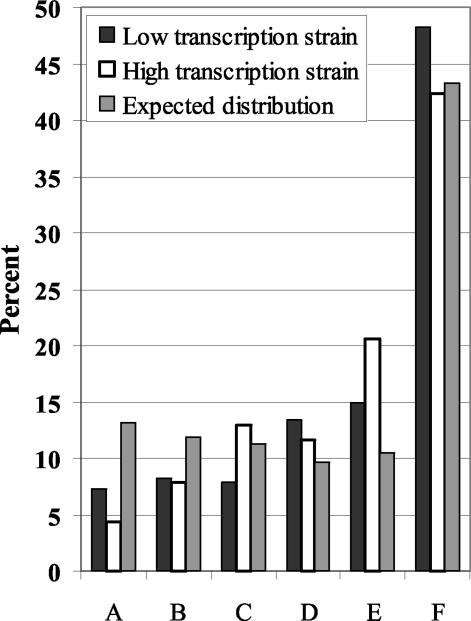
The percentages of 328 low- and 368 high-transcription mutations mapping within each of the six subregions (A to F) are illustrated in Fig. 2. The expected percentages based on the size of each subregion relative to the total A-F region are also shown. Both the low- and high-transcription mutation distributions differed significantly (P < 0.001) from random expectations, and these nonrandom distributions may reflect either mutational hot spots or cold spots or functionally important regions of the corresponding protein. Comparison of the low- and high-transcription mutation distributions by contingency chi-square analysis revealed significant differences between them as well (χ2 = 11.8; P = 0.04), with more high- than low-transcription mutations occurring in regions C and E and fewer high- than low-transcription mutations in regions A and F. Although there were subtle differences in mutation distributions, the mutagenic effect of transcription appeared to be more or less constant across the locus.
FIG. 2.
Mutation distributions among LYS2 subregions A to F from low- and high-transcription strains compared to the expected distribution based on random occurrence. Mutation positions in 328 and 368 mutants from the low- and high-transcription strains, respectively, were mapped to LYS2 regions A through F as described in the legend for Fig. 1.
Mutation types differ between low- and high-transcription strains.
We sequenced the LYS2 forward mutations that mapped to target regions D and E and were able to identify the molecular alteration in 73 of 79 low- and 82 of 88 high-transcription mutants. The remaining mutations may represent mapping errors and were not analyzed further. The mutation spectra are displayed in Fig. 3, and the types of mutations in each spectrum are compiled in Table 2.
FIG. 3.
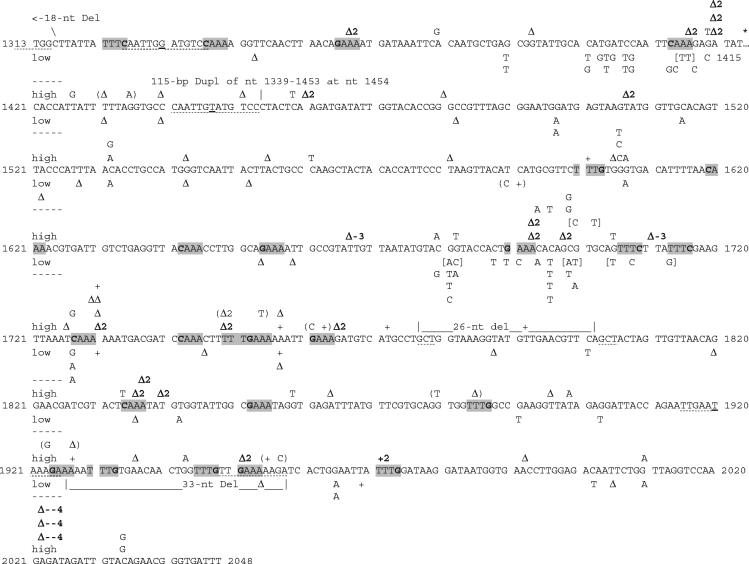
Spectra and distributions of 73 and 82 lys2 forward mutations from the low- and high-transcription strains, respectively. The wild-type LYS2 sequence spanning the forward mutation target region, nt 1313 to 2048, is shown (first nt is translational start), with one exception. An asterisk is placed at position 1415 to identify where a 5-nt stretch, 5′-GTTTA-3′ spanning nt 1416 to 1420, was omitted to improve spacing of distal regions relative to row ends. No mutations occurred there. Throughout the target sequence, spaces separate 10-nt stretches and numbers at row beginnings and ends identify nt positions. Mutations are indicated at points of occurrence above and below the LYS2 wild-type sequence for high- and low-transcription events, respectively. The exact positions for events within repeated sequences are ambiguous. Symbols: +, 1-nt Ins; Δ, 1-nt Del; Δ2, Δ-3, and Δ-4, Del of 2, 3, and 4 nt, respectively; +2, 2-nt Ins. Complex mutations are shown in parentheses, and multiple BS are shown in brackets. The 2- to 4-nt Ins-Del events associated with high transcription are indicated in bold font. An 18-bp deletion of nt 1299 to 1316 extended proximal to the target region. Repeats flanking duplication and deletions are identified by stippled underlines, and any nonmatched nt within imperfect repeats are identified by solid underlines. Gray highlighting identifies sites where the 5′-[G/C]AAA-3′ motif occurs in the TS or NTS (shown), and the bold font at the G/C indicates the motif 5′ end.
