Abstract
How botulinum neurotoxins (BoNTs) cross the host intestinal epithelial barrier in foodborne botulism is poorly understood. Here, we present the crystal structure of a clostridial hemagglutinin (HA) complex of serotype BoNT/A bound to the cell adhesion protein E-cadherin at 2.4 Ångströms. The HA complex recognizes E-cadherin with high specificity involving extensive intermolecular interactions and also binds to carbohydrates on the cell surface. Binding of HA complex sequesters E-cadherin in the monomeric state thereby compromising the E-cadherin-mediated intercellular barrier and facilitating paracellular absorption of BoNT/A. We reconstituted the complete 14-subunit BoNT/A complex using recombinantly-produced components and demonstrated that abolishing either E-cadherin- or carbohydrate-binding of HA complex drastically reduces oral toxicity of BoNT/A complex in vivo. Together, these studies establish the molecular mechanism of how HAs contribute to the oral toxicity of BoNT/A.
Botulinum neurotoxin serotype A (BoNT/A, ~150 kDa) is produced in Clostridial botulinum and together with the non-toxic non-hemagglutinin (NTNHA) protein and three hemagglutinins (HAs: HA70, HA33 and HA17, also known as HA3, HA1 and HA2, respectively) forms a large protein complex (1, 2). NTNHA protects BoNT/A from host gastrointestinal destruction by forming a minimally functional progenitor toxin complex (M-PTC; ~290 kDa) (3). M-PTC associates with the HAs to form the large PTC (L-PTC; ~760 kDa), which exhibits up to 30-fold greater oral toxicity (4-6). Therefore, HAs are critical virulence factors that contribute to the extreme oral toxicity of BoNT/A (7-10). The HAs of BoNT/A (HA/As) assemble into a 3-fold symmetric hetero-dodecameric complex containing three HA70, three HA17, and six HA33 (~470 kDa). The fully assembled HA complex has nine carbohydrate-binding sites, which function by enriching the L-PTC onto cell surfaces in the intestine (Fig. 1A and fig. S1A) (9, 11). Additional interactions of HAs with the cell adhesion protein E-cadherin have been reported for the closely related HAs of BoNT serotype B (HA/Bs) (12), but the molecular mechanism by which HAs exploit E-cadherin and the physiological significance of this interaction in vivo remain poorly defined.
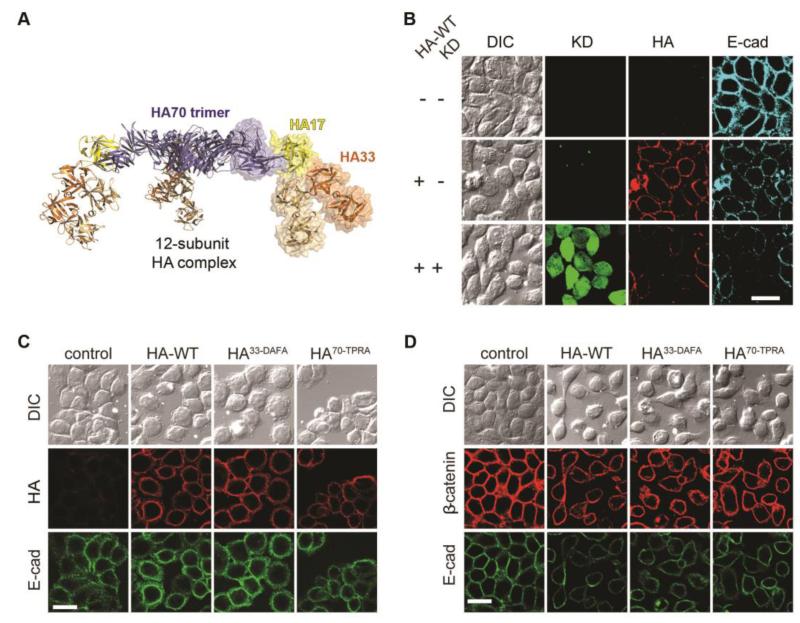
Fig. 1. The HA complex binds to E-cadherin and disrupts cell-cell adhesion.
