Abstract
Objective
To perform a comprehensive molecular pathways analysis of genes identified through genome-wide expression profiling and the published literature for chronic sinusitis with polyps.
Study Design
Molecular pathways analysis.
Setting
Academic medical center.
Methods
A molecular pathways analysis of gene biomarkers discovered through hypothesis-driven and high-throughput molecular studies was performed. Genes identified with a PubMed literature search were analyzed with Ingenuity Pathways Analysis software to identify central molecules implicated in the pathogenesis of chronic sinusitis with polyps. The central pathways were then compared with those identified through genome-wide expression profiling of ethmoid polyps.
Results
A total of 97 molecules were investigated with Ingenuity Pathways Analysis based on 55 studies that evaluated differences in gene expression (39), genetic variation (12), or proteomics (4). The analysis revealed 9 statistically significant molecular networks containing central nodes that included transcription factors, protein kinases, cytokines, and growth factors/receptors. The highest scoring networks implicated nuclear factor kappa-B, tumor necrosis factor, and mitogen-activated protein kinases. The majority of pathways in the literature review analysis overlapped with those identified through a single genome-wide expression study.
Conclusions
Chronic sinusitis with polyps is a complex disease with suspected contribution of multiple genetic and environmental factors. The search for causative genes has led to the discovery of numerous candidates. Pathways analysis applied to these candidate genes identified common central molecules that are likely to be key mediators of the disease process. Novel therapies targeting these molecules may be applicable for the treatment of chronic sinusitis with polyps.
Keywords: nasal polyps, nasal polyposis, polyps, sinusitis, pathways, genetics, gene, bioinformatics, chronic sinusitis
Chronic sinusitis with polyps affects more than 6 million people in the United States.1,2 A significant research effort has sought to understand the etiology of polyp formation, with published studies having identified a large number of molecules that are differentially expressed in polyp tissue compared with control specimens (see Appendix A at otojournal.org ). Recently, investigators have found genetic variations that appear to be associated with polyp formation (see Appendix A online). None of these individual genes or gene products has been proven to be causative. Moreover, any one of these genes is unlikely, by itself, to fully account for the spectrum of clinical manifestations associated with chronic sinusitis—a multifactorial, complex disease. In addition, the immensity of published research and complexity of the involved molecular interactions present a challenge to understand how an individual factor interrelates within a complex process. The end result has been a lack of translation of basic science findings into new therapeutic interventions for patients suffering from chronic sinusitis with polyps.
Pathways analysis attempts to identify meaningful relationships among complex data sets or isolated research findings. This analysis uses large bioinformatic databases, which abstract and organize known relationships between genes, proteins, biological molecules, and disease processes. By leveraging a comprehensive database, critical pathways or critical steps of a single pathway can be identified, which have a high likelihood of biologic or clinical relevance. The primary purpose of this study was to apply pathways analysis to genes and gene products (RNA and protein) that have previously been associated with chronic sinusitis with polyps in the published literature. The secondary aim was to determine whether similar pathways are identified when comparing these reported markers of sinusitis to those found in a previous study of genome-wide expression data from ethmoid polyps, which contained the largest number of transcripts to date.3
Methods
Literature Review
A PubMed literature search (from January 1950 to July 2010) was performed to identify candidate molecular markers associated with chronic sinusitis with polyps. Search terms included sinusitis or nasal polyp, combined with gene, genetic, genome-wide, microarray, proteomic, DNA, or RNA. Abstracts were reviewed to ensure that all studies compared human subjects with sinus polyps with normal controls. Studies of patients with sinusitis in which no specific mention of polyps was made were excluded. In addition, studies of cystic fibrosis and those focusing on isolated antrochoanal polyps were excluded. Abstracted studies were classified according to whether they were hypothesis driven or hypothesis generating (high throughput) and whether the marker of interest was identified in the genome (DNA), transcriptome (RNA), or proteome (protein). Gene products (RNA and proteins) that were significantly up- or downregulated in human polyp tissue compared with control tissues were included. Individual genes whose variations (single-nucleotide polymorphisms) were significantly associated with polyps were also included. Several high-throughput studies identified a large number of differentially expressed genes, all of which were not provided in the manuscript. In these instances, only genes specifically mentioned and believed by the authors to be most important were included.
Genome-Wide Expression Data
Raw data from a genome-wide expression profiling study, previously reported by our group, that compared ethmoid polyp tissue from patients with chronic sinusitis (n = 10) to healthy mucosa from nonsinusitis control subjects (n = 10) were used for this aspect of study.3 Gene expression of over 47,000 transcripts was evaluated using an Affymetrix HG-U133 plus 2.0 GeneChip (Affymetrix, Santa Clara, California). Differentially expressed genes were recorded by name, fold change, and false discovery rate (FDR) for input into pathways analysis software.