In the low-transcription strain, BS comprised the majority (64%) of mutations, with most of the remaining mutations (27%) being 1-nt insertion-deletion (Ins-Del) events. In contrast, short Ins-Del events comprised a majority (56%) of the high-transcription mutations, and only 32% of the events were BS. The low- and high-transcription spectra thus clearly differed, with a preponderance of BS mutations in the former and frameshifts in the latter (P < 0.001 when mutations were categorized as BS, Ins-Del, or other). Although the proportions of BS events differed in the low- and high-transcription strains, the types of changes observed were similar. In both the low- and high-transcription strains, BS occurred much more often (approximately eightfold more often) at GC base pairs than at AT base pairs, despite the fact that the GC content of the target region sequence was only 38%. In addition, the relative proportions of transition versus transversion mutations were not statistically different in the low- and high-transcription spectra. The difference between spectra was not due to a general shift towards frameshift mutagenesis in the high-transcription strain, as the proportions of single-nucleotide Ins-Del events were almost identical (27 and 28% in the low- and high-transcription spectra, respectively), and both spectra exhibited a strong bias for −1 events. The major difference corresponded instead to the large proportion of 2-nt deletions (21%) in the high-transcription spectrum, whereas none of these events was observed among the 73 low-transcription mutations.
In addition to simple BS and short Ins-Del events, we also recovered four multiple BS mutations (5.5%), a single complex mutation (TC→CCC), and a single 33-nt deletion flanked by imperfect (10 of 11) direct repeats from the low-transcription strain. From the high-transcription strain, we recovered one 2-nt insertion, two 3-nt deletions, and three identical 4-nt deletions, as well as six 1-nt insertions. Finally, two deletions of 18 and 26 nt flanked by 5- and 3-nt direct repeats, respectively, one 115-nt duplication flanked by imperfect 13-nt direct repeats, six complex mutations involving a frameshift and closely spaced BS, and a multiple BS were recovered from the high-transcription strain. The high-transcription complex mutations contained a BS that was near but not adjacent to the frameshift, in contrast to the single low-transcription complex event, which involved adjacent nucleotides. As these noncanonical types of mutations were present in low numbers, it was not possible to demonstrate a significant difference between the low- and high-transcription strains with respect to them.
Mutation distributions within the target region.