(A) Structural presentation of the HA complex comprising three HA70 (blue), three HA17 (yellow), and six HA33 (orange) (9). The moiety of the mini-HA complex is shown as a transparent surface representation. (B) Knocking down (KD) E-cadherin (E-cad) in HT29 cells via a lenti-virus mediated shRNA expression reduced cell binding of the HA complex (25 nM, 90 min). Lenti-shRNA infected cells were marked with GFP. (C) Binding of HA-WT, HA33-DAFA, and HA70-TPRA complexes to HT29 cells (10 nM, 30 min). (D) HA-WT, HA33-DAFA, and HA70-TPRA all disrupted cell-cell adhesion in HT29 cells (10 nM, 24 hrs), marked by the separation of cells in both DIC images (upper panel) and immunofluorescence images of E-cadherin (lower panel) and β-catenin (middle panel). Scale bar: 20 μm.
We found that the binding of HA/As to the well-characterized human colorectal adenocarcinoma HT29 cells was greatly reduced when endogenous E-cadherin was knocked down, indicating that the cell-surface binding of HAs requires E-cadherin (Fig. 1B and fig. S1B). To distinguish E-cadherin binding versus carbohydrate binding, we next tested two HA complexes with point mutations that abolish their ability to bind carbohydrates (HA33-D263A/F278A and HA70-T527P/R528A, hereinafter referred to as HA33-DAFA and HA70-TPRA) (9). Binding of these two HA mutants to HT29 cells was similar to that of the wild-type HA complex (HA-WT), further supporting that E-cadherin is largely responsible for the observed cell-surface binding of HAs (Fig. 1C).
Binding of HAs to cells induces disruption of cell-cell adhesions. This may create a paracellular route for absorption of BoNTs across the intestinal epithelial barriers (12). Indeed, we observed that binding of HA-WT to HT29 cells for 24 hours severely disrupted cell-cell adhesions, shown by separation of cells (Fig. 1D). Consistent with their ability to bind HT29 cells, the two carbohydrate-binding deficient mutant HAs were still capable of disrupting cell-cell junctions of HT29 cells within 24 hours. Thus carbohydrate binding is not essential for HAs to disrupt cell-cell junctions under these experimental conditions. On the other hand, experiments with a much shorter incubation time (150 min) revealed that the two carbohydrate-binding deficient HA complexes disrupted cell-cell junctions much less than HA-WT (fig. S1C–D), suggesting that carbohydrate binding might enhance disrupting cell-cell junctions, likely by enriching HAs onto cell surfaces and increasing the overall binding avidity. Similar results on cell-surface binding of HAs and HA-induced cell-cell dissociation were observed on Caco-2 cells, another well-established human epithelial colorectal adenocarcinoma cell line (fig. S2).
How HAs recognize E-cadherin is not known. The ectodomain of E-cadherin mediates self-assembly in adherens junctions, which are crucial intercellular structures maintaining physical association between parallel apposed cells (13-15). E-cadherin consists of five tandem extracellular cadherin (EC) domains (EC1–5) (16), and the most amino-terminal EC1–2 domain binds to HA/Bs (12). We found that the EC1–2 of E-cadherin also binds to HA/As with a dissociation constant (Kd) of ~2.3 μM and 3:1 stoichiometry, indicating that each of the three arms of the HA complex likely binds one E-cadherin (fig. S3A). Consistent with this finding, a monomeric HA sub-complex (mini-HA), which represents one of the 3 identical arms of the HA complex comprising the D3 domain of HA70 (HA70D3, residues Pro378–Asn626), HA17, and two HA33 (Fig. 1A), is sufficient to bind EC1–2 in a 1:1 ratio (fig. S3B). The binding affinity of EC1–2 to the mini-HA complex (Kd ~2.7 μM) is similar to that of the 3-arm HA complex, indicating that the mini-HA complex is the minimum E-cadherin-binding unit.