Pathway Analysis
Two separate pathway analyses were performed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, California). The first analysis used input genes identified through the literature review. Given the heterogeneity of statistical data found in the literature, each identified gene was assigned equivalent weight for the purposes of this study. The second analysis used raw data from the genome-wide expression study.3 Stringent statistical filters were used—only genes with a fold change >3 and an FDR <0.1 were included. The importance of each gene was weighed by its fold change and FDR, per the IPA algorithm. In both analyses, IPA generated networks involving 35 molecules or less, using known direct and indirect molecular interactions. Resulting molecular networks were classified by biologic function. The top 10 individual networks were reviewed to identify the specific gene or genes that were most interconnected within the network (central node).
Statistical analysis of biologic functions and networks was done as part of the overall IPA using the right-tailed Fisher exact test. The significance value obtained is a measure of the likelihood that the association between the input genes of the experiment and a given function, pathway, or network is owing to random chance. The smaller the P value, the less likely it is that the association is random and thus the greater significance. Network scores are given a numerical value that is equal to the negative exponent of the P value for that network. For example, a network with a P value of .01 (10−2) would have a score of 2. For all analyses, P values less than .05 were considered statistically significant.
Results
Literature Review
A total of 667 studies were identified by the search terms and reviewed for inclusion and exclusion measures. Of these studies, 55 met inclusion criteria and were included in the final analysis. Most studies assessed gene expression, with 25 being focused, hypothesis-driven protocols and 13 being hypothesis-generating high-throughput microarray studies (see Appendix A). An association between polyps and a specific, hypothesis-driven genetic variation (DNA level) was shown in 13 studies (see Appendix A). Four studies employed proteomic methodologies (see Appendix A), and no comprehensive genome-wide association studies were identified linking polyps to a specific polymorphism. After accounting for overlap between studies, a total of 97 unique genes or gene products were available for pathways analysis (see Appendix A).
Genome-Wide Expression Data
The original findings of the genome-wide expression study included 2724 genes whose expression was upregulated and 808 whose expression was downregulated compared with controls.3 Using a cutoff that included only genes with a fold change >3 and an FDR <0.1, a total of 554 genes were available for pathways analysis, including 365 that were upregulated and 189 that were downregulated.
Pathways Analysis
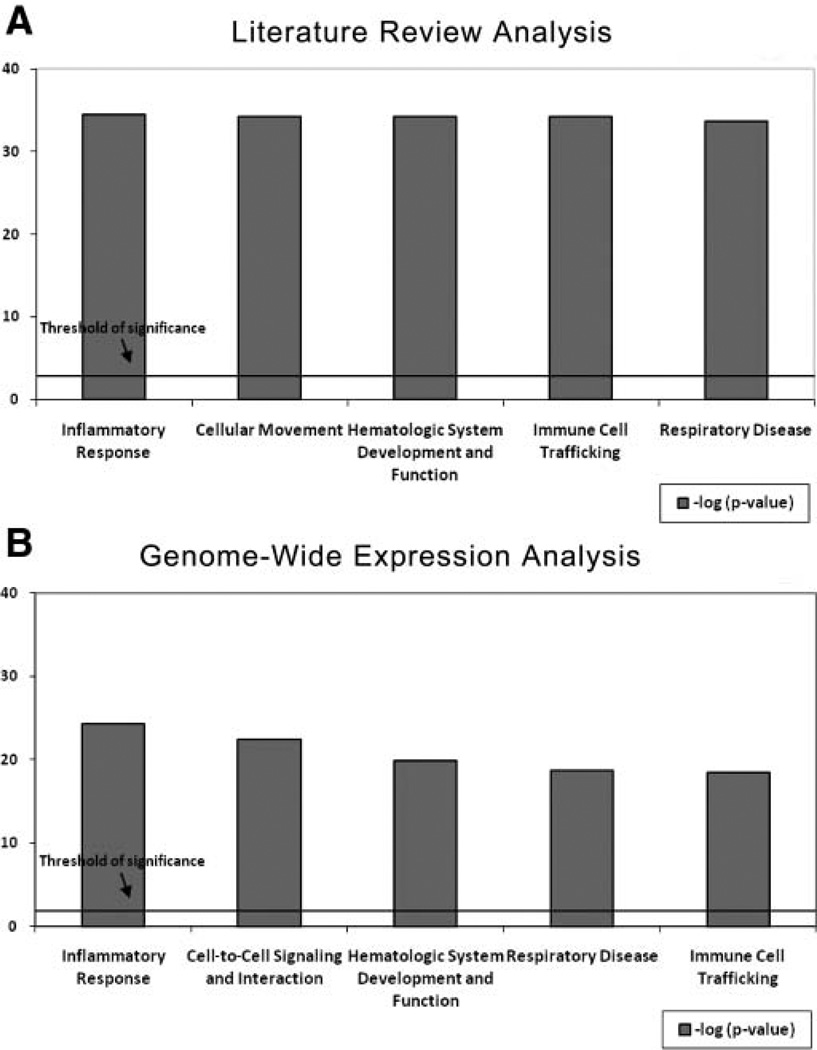
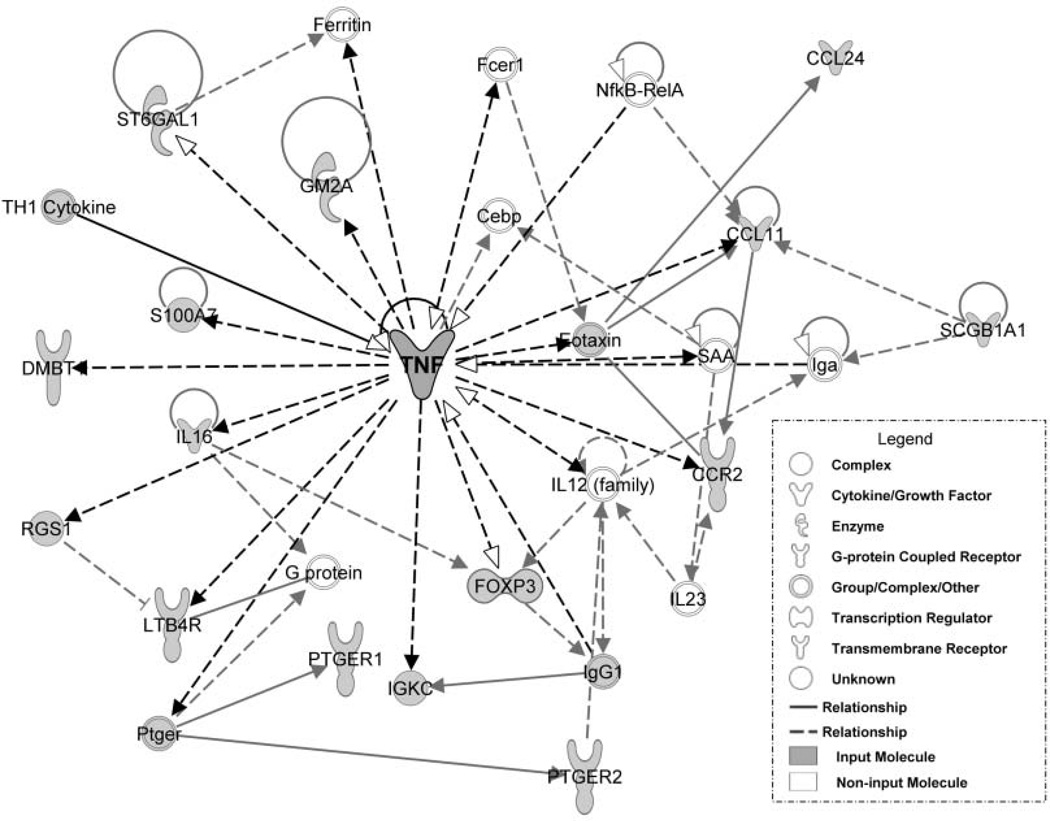
The IPA of genes and gene products identified by literature review revealed pathways with highly significant biologic functions that included inflammatory response, cellular movement, hematological system development and function, immune cell trafficking, and respiratory disease (Figure 1A). A total of 9 molecular networks were identified, with significance scores ranging from 3 to 41. Network 1, which had the highest score of 41, is shown in Figure 2 as an example of a typical network. The most interconnected genes (ie, nodal genes) were considered critical to each of these molecular networks.4 The nodal genes included transcription factors, protein kinases, cytokines, enzymes, and growth factors/receptors (Table 1).
Figure 1.
The highest ranked biologic functions of molecular markers identified through (A) literature review pathways analysis and (B) genome-wide expression pathways analysis. Threshold of significance: P < .05.
Figure 2.