Of 20 Ins-Del of a single nucleotide from the low-transcription spectrum, 18 occurred at independent positions spread over the target region. The remaining 1-nt insertion and 1-nt deletion occurred at the 6-A run beginning at nt position 1753 (Fig. 3). Similarly, the 23 1-nt Ins-Del mutations in the high-transcription spectrum were spread across the target region, with the majority (17) occurring at unique sites. Of the remaining events, three −1 and one +1 frameshift occurred at the 6-A run beginning at nt position 1728, and single −1 and +1 frameshifts occurred at the 6-A run beginning at nt position 1753. In contrast to the 1-nt frameshifts, the majority of 2- and 4-nt Ins-Del events in the high-transcription strain were clustered, with 10 of the 17 2-nt deletions occurring in three short regions. Four of the 2-nt deletions occurred near nt position 1410, and three of these were identical deletions of an AT or TA dinucleotide. Two additional clusters of three 2-nt deletions occurred near nt positions 1696 and 1838, and three identical 4-nt deletions occurred near nt position 2022 at a 4-nt tandem repeat (AGATAGAT).
In contrast to the 1-nt Ins-Del in the low-transcription spectrum, many of the BS appeared to be clustered. Two notable clusters were centered around positions 1395 and 1695 and presumably correspond to functionally important regions of the Lys2 protein. The latter cluster contained five BS at nt 1680 to 1682, four C-to-T transitions at nt 1696, and three of the four multiple BS. This cluster was also evident in the high-transcription spectrum. In the cluster centered at nt position 1395, however, 10 BS occurred in the low-transcription spectrum but there were no high-transcription mutations. It is possible that mutations in this region lead to growth on α-AA when a normal amount of protein is produced, but not when large quantities of a protein with partial function are produced under the high-transcription conditions. To explore this possibility, we inserted the pGAL promoter in front of three of the lys2 BS: a G→T transversion at position 1382 (G1382T), a C1399T transition, and a C1400G transversion. The resulting high-transcription and parental low-transcription strains were plated on α-AA medium together with background cells to mimic the original mutant isolation conditions. There was no difference in the sizes of the low- versus high-transcription Lys− mutant colonies (data not shown).
Sequence context of 2- to 4-nt Ins-Del mutations in the high-transcription spectrum.
Table 3 presents sequence characteristics of sites where the 2- to 4-nt Ins-Del occurred. The most common characteristic at the 2-nt deletion sites is a repeated dinucleotide. For example, four AT or TA deletion events occurred in 5′-ATAT-3′ sequences beginning at nt 1412 and 1837. Similarly, a deletion of AG or GA occurred at a 5′-AGAGA-3′ sequence beginning at nt 1408, and two AC or CA deletions occurred at a 5′-ACACA-3′ sequence beginning at nt 1693. Of the 14 sites where the 17 2-nt deletions occurred, 12 contained 1.5, 2, or 2.5 copies of the deleted dinucleotide. Only a minority (4 of 17) of 2-nt deletions occurred at homopolymer runs, one each in runs of 3-A, 4-A, 5-T, and 6-A, beginning at nt 1835, 1358, 1747, and 1728, respectively. The two 3-nt deletions occurred at 5′-TTGTT-3′ and 5′-TTATT-3′ sequences beginning at nt 1667 and 1710, respectively, each of which contained a nontandem TT dinucleotide. Finally, the 4-nt deletions occurred at a tandem repeat of the sequence 5′-AGAT-3′ beginning at nt 2022. In a total of 23 short 2- to 4-nt Ins-Del mutations, the positions of only two were unambiguous: a deletion of two As and a GA deletion at nt 1460 to 1461 and 1951 to 1952, respectively.
TABLE 3.