To understand the molecular mechanism underlying the HA–E-cadherin interaction, we determined a 2.4 Å resolution crystal structure of the mini-HA complex bound to EC1–2 of mouse E-cadherin (Fig. 2A–B and table S1). Two sets of nearly identical hetero-pentameric complexes are observed in the asymmetric unit (root mean square deviation, rmsd, ~1.01 Å over 1,149 Cα pairs), in which EC1–2 binds to HA70D3, HA17, and one of the two HA33. No conformational change is observed for the mini-HA complex upon E-cadherin binding (9). The EC1 and EC2 domains each adopts a seven-stranded β-barrel fold and are connected by a short linker that is maintained in a rigid conformation by the interactions with three calcium atoms (Fig. 2A–B). Superimposing HA-bound EC1–2 to the structure of mouse E-cadherin EC1–5 (15) (rmsd ~1.57 Å for 206 Cα pairs) shows that the EC3–5 extends away from the HA complex (9), thus is dispensable for HA binding.
Fig. 2. Structure of the HA–E-cadherin complex.
(A) Structure of EC1–2 (red) bound to the mini-HA complex. Three E-cadherin-bound calcium atoms are depicted as green balls. The view direction is similar to that shown in Fig. 1A. (B) A 90° rotation of the complex about a horizontal axis. (C) The HA-bound E-cadherin (red) is superimposed with an E-cadherin (cyan) in the context of a trans dimer (its binding partner is shown as a surface representation). Large conformational changes are observed in the N-terminus of EC1 (residues 2–8 are shown as sticks). The residue Trp2 of the HA-bound E-cadherin occupies a pocket that otherwise accommodates the swapped Trp2 (dots representation) of its dimeric binding partner. (D) An open-book view of the HA70D3/HA17–E-cadherin interface highlighted in the box in panel B. HA33 is omitted for clarity. Residues on HA70/17 that form hydrogen bonds or salt bridges with E-cadherin are red, whereas the E-cadherin residues that bind to HA70 and HA17 are blue and yellow, respectively. Residues involved in hydrophobic interaction are in gray. (E–F) The mini-HA complex is shown as a transparent surface representation. When an E-cadherin molecule is modeled to form a trans dimer (orange ribbon in panel E) or an X-dimer (green ribbon in panel F) with a HA-bound E-cadherin (red ribbon), it severely clashes with the HA complex.
Large conformational changes are observed in the amino-terminal region of EC1 upon HA binding (Fig. 2C). EC1 dimerizes in trans with EC1 of E-cadherin from apposing cells (16). Specifically, residue Trp2 acts as an anchor docking into a complementary Trp-binding pocket in the partner E-cadherin molecule (14, 15, 17, 18), leading to a three-dimensional domain swap within an E-cadherin homodimer (19). Using analytical ultracentrifugation, we found that EC1–2 forms a dimer in solution with a Kd of ~175.2 μM, which is consistent with previous reports (15, 20), but is significantly weaker than its binding affinity to the HA complex.
Strand-swapping in EC1 is believed to be driven by conformational strain in its first ten amino-terminal residues (termed the A*/A strand), which only arises in its monomeric form (21). Upon HA binding, the A*/A strand of E-cadherin is pushed back to its monomeric conformation through hydrogen bond interactions between Ser8/Pro6E-cad and Glu501/Arg505HA70. This conformation is further stabilized by hydrophobic interactions involving Ile417/Leu473/Met508HA70 and Val3/Pro5/Pro6 E-cad, whereas Pro5/Pro6E-cad serves as a hinge that mediates conformational changes necessary for strand swapping (Fig. 2C–D and fig. S4) (14). Therefore, the HA complex stabilizes E-cadherin in the strained monomeric conformation with Trp2 binding intramolecularly into its own Trp-binding pocket (Fig. 2C and fig. S4A). Meanwhile, it physically blocks the E-cadherin trans dimerization by occupying the E-cadherin dimer interface (Fig. 2E). Moreover, the HA complex also prevents E-cadherin from forming the X-dimer, which is a dimeric intermediate required for the formation of the strand-swapped trans dimer (Fig. 2F) (14, 22).