Network 1 of pathway analysis generated from literature review. Tumor necrosis factor (TNF) was the central, nodal molecule of the highest scoring network (P = 1 × 10^–41). Shaded molecules were specifically identified by literature review and input into the pathways analysis. See Appendix B at otojournal.org for complete list of gene abbreviations.
Table 1.
Central Nodes Identified by Pathways Analysis
| Literature Review Data |
Genome-Wide Expression Data |
||
|---|---|---|---|
| Genes | Score | Genes | Score |
| AKT | 3 | AKT | 37 |
| AP-1 | 9 | AP-1 | 28 |
| EGFR | 9 | EGFR | 26 |
| ERK 1/2 | 16 | ERK 1/2 | 35 |
| IFN-alpha | 9 | IFN-alpha | 32 |
| JNK | 3 | JNK | 28 |
| NFκB | 17 | NFκB | 41 |
| P38MAPK | 3 | P38MAPK | 25 |
| PKC | 9 | PKC | 26 |
| VEGF | 9 | VEGF | 28 |
| ____ | ____ | ||
| CREB | 9 | Histone h3 | 28 |
| IFNG | 14 | IgG | 25 |
| IL6 | 31 | IL10 | 32 |
| Peptidase | 4 | IL12 | 32 |
| PI3K | 3 | NOS2 | 26 |
| TGFB1 | 31 | SOCS3* | 32 |
| TNF | 41 | ||
Nodes are listed in alphabetical order with genes identified in both analyses listed first. The network score is equal to the negative exponent of the P value for that network (higher network score = lower P value). See Appendix B at otojournal.org for complete list of gene abbreviations.
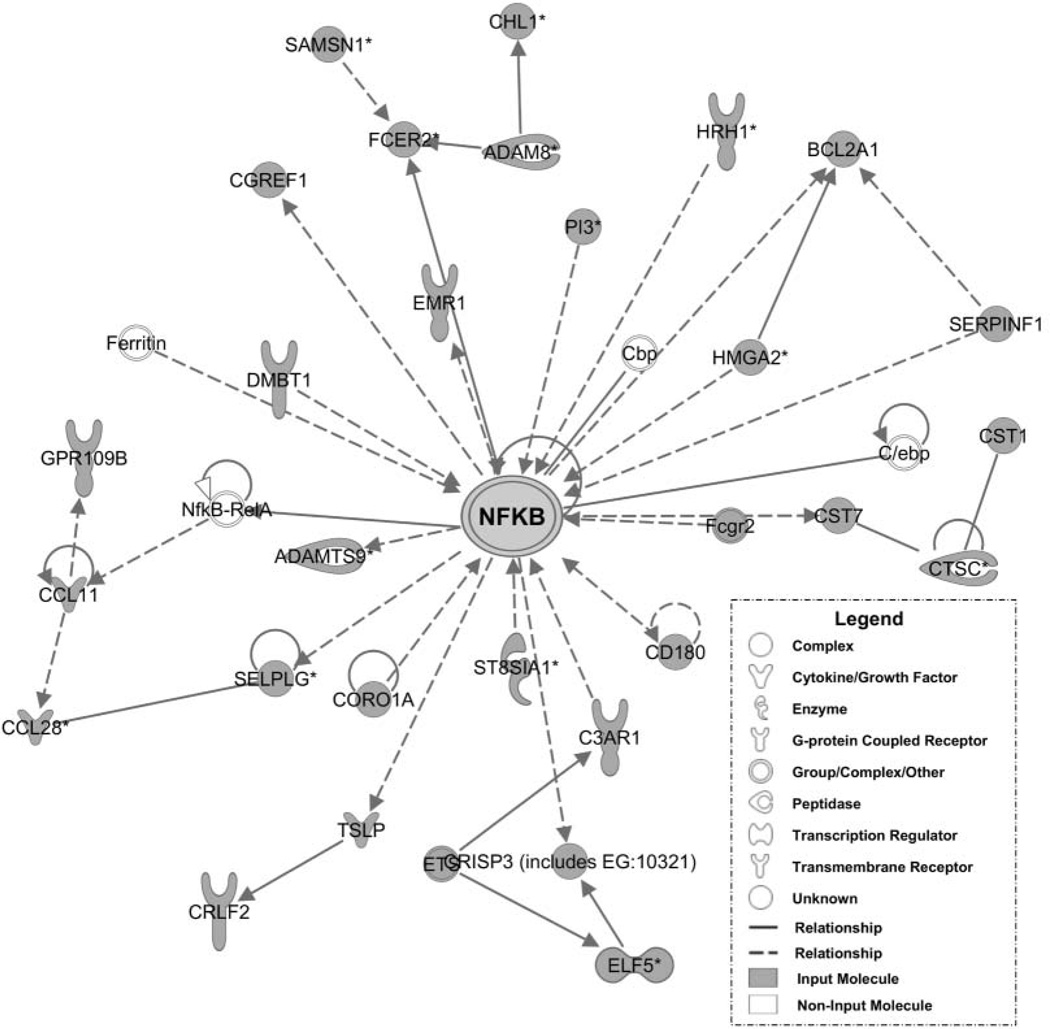
The second pathways analysis of genes identified in the genome-wide expression data set3 showed similar top biologic functions, including inflammatory response, cell-cell signaling and interaction, hematological system development and function, respiratory disease, and immune cell trafficking (Figure 1B). The top 10 identified pathways had scores ranging from 25 to 41. Network 1, with a score of 41, is shown in Figure 3. Genes believed to be central to each of these molecular networks included transcription factors, protein kinases, cytokines, enzymes, and growth factors or receptors, as listed in Table 1.