Sequence characteristics at sites of 2- 4-nt Ins-Del mutations in the high-transcription strain
| Position on NTSa | No. | Mutation | Context (5′→3′) | Repeat units | NTS sequence context (5′→3′) aligned by the 5′ G/C closest to the mutation | A-rich strand aligned by 5′ G/C closest to mutation (TS or NTS) |
|---|---|---|---|---|---|---|
| 1358 | 1 | −AA | 4-A run | 2 | G-AAAAT-G | A G* A A A A T G A T A (NTS) |
| 1408 | 1 | −AG/GA | AGAGA | 2.5 | C-AAAGAGATAT-G | T C* A A A G A G AT A T (NTS) |
| 1412 | 3 | −AT/TA | ATAT | 2 | G-ATAT-G | T C* A A A G A G AT A T (NTS) |
| 1460 | 1 | −AA | AA | 1 | C-AA-G | T C* A A G A T G A T A (NTS) |
| 1507 | 1 | −AT/TA | TAT | 1.5 | G-TAT-G | C C* A T A C T T A C T (TS) |
| 1667 | 1 | −3 | TTGTT | G-TATTGTTAATAT-G | A A C* A A T A C G G C A (TS) | |
| 1693 | 2 | −AC/CA | ACACA | 2.5 | G-AAACACA-G | T G* A A A C A C AG C G (NTS) |
| 1698 | 1 | −GC/CG | GCG | 1.5 | C-AGCGT-G | T G* A A A C A C AG C G (NTS) |
| 1710 | 1 | −3 | TTATT | C-TTATTT-C | C G* A A A T A A G A A (TS) | |
| 1728 | 1 | −AA | 6-A run | 3 | C-AAAAAAT-G | T C* A A A A A A T G A (NTS) |
| 1747 | 1 | −TT | 5-T run | 2.5 | C-TTTTT-G | T C* A A A A A G T T T (TS) |
| 1764 | 1 | −AG/GA | AGA | 1.5 | G-AAAGAT-G | T G* A A A G A T G T C (NTS) |
| 1835 | 1 | −AA | 3-A run | 1.5 | C-AAATAT-G | T C* A A A T A T G T G (NTS) |
| 1837 | 1 | −AT/TA | ATAT | 2 | C-AAATAT-G | T C* A A A T A T G T G (NTS) |
| 1840 | 1 | −TG/GT | TGTG | 2 | C-AAATATGTG-G | T C* A A A T A T G T C (NTS) |
| 1951 | 1 | −GA | GA | 1 | G-AAAAA-G | T G* A A A A A G A T G (NTS) |
| 1971 | 1 | +TT | 3-T run | 1.5 | G-AATTATTT-G | C C* A A A T A A T T C (TS) |
| 2022 | 3 | −4 | AGATAGAT | 2 | G-AGATAGATT-G | A G* A G A T A G A T T (NTS) |
Positions are relative to +1 of coding sequence and correspond to numbering in Fig. 3. In the last two columns, underlined characters represent the sequence where the Ins-Del occurred. Bold is used in the final column to indicate where separate mutations within clusters occurred.
We observed two patterns in the sequence contexts adjacent to sites of 2- to 4-nt Ins-Del mutations in short repeats. First, they were AT rich. Because the mutational target is 62% AT, however, the relevance of this observation is unclear. Second, the 2- to 4-nt Ins-Del events occurred commonly, but not exclusively, distal to a G or C followed by a stretch of As. These patterns are evident in column 6 of Table 3, where the sequence contexts are arbitrarily aligned by the proximal G/C. Examples include the 2-nt deletions at the 5′-GAAAAT-3′ sequence beginning at nt 1358 and the 5′-CAAAGAGA-3′ sequence beginning at nt 1408. The alignment of the Ins-Del contexts was examined in two other ways, as illustrated in the final column of Table 3. First, using the G/C nucleotide that precedes the run of As as the fixed point for aligning the sites, 10 nt of the nontranscribed strand (NTS) sequence were aligned, beginning with the nucleotide just 5′ to the G/C and extending 8 nt in the 3′ direction. An effect of this was that mutations comprising clusters collapsed into single sites. Second, we noticed that several of the NTS mutation contexts did not seem to fit the emerging pattern, specifically, the sites where 2-nt deletions occurred at positions 1747 and 1507, the 2-nt insertion at 1971, and the two 3-nt deletions at positions 1667 and 1710 (see column 6 in Table 3). For these five mutation contexts, there was no run of As present. After closer inspection, however, we identified 5′-T(2-5)-[G/C]-3′ motifs in four of those mutation contexts (excepting the 2-nt deletion at position 1507), indicating the presence of the 5′-[G/C]-AA…-3′ pattern in the complementary or transcribed strand (TS) at these sites. For these mutation sites, we therefore aligned TS sequence rather than NTS sequence (final column of Table 3).