Complementing the HA70–EC1 interactions, HA17 interacts with both EC1 and EC2, as well as the Ca2+-binding linker (Fig. 2 and fig. S4D), and one HA33 buries a small hydrophobic interface with EC1. The HA–E-cadherin interaction was abolished when Ca2+ was chelated by EDTA (fig. S3C) (12), suggesting that the intertwined interactions among three HAs and two EC domains rely on Ca2+-coordination in the EC1–EC2 linker that fixes the relative positioning of the two EC domains. This binding mode contrasts the listerial protein internalin (InlA), which only binds to EC1 in a one-on-one manner (23). Since E-cadherin’s adhesive function depends on Ca2+-rigidified connections between successive EC domains (14), this is yet another example of a pathogen hijacking host cellular function.
To fine map the HA–E-cadherin binding specificity, we examined binding of the carbohydrate-binding deficient HA33-DAFA complex to wild-type or mutant mouse E-cadherin ectopically expressed in CHO or HEK293 cells, which physiologically do not express E-cadherin (Fig. 3A and fig. S5). The exogenous, wild-type E-cadherin mediated robust binding of the HA33-DAFA complex to transfected cells, which further demonstrates their direct interaction (Fig. 3A). When key HA-binding residues on E-cadherin conserved across different species were mutated (e.g., K14S and Q23A), the cell-surface binding of the HA33-DAFA complex was greatly diminished (Fig. 3A). Residues Lys19 and Asn20 of mouse E-cadherin contribute crucial interactions with HA70, which are likely weakened by their equivalent residues on rat (Gln19 and Arg20) or chicken (Met19 and Arg20) (fig. S4B–C). We found that mutating these two residues (e.g., K19M or N20R) clearly reduced binding of the HA33-DAFA complex onto cell surfaces (Fig. 3A). Consistently, the WT mini-HA complex did not bind EC1–2 that carried these E-cadherin mutants (fig. S6A). These mutagenesis studies further confirm the physiological relevance of the HA–E-cadherin interactions observed in the crystal structure.
Fig. 3. The HA complex recognizes E-cadherin with high specificity to disrupt cell-cell adhesion.
(A) Binding of the HA33-DAFA complex to the surface of CHO cells over-expressing WT or mutant E-cadherin as indicated. (B) Binding of HA-WT or HA-RSDA to HT29 cells (10 nM, 30 min). (C) HA-RSDA failed to disrupt cell-cell adhesions (10 nM, 24 hrs). Scale bar: 20 μm. (D) TER of polarized Caco-2 monolayers was measured after application of HA-WT or HA-RSDA (n≥3 +SD). (E) In contrast to HA-WT, HA-RSDA could not facilitate the flux of FITC-dextran from apical to basolateral compartment (n≥3 +SD). (F) Survival of mice (n=10, combination of two experiments with 5 mice each) after being given by intragastric gavage the reconstituted wild-type L-PTC (rL-PTC WT), rL-PTC RSDA that contains the E-cadherin-binding deficient HAs, or rL-PTC DAFA/TPRA that contains the carbohydrate-binding deficient HAs.
In contrast to carbohydrate-binding deficient HA33-DAFA mutant, the HA-WT complex was still able to bind to cells that expressed E-cadherin mutants tested above (fig. S5B and S5D). This suggests that the ability of HA-WT complex to bind carbohydrates enhances the overall binding avidity of the HA complex to cells, which compensates for the reduced binding to E-cadherin mutants. E-cadherin is known to be glycosylated, but the O-linked carbohydrates observed in the structure of mouse EC1–5 are distant from the carbohydrate-binding sites on HAs – the shortest distances are ~62 Å and ~69 Å to HA70 and HA33, respectively (15). Indeed, a low level binding of HA-WT to untransfected CHO or HEK293 cells was mediated by carbohydrates since such binding was not detected for HA33-DAFA (Fig. 3A and fig. S5).