Figure 3.
Network 1 of pathway analysis generated from genome-wide expression pathway analysis. Nuclear factor kappa B (NFκB) was the central, nodal molecule of the highest scoring network (P = 1 × 10^–41). Shaded molecules were differentially expressed in the genome-wide data set and input into the pathways analysis. See Appendix B at otojournal.org for complete list of gene abbreviations.
The majority of central pathway genes identified in the literature review analysis were also seen in the genome-wide expression analysis. This overlap included the cytokine interferon alpha (IFN-alpha), the transcription regulators nuclear factor kappa B (NFκB), and activator protein-1 (AP-1), as well as the protein kinases p38MAPK, ERK 1/2, JNK, AKT, and PKC. Several growth factors/receptors were also identified in both analyses, including vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR).
Discussion
With a complex, chronic disease such as sinusitis, the synthesis of isolated research findings into a comprehensive and meaningful model can be challenging. This study is the first to perform an integrated bioinformatic pathway analysis of previously identified genes associated with chronic sinusitis with polyps. Functional and network pathways were identified, which primarily involved inflammatory response and immune cell movement. Individual networks were found to contain a number of central, nodal molecules that may be of critical importance in the pathophysiology of sinus polyps. A secondary analysis of data from a genome-wide expression study closely mirrored results of the initial analysis, further strengthening the findings.
The power of the pathways analysis largely rests on the completeness and validity of the IPA platform. Traditionally, researchers in a particular field interpreted individual findings based on their own accumulated knowledge, usually supplemented by a targeted literature review. However, the exponential rise in research data generated worldwide limits this approach, particularly with respect to complex molecular interactions. IPA is a proprietary program based on a knowledge bank of millions of individually modeled molecular interactions. The database is continually updated with studies from more than 3600 peer-reviewed scientific journals.4 Significant findings are manually reviewed by PhD-level researchers, resolved for synonyms, and curated based on involved molecular interactions and disease context. To date, the IPA database has been cited in close to 3000 scientific publications.4
Application of the IPA platform revealed a remarkable overlap of findings in the literature review analysis and the genome-wide expression analysis. This result is important because the 2 analyses approached the question of interest from 2 very different perspectives. The literature review, in essence, is a meta-analysis of published findings, which incorporates a wide range of investigative methodologies, study populations, and research groups. Only genes previously shown to be associated with nasal polyps were analyzed. Conversely, the genome-wide expression analysis can be considered an impartial, hypothesis-generating analysis because data from over 47,000 known gene transcripts were uploaded into the pathways analysis, with no preconceived notion as to which may or may not be relevant. For 2 independent analyses to identify similar pathways lends support to the validity of the pathway findings.