In order to explore whether the 5′-[G/C]-A(2-6)-3′ motif identified in the above analysis is associated with other mutation types, we compared the occurrence of 2-nt deletions, 1-nt frameshifts, and simple BS at the motif sites. First we defined a more specific test motif. The sequence 5′-[G/C]-AAA-3′ was chosen because a majority of 2- to 4-nt Ins-Del events fit this pattern and because this motif occurs 24 times when both strands of the target region are considered. Fifteen of the motifs occur in the NTS, and nine occur in the TS. In Fig. 3, motifs in the TS (not shown) are identified as 5′-TTT[C/G]-3′ sites in the NTS. In addition to defining a more specific test motif, we included in the analysis only those events that could have occurred at or within 4 nt 3′ of the motif G/C at each site. By imposing these restrictions, 11 of the 23 Ins-Del of 2 to 4 nt did not fit the more strict pattern, leaving 12 (or 52%). In contrast to 2- to 4-nt Ins-Del, only 4 of 26 (15.4%) simple BS occurred at motifs. The distributions of mutations within versus outside of the motifs were significantly different for the 2- to 4-nt Ins-Del versus the BS (P = 0.015). A similar comparison between the occurrence of 2- to 4-nt Ins-Del at motif sites to the occurrence of 1-nt frameshifts at motif sites (8 of 23, or 35%) did not yield a significant difference (P = 0.37). Six of the eight 1-nt frameshifts occurred at two 6-A runs (nt 1728 and 1753), however, and homopolymer runs are known hot spots for 1-nt frameshifts. If the run-associated 1-nt frameshifts were eliminated from the analysis, then the 1-nt frameshift occurrence at motif sequences decreased to approximately 12% (2 of 17). In contrast, 2- to 4-nt Ins-Del occurrence at motifs was not affected (11 of 22, or 50%) when run-associated 2-nt deletions were eliminated. Chi-square analysis comparing the 11 of 22 versus 2 of 17 was significant (P = 0.03).
DISCUSSION
High levels of transcription through the LYS2 gene have been shown to stimulate forward mutations as well as reversion of a +4 frameshift allele (8). Determining the spectrum of transcription-associated frameshift revertants was straightforward but uninformative, presumably because of the limited types of events detected (32). While a forward mutation assay affords the opportunity to examine a wide range of mutation types, the large size of the LYS2 locus has limited its utility for this type of analysis. It not only is difficult to identify unique mutations within a large target, but one also must sequence large numbers of mutations in order to uncover mutagenesis patterns. To minimize these problems, we developed a series of deletion tester strains that allowed the rapid mapping of forward mutations to specific LYS2 subregions (Fig. 1) and then focused our efforts on sequencing forward mutations mapping to a defined interval.
There were distinct differences in the types of forward mutations that accumulated under high- versus low-transcription conditions (Table 2). Whereas BS accounted for 64% of the mutations identified under low-transcription conditions, they comprised only 32% of the events that arose under high-transcription conditions. Among the BS identified under high- or low-transcription conditions, transversions were more frequent than transitions, with the overall BS pattern being similar to that reported at CAN1 (20). In spite of the similarities between the overall BS patterns in the low- and high-transcription spectra, the distributions of BS were very different. In particular, a cluster of BS mutations near nt 1395 in the low-transcription spectrum was absent from the high-transcription spectrum. A rate for the BS near nt 1395 can be estimated by multiplying the total forward mutation rate by the proportion of total events that are of this type. In the low-transcription strain, the proportion of BS near nt 1395 was 10 of 73, giving an estimated BS rate of 6.6 × 10−8 (95%CI, 6.0 × 10−8 to 7.6 × 10−8). In the high-transcription strain, no BS near nt 1395 was seen among the 82 mutations sequenced. If it were assumed that one event was detected, the estimated rate would be 2.1 × 10−8 (CI of 1.8 × 10−8 to 2.6 × 10−8), which is significantly lower than that in the low-transcription strain. One trivial explanation for the rate difference is that high-level expression of hypomorphic BS alleles produces enough Lys2 enzymatic activity to prevent growth on α-AA. This possibility was eliminated, however, by demonstrating that some of the relevant BS mutant alleles that had been recovered from the low-transcription strain were able to support growth on α-AA when expressed at high levels (see Results). These data suggest that high levels of transcription do not simply impose additional mutations on top of a basal pattern of mutagenesis, but rather that high transcription alters the basic mechanism(s) of mutagenesis.