Sequence alignment reveals that all the E-cadherin-binding residues on HA70 and HA17 are conserved between the HAs of BoNT/A and B that mediate foodborne botulism in human, but few are conserved on HAs of BoNT serotypes C and D (HA/C and HA/D), which predominantly cause botulism in birds and cattle. As predicted by our structure, mutating an E-cadherin-binding residue strictly conserved across HA/A–D, such as D109AHA17/A, abolished its interaction with mouse E-cadherin due to the loss of an intermolecular salt bridge (fig. S6B). Arg505HA70/A, Asn586HA70/A, and Lys104HA17/A of HA/A bind to mouse E-cadherin, but the corresponding residues of HA/C and D (Ser, Tyr, and Ile, respectively) are unlikely to mediate binding. Mutating residue Arg505HA70/A, which is crucial for binding and stabilization of the A*/A strand of E-cadherin in the monomeric form, to a serine (R505SHA70/A) diminished the binding of the HA complex to mouse E-cadherin (fig. S6B). These data might explain why BoNT/C and D have hardly caused human foodborne botulism. However, some E-cadherin residues (e.g., Ser8, Glu54, and Gln101), which favorably bind to HA/A–B-like residues, but not the HA/C–D-like residues, are also conserved in chicken and bovine that are susceptible to BoNT/C and D. Therefore, while the HA–E-cadherin binding mode is likely conserved in HA/A–B, it is unclear whether or how HA/C–D might act through E-cadherin.
Based on the structural analysis we created a specific mutant HA complex deficient in E-cadherin binding (HA-RSDA) by combining HA70R505S, HA17D109A, and the wild-type HA33. Binding of HA-RSDA to HT29 or Caco-2 cells was greatly reduced, and it failed to disrupt cell-cell junctions (Fig. 3B–C and fig. S2). Consistently, the HA-RSDA barely decreased the transepithelial electrical resistance (TER) of the differentiated Caco-2 cell monolayers when applied to the apical or basolateral side (Fig. 3D) and did not facilitate the paracellular transport of FITC-dextran from apical to basolateral compartments as the HA-WT complex did (Fig. 3E). Together, these data demonstrate that the HA complex disrupts cell-cell junctions through binding to E-cadherin.
Based on the structures of the HA complex and the full-length ectodomain of mouse E-cadherin (9, 15), we built a complete structural model of the HA–E-cadherin complex, where three molecules of E-cadherin bind the HA complex (Fig. 4A). The model shows that the C-terminal EC5 of E-cadherin and the carbohydrate-binding sites on HA33 both point towards the proposed plane of the plasma membrane (Fig. 4B). Since the crescent shape of EC1–5 has considerable conformational flexibility to adopt different curvatures (14, 15) and HA33 also displays a large structural flexibility (9), the HA complex is able to bind to its carbohydrate receptor and protein receptor on the same cell surface (Fig. 4B). Therefore, these two different types of receptors likely act synergistically to achieve high binding avidity and specificity on the cell surface.
Fig. 4. The HA complex disrupts the ordered assembly of E-cadherin in the adherens junctions.
(A) Structural model of the HA complex (pale cyan) bound with three E-cadherin ectodomains (red). The lactose molecules bound to HA33 are in sphere models (9). (B) Proposed binding mode of the HA complex on the cell surface in adherens junctions, viewed along the proposed plane of the membrane. The HA complex could simultaneously bind to membrane-anchored E-cadherin and carbohydrates. (C) Proposed model for transepithelial delivery of the L-PTC. A small amount of L-PTC first crosses epithelial cells by transcytosis (1). On basolateral side, the HA complex binds to E-cadherin and carbohydrates and disrupts cell-to-cell adhesion (2). Finally, more L-PTCs are absorbed via the paracellular route (3).