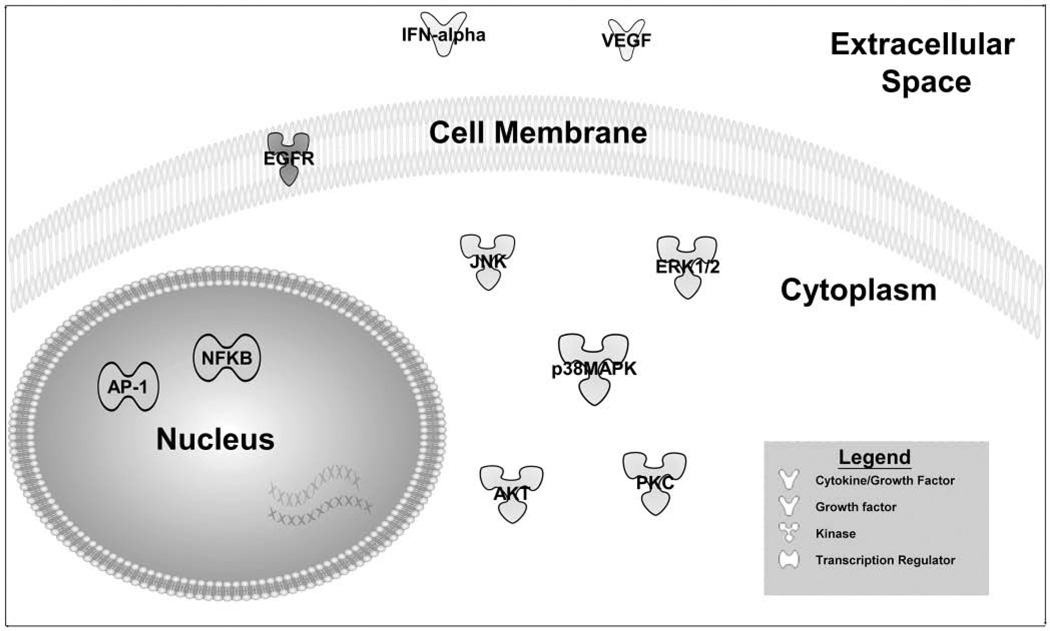
The networks identified in this study contain central, nodal genes that are likely to be of critical importance to polyp development and/or maintenance. These nodal molecules serve as hubs through which a great number of other molecules are affected either directly or indirectly. The identified nodal genes (Figure 4) have a high degree of biological potency based on their ability to amplify downstream responses, particularly with respect to inflammation and cell growth, processes central to polyp development. These genes should be considered potential targets for therapeutic interventions aiming to arrest polyp development and growth.
Figure 4.
Location and function of nodal genes found in both literature review and genome-wide expression pathways analyses. See Appendix B at otojournal.org for complete list of gene abbreviations.
Alterations in protein kinases have been identified in a number of disorders, including inflammatory conditions such as asthma and chronic obstructive pulmonary disease.5,6 Kinases are known to play an important role in posttranscriptional control of inflammatory gene expression, T cell differentiation, and cell apoptosis.7 Of the protein kinases highlighted in this analysis, individual studies have linked P38 MAP kinase8,9 and ERK1/2 kinase10 to sinusitis, whereas AKT, PKC, and JNK have not been studied specifically. Although there is considerable interest in using targeted kinase inhibitors to treat asthma and chronic obstructive pulmonary disease (COPD),11,12 a similar strategy has not been widely proposed for sinusitis with polyps.
The transcription factors NFκB and AP-1 have been shown to be activated in sinusitis and many other mucosal inflammatory conditions.13–18 By binding on or near specific DNA sequences, transcription factors regulate the expression of individual genes or sets of genes. NFκB is known to regulate genes responsible for cell proliferation and survival and has been linked to inflammation in arthritis,15 inflammatory bowel disease (IBD),16 and asthma.17 The AP-1 pathway, consisting of c-Fos and c-Jun, also regulates the transcription of genes related to cellular differentiation and proliferation, particularly in response to cytokines, growth factors, stress, and infectious pathogens.19 Downregulation of the AP-1 pathway has been proposed as a mechanism by which glucocorticoids decrease nasal polyp formation.20 Pharmaceutical agents specifically targeting the NFκB21 and AP-119 pathways are being studied for a number of mucosal inflammatory conditions, including asthma, COPD, and IBD.17
The finding that growth factors and cytokines might be important to polyp formation is not unexpected. Several prior studies have also highlighted VEGF as a significant factor in the development of polyps.22 EGFR is a cell surface receptor that transmits transmembrane signals through its intrinsic tyrosine kinase function. This auto-phosphorylation elicits downstream activation and signaling by several other transduction cascades, including MAPK, AKT, and JNK. The cytokines highlighted by this analysis include IFN-alpha and tumor necrosis factor (TNF), both of which have been extensively studied in chronic sinusitis.23 These cytokines have well-established roles in the innate and adaptive immune responses and have been successfully targeted to inhibit inflammatory conditions such as psoriasis, rheumatoid arthritis, and IBD.24–26
It should not be surprising that the same molecular pathways critical to other chronic inflammatory conditions such as asthma and IBD would also be critical to sinusitis with polyps. Although clinically and anatomically diverse, these inflammatory conditions share many of the same redundant molecular pathways. Therapeutics such as glucocorticoids have proven efficacious in each condition, despite the fact that the individual diseases have a distinct etiology and pathophysiology. A number of bioactive compounds that target these transcription factors, growth factors, and protein kinases are currently in multiple stages of development for the treatment of other inflammatory diseases and neoplastic processes. Although side effects may limit the systemic application of some drugs, the potential topical application of medications directly onto the sinonasal mucosa may allow for the targeted treatment of polyps without unwanted systemic effects.