Concomitant with a transcription-associated decrease in the measured percentage of BS, short Ins-Del mutations comprised a larger percentage of the high-transcription mutations than of the low-transcription mutations (56 and 27%, respectively). All of the short Ins-Del events isolated under low-transcription conditions were single-nucleotide events and, as seen in other assays, deletions greatly outnumbered insertions (13, 20, 26). In contrast to the presence of only 1-nt Ins-Del events in the low-transcription spectrum, there were many 2-nt Del in the high-transcription spectrum. Not only are 2-nt Del rarely seen among frameshift mutations in yeast, but when they have been observed, they have generally been in long mononucleotide or dinucleotide runs (17, 42). The −2 events associated with high levels of transcription generally occurred where there were only 1.5 to 2.5 copies of a dinucleotide repeat, a repeat number that likely is too small to promote slippage of the replicative DNA polymerases.
In the high-transcription spectrum, the 2-nt Del and the 4-nt Del did not occur at random locations but appeared to cluster at specific sites in the LYS2 target region. By aligning these sites, we were able to identify a motif (5′-[G/C]AAA-3′) that may be associated with these events. In considering the significance of this motif with regard to TAM, it should be noted that this motif is likely overrepresented in the yeast genome, both because the genome is AT rich and because it has an underrepresentation of TA dinucleotides (23). In addition, the motif was not present at 4 of the 14 sites where we recovered 2-nt Del events, and no 2-nt Del events were recovered at 16 of the 24 sites where the motif was present. Thus, although the motif appears to be associated with −2 events, its presence or absence is not highly predictive of where these mutations will occur. Clearly, additional experiments using an assay that is designed to detect −2 events will be necessary to determine the relevance of the motif to TAM.
A possible reason for the apparent clustering of the 2-nt Del events is that the cluster sites represent sequence contexts (or structures) that are susceptible to mutation-initiating events or that are repaired inefficiently. It has been shown, for example, that a high level of transcription through a plasmid-based microsatellite both increases polymerase errors and reduces mismatch repair (MMR) efficiency (43). As noted above, however, the 2-nt Del observed here do not occur in the type of extended dinucleotide repeat that facilitates slippage of replicative DNA polymerases (17, 42). In addition, the high-transcription LYS2 forward mutation spectrum is very different from the CAN1 forward mutation spectra obtained in various MMR-deficient strains (30), which argues against a primary role of MMR inhibition in the generation of the 2-nt Del events. Given that the 2-nt Del events are not characteristic of the types of mutations associated with DNA replication, the most likely explanation for clustering of these events is susceptibility of specific sites to mutation-initiating DNA damage. The question remains as to how the level of transcription might influence mutagenesis in a sequence-dependent manner. Because a direct correlation between transcription level and topoisomerase activity at the LYS2 locus would be expected (28), we suggest that the 2-nt Del events might be generated in response to topoisomerase-generated nicks.
Although the most striking difference between the high- and low-transcription spectra was the presence of the novel transcription-associated −2 events, it should be noted that most other types of mutations were also stimulated by high levels of transcription, but to a lesser extent. The overall rate of BS occurrence was slightly higher (1.8-fold [Table 2]) in the high-transcription than in the low-transcription strain. The small effect of high levels of transcription on BS mutagenesis detected at LYS2 is consistent with the observed increase in BS in response to transcriptional induction in E. coli (3, 24). In the E. coli studies, a specific increase in C→T mutations has been attributed to enhanced deamination of cytosines located on the NTS, presumably because of the transcription-associated single-stranded character of this strand (3). It has recently been demonstrated that cytosines on the NTS are preferred targets for AID, a deaminase involved in initiating somatic hypermutation of immunoglobulin genes (35, 37). In addition to enhancing BS, an increase in large deletions that inactivate a plasmid-based lacZ target has been reported when high levels of transcription oppose the direction of replication fork movement in E. coli (40). In this case, it is thought that the deletions result from replication fork collapse. Although we did not detect any deletions larger than 33 nt in the target region analyzed here, larger deletions may have occurred in other LYS2 regions that were not analyzed. Finally, as reported previously using a frameshift reversion assay (8), transcription stimulated the occurrence of 1-nt deletions.