The assembly of the E-cadherin network in adherens junctions (~150–300 Å in width) is achieved by a combination of trans-dimerization between cadherins from opposing cell surfaces and cis-dimerization between molecules from the same cell surface (fig. S7) (14, 15). We propose that the condensed array of E-cadherin clustering between cells is disrupted by the bulky HA complexes (~260 Å wide and ~100 Å height) upon binding. Interestingly, the mini-HA complex bound to HT29 cells weakly and failed to disrupt cell-cell adhesion (fig. S8), and neither apical nor basolateral addition of the mini-HA complex disrupted the tight junctions of polarized Caco-2 cell monolayer (9). These results suggest that multivalent binding to carbohydrates and E-cadherin in the context of the triskelion-shape of the full HA complex is necessary to ensure its function. Furthermore, the HA complex may destabilize adherens junctions by affecting E-cadherin homeostasis because the organization of individual adherens junction is highly dynamic where E-cadherin is actively turned over (24).
Given the observed cellular consequences of HA–E-cadherin interactions, we determined the physiological significance of HA–E-cadherin and HA–carbohydrate interactions to the oral toxicity of BoNT/A in vivo. This question was directly addressed by comparing the oral toxicity of BoNT/A L-PTC (L-PTC/A) containing HA-WT versus mutant HA complex. To do so, we developed a protocol to reconstitute the complete L-PTC/A, composed of a total of 14 proteins (~760 kDa), using recombinant components expressed in E. coli (Fig. S9). This allowed us to compare L-PTC/As that were all constructed in the same way and contained the same active BoNT/A, with either HA-WT or mutant HA complex. The mouse oral toxicity of the reconstituted wild-type L-PTC/A (rL-PTC-WT) is similar to native L-PTC/A purified from C. botulinum (10). The rL-PTC-RSDA containing mutated HAs deficient in E-cadherin binding (HA-RSDA) showed a greatly reduced oral toxicity as compared to the rL-PTC-WT under the same assay conditions (Fig. 3F). Thus the ability of HA complex to bind E-cadherin and disrupt cell-cell junctions is critical for the oral toxicity of BoNT/A in vivo. We also observed a significant reduction in oral toxicity for the rL-PTC-DAFA/TPRA comprising a HA complex deficient in carbohydrate binding (a combination of HA33-DAFA and HA70-TPRA) (Fig. 3F) (9), demonstrating that binding of HA complex to carbohydrates is also important to facilitate BoNT/A absorption in vivo, likely by enriching L-PTC/A onto cell surfaces. Together, these results establish the physiological relevance of HA–E-cadherin and HA–carbohydrate interactions to the extreme oral toxicity of BoNT/A. Our study also provides a framework to investigate the oral intoxication of some HA-less BoNTs (e.g., BoNT/A2, E and F), which instead contain several orfX proteins, whose expression and function are still unknown (2, 25).
Based on work described previously and presented here, we suggest the following model (Fig. 4C). The abundant carbohydrates accumulate L-PTCs on the surface of the small intestine to initiate absorption (9). Then some L-PTC may cross the epithelial cells by transcytosis without interfering with the epithelial barrier, which could be mediated either by BoNT or HAs although the underlying mechanism is not fully understood (26-28) (Fig. 4C, step 1). Once the HA complexes gain access to the basolateral surface, they specifically bind to E-cadherin and carbohydrates and disrupt E-cadherin-mediated cell-to-cell adhesion (Fig. 4C, step 2), thereby opening up the paracellular route for BoNT absorption (Fig. 4C, step 3).
BoNTs are the most potent bacterial toxins known. Our findings reveal the targeting of a host protein by a component of the toxin complex to achieve efficient absorption through possibly the most challenging route of entry into the circulation of humans or other animals. Understanding this remarkable molecular machinery not only provides new targets for therapeutic intervention against oral BoNT intoxication, but also could potentially guide the development of new adherens junction modulators to enhance the permeability of the intestinal epithelium to facilitate drug delivery.
Supplementary Material
Acknowledgments
We thank Andrey Bobkov for technical assistance. This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) grant R01AI091823 to R.J.; by the NIH Grant 8P51OD011103-51 (to the New England Primate Research Center), 1R56AI097834-01, and 1R01NS080833-01 to M.D.; and by the Swiss Federal Office for Civil Protection BABS #353003325 to A.R.. NE-CAT at the Advanced Photon Source (APS) is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403). Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. The atomic coordinates and structure factors of the mini-HA–EC1–2 complex have been deposited in the Protein Data Bank under the accession code 4QD2.