The current analysis represents a snapshot-in-time rather than a final conclusion. As future research is published—both related to nasal polyps and molecular science—the knowledge base underlying this analysis will become more robust and the involved pathways more fully developed. The nodal genes identified in this article thus serve as likely starting points to generate mechanistic hypotheses and focus future research. Further basic science investigation is necessary to determine whether these modeled pathways are in fact contributing to polyp development and, more important, whether targeted manipulation of nodal molecules has measured therapeutic effects. As with any synthesis of previously published findings, this analysis is limited in part by the diversity of the included studies. Each study in the literature review analysis was essentially weighted equally, irrespective of the soundness of the methodology or strength of the statistical findings (as long as the finding had a P value <.05). In addition, although each individual study included patients with polyps, information was not available to control for other comorbidities such as asthma, aspirin intolerance, or allergic fungal sinusitis, which may impart unique findings.
Conclusion
Many genes and gene products have been implicated in chronic sinusitis with polyps. This analysis provides a conceptual framework of central pathways connecting these molecules. Identified nodal genes included transcription factors, protein kinases, cytokines, and growth factors/receptors. Novel therapeutic interventions targeting these nodal genes and their products may produce tangible clinical effects, especially if applied topically to avoid systemic side effects.
Supplementary Material
Acknowledgments
Funding sources: This project was funded by Massachusetts Eye and Ear Infirmary Research Foundation. KMS is supported by grants from NIH-NIDCD K08DC010419-01, Massachusetts Life Sciences Center, the Boston Foundation and the Shore Fellowship at Harvard Medical School. MPP received support from NIH grant UL1RR025771 and the BU-Bridge CTSI.
Footnotes
This article was presented at the 2010 AAO-HNSF Annual Meeting & OTO EXPO; September 26–29, 2010; Boston, Massachusetts.
The study protocol received approval from the Human Studies Committee of the Massachusetts Eye and Ear Infirmary.
Author Contributions
Michael P. Platt, conception and design, acquisition of data, analysis and interpretation of data, drafting the article, revising critically for important intellectual content, final approval; Zachary Soler, design, acquisition of data, analysis and interpretation of data, drafting the article, revising the manuscript critically for important intellectual content, final approval; Ralph Metson, conception, acquisition of data, revising the manuscript critically for important intellectual content, final approval; Konstantina M. Stankovic, conception and design, acquisition of data, analysis and interpretation of data, revising the article critically for important intellectual content, final approval.
Disclosures
Competing interests: None.
Sponsorships: None.
Supplemental Material
Additional supported information may be found at http://oto.sagepub.com/content/by/supplemental-data
References
- 1.Centers for Disease Control and Prevention (CDC) CDC Summary Health Statistics for US Adults: National Health Information Survey. Atlanta, GA: CDC; 2006. [Google Scholar]
- 2.Settipane GA. Epidemiology of nasal polyps. Allergy Asthma Proc. 1996;17:231–236. doi: 10.2500/108854196778662246. [DOI] [PubMed] [Google Scholar]
- 3.Stankovic KM, Goldsztein H, Reh DD, Platt MP, Metson R. Gene expression profiling of nasal polyps associated with chronic sinusitis and aspirin-sensitive asthma. Laryngoscope. 2008;118:881–889. doi: 10.1097/MLG.0b013e31816b4b6f. [DOI] [PubMed] [Google Scholar]
- 4.Ingenuity Systems. [Accessed September 19, 2010]; www.Ingenuity.com.