The molecular characterization of forward mutation events at LYS2 has demonstrated that high levels of transcription not only stimulate a wide variety of mutation types but also generate a novel mutation signature in the form of 2-nt Del events. In mammalian cells, −2 events have been reported to comprise 21 or 11.5% of the mutations within a human HPRT cDNA target integrated into a mouse or human cell line, respectively, as part of a retroviral vector (22, 27). In contrast, −2 events constitute only 1.5% of mutations occurring at the endogenous human HPRT locus (http://info.med.yale.edu/mutbase/) (6). Although the source of this discrepancy is not known, we suggest that elevated transcription may be the cause, as the cDNA targets were regulated by the strong Moloney murine leukemia virus long terminal repeat promoter (22, 27). If correct, this would indicate that TAM is a general phenomenon that occurs in mammalian cells as well as yeast. Based on our previous genetic analyses of TAM associated with reversion of a +4 frameshift mutation in yeast, it is likely that many of the TAMs examined here involve the damage bypass activity of the Pol ζ translesion DNA polymerase (8). It will be interesting to determine whether the 2-nt Del events characterized in this study also involve the activity of Pol ζ. Future studies will focus on determining the genetic requirements of the novel, transcription-associated 2-nt Del and exploring the role of sequence context in TAM.
Acknowledgments
We thank Sarah Musella for technical assistance, Nayun Kim for quantifying RNA, Douglas Johnson (University of Vermont) for his gift of strain SLY186, Mark Longtine (Oklahoma State University) for his gift of plasmid pFA6a-kanMX6-PGAL1, and George Hoffman (College of the Holy Cross) for valuable discussions and helpful comments on the manuscript.
This work was supported by Vermont Experimental Program to Stimulate Competitive Research grant number EPS 0236976 to M. J. Lippert and the Vermont Genetics Network through National Institutes of Health grant number 1 P20 RR16462 from the BRIN Program of the National Center for Research Resources to M. J. Lippert. In addition, this work was supported by National Institutes of Health grant GM-38464 to S. Jinks-Robertson. J. A. Freedman was partially supported by the Graduate Division of Biological and Biomedical Sciences of Emory University. M. A. Barber was supported by a Summer Undergraduate Research Fellowship from Pfizer, Inc.
REFERENCES
- 1.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beletskii, A., and A. S. Bhagwat. 1996. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beletskii, A., A. Grigoriev, S. Joyce, and A. S. Bhagwat. 2000. Mutations induced by bacteriophage T7 RNA polymerase and their effects on the composition of the T7 genome. J. Mol. Biol. 300:1057-1065. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Cariello, N. F., and T. R. Skopek. 1993. In vivo mutation at the human HPRT locus. Trends Genet. 9:322-326. [DOI] [PubMed] [Google Scholar]
- 7.Chattoo, B. B., F. Sherman, D. A. Azubalis, T. A. Fjellstedt, D. Mehnert, and M. Ogur. 1979. Selection of lys2 mutants of the yeast Saccharomyces cerevisiae by the utilization of α-aminoadipate. Genetics 93:51-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta, A., and S. Jinks-Robertson. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268:1616-1619. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, W. J., and F. J. Massey, Jr. 1969. Introduction to statistical analysis, 3rd ed. McGraw-Hill, New York, N.Y.