References and Notes
- 1.Collins MD, East AK. J Appl Microbiol. 1998;84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- 2.Hill KK, Smith TJ. Curr Top Microbiol Immunol. 2013;364:1–20. doi: 10.1007/978-3-642-33570-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Gu S, et al. Science. 2012;335:977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohishi I, Sugii S, Sakaguchi G. Infect Immun. 1977;16:107–109. doi: 10.1128/iai.16.1.107-109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohishi I. Infect Immun. 1984;43:487–490. doi: 10.1128/iai.43.2.487-490.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakaguchi G. Pharmacol Ther. 1982;19:165–194. doi: 10.1016/0163-7258(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 7.Matsumura T, et al. Cell Microbiol. 2008;10:355–364. doi: 10.1111/j.1462-5822.2007.01048.x. [DOI] [PubMed] [Google Scholar]
- 8.Ito H, et al. FEMS Immunol Med Microbiol. 2011:1–9. doi: 10.1111/j.1574-695X.2011.00825.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee K, et al. PLoS Pathog. 2013;9:e1003690. doi: 10.1371/journal.ppat.1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng LW, et al. Toxicology. 2008;249:123–129. doi: 10.1016/j.tox.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Amatsu S, Sugawara Y, Matsumura T, Kitadokoro K, Fujinaga Y. J Biol Chem. 2013;288:35617–35625. doi: 10.1074/jbc.M113.521179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara Y, et al. J Cell Biol. 2010;189:691–700. doi: 10.1083/jcb.200910119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng W, Takeichi M. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasch J, Harrison OJ, Honig B, Shapiro L. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison OJ, et al. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro L, Weis WI. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boggon TJ, et al. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Eisenberg D. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch AW, Pokutta S, Lustig A, Engel J. Biochemistry. 1997;36:7697–7705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- 21.Vendome J, et al. Nat Struct Mol Biol. 2011;18:693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison OJ, et al. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert WD, et al. Cell. 2002;111:825–836. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 24.Baum B, Georgiou M. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin G, Tepp WH, Pier CL, Jacobson MJ, Johnson EA. Appl Environ Microbiol. 2010;76:40–47. doi: 10.1128/AEM.01882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujinaga Y, Sugawara Y, Matsumura T. Curr Top Microbiol Immunol. 2013;364:45–59. doi: 10.1007/978-3-642-33570-9_3. [DOI] [PubMed] [Google Scholar]
- 27.Maksymowych AB, Simpson LL. J Pharmacol Exp Ther. 2004;310:633–641. doi: 10.1124/jpet.104.066845. [DOI] [PubMed] [Google Scholar]
- 28.Couesnon A, Pereira Y, Popoff MR. Cell Microbiol. 2008;10:375–387. doi: 10.1111/j.1462-5822.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- 29.Rummel A, Mahrhold S, Bigalke H, Binz T. FEBS J. 2011;278:4506–4515. doi: 10.1111/j.1742-4658.2011.08196.x. [DOI] [PubMed] [Google Scholar]
- 30.Holtje M, et al. Toxicon. 2013;75:108–121. doi: 10.1016/j.toxicon.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. Acta Crystallogr D Biol Crystallogr. 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potterton E, Briggs P, Turkenburg M, Dodson E. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger AT. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen VB, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciarlet M, Crawford SE, Estes MK. J Virol. 2001;75:11834–11850. doi: 10.1128/JVI.75.23.11834-11850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, et al. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Courcelle E, Stuart DI, Metoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 41.Sagane Y, et al. Biochem Biophys Res Commun. 2002;292:434–440. doi: 10.1006/bbrc.2002.6689. [DOI] [PubMed] [Google Scholar]
- 42.Inoue K, et al. Infect Immun. 1996;64:1589–1594. doi: 10.1128/iai.64.5.1589-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.