- 5.Pouliot P, Olivier M. Opposing forces in asthma: regulation of signaling pathways by kinases and phosphatases. Crit Rev Immunol. 2009;29:419–442. doi: 10.1615/critrevimmunol.v29.i5.40. [DOI] [PubMed] [Google Scholar]
- 6.Chung KF, Marwick JA. Molecular mechanisms of oxidative stress in airways and lungs with reference to asthma and chronic obstructive pulmonary disease. Ann N Y Acad Sci. 2010;1203:85–91. doi: 10.1111/j.1749-6632.2010.05600.x. [DOI] [PubMed] [Google Scholar]
- 7.Teixeiro E, Daniels MA. ERK and cell death: ERK location and T cell selection. FEBS J. 2010;277:30–38. doi: 10.1111/j.1742-4658.2009.07368.x. [DOI] [PubMed] [Google Scholar]
- 8.Lin SK, Kok SH, Shun CT, et al. Tumor necrosis factor-alpha stimulates the expression of C-C chemokine ligand 2 gene in fibroblasts from the human nasal polyp through the pathways of mitogen-activated protein kinase. Am J Rhinol. 2007;21:251–255. doi: 10.2500/ajr.2007.21.2958. [DOI] [PubMed] [Google Scholar]
- 9.Raia V, Maiuri L, Ciacci C, et al. Inhibition of p38 mitogen activated protein kinase controls airway inflammation in cystic fibrosis. Thorax. 2005;60:773–780. doi: 10.1136/thx.2005.042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Zhang Q, Li Y, et al. Involvement of mitogen-activated protein kinases and nuclear factor kappa B pathways in signaling COX-2 expression in chronic rhinosinusitis. Inflamm Res. 2009;58:649–658. doi: 10.1007/s00011-009-0030-x. [DOI] [PubMed] [Google Scholar]
- 11.Adcock IM, Chung KF, Caramori G, Ito K. Kinase inhibitors and airway inflammation. Eur J Pharmacol. 2006;533:118–132. doi: 10.1016/j.ejphar.2005.12.054. [DOI] [PubMed] [Google Scholar]
- 12.Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:693–696. doi: 10.1513/pats.200907-071DP. [DOI] [PubMed] [Google Scholar]
- 13.Valera FC, Queiroz R, Scrideli C. Expression of transcription factors NF-kappaB and AP-1 in nasal polyposis. Clin Exp Allergy. 2008;8:579–585. doi: 10.1111/j.1365-2222.2007.02929.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, Xu G, Shi J, Wen W. A correlative study of NF-kappaB activity and cytokines expression in human chronic nasal sinusitis. J Laryngol Otol. 2007;121:644–649. doi: 10.1017/S0022215106001824. [DOI] [PubMed] [Google Scholar]
- 15.Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the ‘epimmunome’. Nat Immunol. 2010;11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce BF, Yao Z, Xing L. Functions of nuclear factor kappaB in bone. Ann N Y Acad Sci. 2010;1192:367–375. doi: 10.1111/j.1749-6632.2009.05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wullaert A. Role of NF-kappaB activation in intestinal immune homeostasis. Int J Med Microbiol. 2010;300:49–56. doi: 10.1016/j.ijmm.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Janssen-Heininger YM, Poynter ME, Aesif SW, et al. Nuclear factor kappaB, airway epithelium, and asthma: avenues for redox control. Proc Am Thorac Soc. 2009;6:249–255. doi: 10.1513/pats.200806-054RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 20.Li CW, Cheung W, Lin ZB, Lim JT, Wang DY. Oral steroids enhance repair in nasal polyposis via upregulation of the AP-1 gene network. Thorax. 2009;64:306–312. doi: 10.1136/thx.2008.106096. [DOI] [PubMed] [Google Scholar]
- 21.Edwards MR, Bartlett NW, Clarke D, et al. Targeting the NF-kappaB pathway in asthma and chronic obstructive pulmonary disease. Pharmacol Ther. 2009;121:1–13. doi: 10.1016/j.pharmthera.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HS, Myers A, Kim J. Vascular endothelial growth factor drives autocrine epithelial cell proliferation and survival in chronic rhinosinusitis with nasal polyposis. Am J Resp Crit Care Med. 2009;180:1056–1067. doi: 10.1164/rccm.200905-0740OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto BA, Wenzel SE. The role of cytokines in chronic rhinosinusitis with nasal polyps. Curr Opin Otolaryngol Head Neck Surg. 2008;16:270–274. doi: 10.1097/MOO.0b013e3282fb2885. [DOI] [PubMed] [Google Scholar]
- 24.Furst DE. Development of TNF inhibitor therapies for the treatment of rheumatoid arthritis. Clin Exp Rheumatol. 2010;28(3) suppl 59:S5–S12. [PubMed] [Google Scholar]
- 25.Miteva M. Psoriasis: targeting therapy towards the inflammatory cascade. Am J Clin Dermatol. 2010;11(suppl 1):11–13. doi: 10.2165/1153414-S0-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 26.Monteleone G, Caprioli F. T-cell-directed therapies in inflammatory bowel diseases. Clin Sci. 2010;118:707–715. doi: 10.1042/CS20100027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.