- 10.Fleig, U. N., R. D. Pridmore, and P. Philippsen. 1986. Construction of LYS2 cartridges for use in genetic manipulations of Saccharomyces cerevisiae. Gene 46:237-245. [DOI] [PubMed] [Google Scholar]
- 11.Freedman, J. A., and S. Jinks-Robertson. 2002. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics 162:15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Rubio, M., P. Huertas, S. Gonzalez-Barrera, and A. Aguilera. 2003. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics 165:457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giroux, C. N., J. R. Mis, M. K. Pierce, S. E. Kohalmi, and B. A. Kunz. 1988. DNA sequence analysis of spontaneous mutations in the SUP4-o gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:978-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene, C. N., and S. Jinks-Robertson. 1997. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol. Cell. Biol. 17:2844-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, C., P. Schaer, P. Munz, and J. Kohli. 1991. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol. Cell. Biol. 11:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 21:8949-8956. [DOI] [PubMed] [Google Scholar]
- 17.Harfe, B. D., and S. Jinks-Robertson. 1999. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4766-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff, S. 1984. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351-359. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 20.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huertas, P., and A. Aguilera. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12:711-721. [DOI] [PubMed] [Google Scholar]
- 22.Ikehata, H., T. Akagi, H. Kimura, S. Akasaka, and T. Kato. 1989. Spectrum of spontaneous mutations in a cDNA of the human hprt gene integrated in chromosomal DNA. Mol. Gen. Genet. 219:349-358. [DOI] [PubMed] [Google Scholar]
- 23.Karlin, S., I. Ladunga, and B. E. Blaisdell. 1994. Heterogeneity of genomes: measures and values. Proc. Natl. Acad. Sci. USA 91:12837-12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klapacz, J., and A. S. Bhagwat. 2002. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J. Bacteriol. 184:6866-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lea, D. E., and C. A. Coulson. 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 26.Lee, G. S., E. A. Savage, R. G. Ritzel, and R. C. von Borstel. 1988. The base-alteration spectrum of spontaneous and ultraviolet radiation-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol. Gen. Genet. 214:396-404. [DOI] [PubMed] [Google Scholar]
- 27.Lichtenauer-Kaligis, E. G., J. Thijssen, H. den Dulk, P. van de Putte, J. G. Tasseron-de Jong, and M. Giphart-Gassler. 1993. Genome wide spontaneous mutation in human cells determined by the spectrum of mutations in hprt cDNA genes. Mutagenesis 8:207-220. [DOI] [PubMed] [Google Scholar]
- 28.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 30.Marsischky, G. T., N. Filosi, M. F. Kane, and R. Kolodner. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407-420. [DOI] [PubMed] [Google Scholar]
- 31.Morales, V., C. Giamarchi, C. Chailleux, F. Moro, V. Marsaud, S. Le Ricousse, and H. Richard-Foy. 2001. Chromatin structure and dynamics: functional implications. Biochimie 83:1029-1039. [DOI] [PubMed] [Google Scholar]
- 32.Morey, N. J., C. N. Greene, and S. Jinks-Robertson. 2000. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics 154:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevo-Caspi, Y., and M. Kupiec. 1994. Transcriptional induction of Ty recombination in yeast. Proc. Natl. Acad. Sci. USA 91:12711-12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickoloff, J. A. 1992. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 12:5311-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham, P., R. Bransteitter, J. Petruska, and M. F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103-107. [DOI] [PubMed] [Google Scholar]
- 36.Saxe, D., A. Datta, and S. Jinks-Robertson. 2000. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 20:5404-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A. S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svejstrup, J. Q. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116:447-451. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 40.Vilette, D., S. D. Ehrlich, and B. Michel. 1995. Transcription-induced deletions in Escherichia coli plasmids. Mol. Microbiol. 17:493-504. [DOI] [PubMed] [Google Scholar]
- 41.Voelkel-Meiman, K., R. L. Keil, and G. S. Roeder. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48:1071-1079. [DOI] [PubMed] [Google Scholar]
- 42.Wierdl, M., M. Dominska, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics 146:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wierdl, M., C. N. Greene, A. Datta, S. Jinks-Robertson, and T. D. Petes. 1996. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics 143:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshiyama, K., and H. Maki. 2003. Spontaneous hotspot mutations resistant to mismatch correction in Escherichia coli: transcription-dependent mutagenesis involving template-switching mechanisms. J. Mol. Biol. 327:7-18. [DOI] [PubMed] [Google Scholar]