Abstract
A ∼2.4-kb imprinting control region (ICR) regulates somatic monoallelic expression of the Igf2 and H19 genes. This is achieved through DNA methylation-dependent chromatin insulator and promoter silencing activities on the maternal and paternal chromosomes, respectively. In somatic cells, the hypomethylated maternally inherited ICR binds the insulator protein CTCF at four sites and blocks activity of the proximal Igf2 promoter by insulating it from its distal enhancers. CTCF binding is thought to play a direct role in inhibiting methylation of the ICR in female germ cells and in somatic cells and, therefore, in establishing and maintaining imprinting of the Igf2/H19 region. Here, we report on the effects of eliminating ICR CTCF binding by severely mutating all four sites in mice. We found that in the female and male germ lines, the mutant ICR remained hypomethylated and hypermethylated, respectively, showing that the CTCF binding sites are dispensable for imprinting establishment. Postfertilization, the maternal mutant ICR acquired methylation, which could be explained by loss of methylation inhibition, which is normally provided by CTCF binding. Adjacent regions in cis—the H19 promoter and gene—also acquired methylation, accompanied by downregulation of H19. This could be the result of a silencing effect of the methylated maternal ICR.
Imprinted genes exhibit parent-of-origin-specific monoallelic expression (1, 9, 29). In the mouse, two imprinted genes—insulin-like growth factor 2 (Igf2), encoding an embryonic mitogen, and H19, producing a noncoding RNA of unknown function—are located ∼80 kb apart and are expressed only from the paternal and maternal alleles, respectively. Coordinate expression of these genes in tissues of mesoderm and definitive endoderm origin is due to the sharing of a set of enhancers located downstream of H19 (2, 19, 51). Monoallelic expression of the two genes is regulated by an imprinting control element (ICR) located −2 kb to −4.4 kb relative to the transcription start site of H19 (18, 30, 43).
On the maternal chromosome, the ICR mediates silencing of Igf2 in cis through chromatin insulator function. Gene targeting experiments, involving complete or partial deletion of the maternal ICR, result in expression of the maternal Igf2 allele (15, 18, 33, 43, 44). The maternal ICR contains several binding sites for the vertebrate insulator protein CTCF. Using chromatin immunoprecipitation and in vivo footprinting, it has been shown that the ICR binds CTCF in vivo (16, 38). The ICR functions as an insulator in enhancer blocking assays in cell culture and in transgenic mice (3, 10, 16, 33). The presence of CTCF binding sites is sufficient to confer insulator activity at this locus: two copies of the chicken β-globin insulator (ChβGI)2, which contains CTCF binding sites but otherwise lacks homology to the ICR, completely substituted for the chromatin insulator property of the ICR in vivo (41).
On the paternal chromosome, the ICR is methylated (45, 46). CpG methylation inhibits CTCF binding (3, 10, 12, 16, 28, 38); therefore, the methylated paternal ICR lacks insulator activity and the paternal Igf2 promoter interacts with the downstream enhancers. The hypermethylated paternal ICR, while lacking insulator activity, directs postzygotic silencing of the H19 promoter in cis (33). The promoter becomes hypermethylated and packaged into a closed chromatin structure (2, 8, 38).
Differential methylation of the Igf2-H19 ICR, the establishment of which could be viewed as the imprinting mechanism itself, is laid down in the male and female germ lines (6, 45-47). It was shown that establishment of differential methylation is a function autonomous to the ICR (15), but the germ line-specific processes that establish this differential ICR methylation are undefined. Based on the paradigm that transcription factor binding can inhibit DNA methylation, possibly by physically blocking access of DNA methyltransferases (5, 22, 27), it is conceivable that differential protein binding at these sites during germ cell development may play a role in differential acquisition of DNA methylation. This could involve CTCF or another protein with an affinity for the site. In a previous study, we investigated this possibility by replacing the ICR with two copies of the chicken β-globin insulator in mice. Each chicken insulator contained one CTCF binding site of the same consensus sequence as the ICR sites. By contrast to the ICR, the chicken insulator duplex remained hypomethyated in both female and male germ lines, i.e., the duplex lacked the ability to become imprinted. This result suggested that CTCF insulator sites per se are not involved in setting up differential germ line methylation of the ICR (41).
Subsequently, the role of the ICR CTCF binding sites in imprinting has been tested directly by specifically mutating them in mice. In germ cells, differential acquisition of DNA methylation by the mutant ICR in the female and male germ lines was unaffected, again indicating that the CTCF sites are not involved in this process (32). A difficulty with this study was that only approximately one-half of the insulator site core was mutated, and binding of CTCF may not have been completely eliminated. A low level of binding of CTCF was detected in vitro, and it is conceivable that even a much-reduced degree of CTCF binding to the insulator sites may have been sufficient to induce differential methylation in germ cells. Also, if a protein other than CTCF binds to the sites and is involved in differential germ cell methylation, then its binding in germ cells may not have been sufficiently affected by the mutations made. This difficulty has taken on more importance with the results of a recent study: reduction of CTCF in growing oocytes by RNA interference (RNAi) resulted in methylation of the ICR. This result was interpreted to mean that CTCF binding at the ICR—even at low levels, in the case of the previously described mutant ICR—functions to inhibit methylation of the region in oocytes (7). After fertilization, maternal ICRs with mutated insulator sites have been observed to acquire methylation, suggesting that CTCF binding is normally required to maintain the hypomethylation of this region in somatic cells (26, 32). Overall, the results point to a crucial role for the CTCF insulator binding sites, and CTCF binding, in establishing and maintaining imprinting of the Igf2/H19 region.
Here we report on the effect of severely mutating all four ICR CTCF binding sites such that protein binding at these sites is eliminated. We found that female and male germ cells remained hypomethylated and became hypermethylated, respectively, demonstrating that the ICR CTCF insulator sites are not required for the regulation of this process. Further, in somatic cells, the maternally inherited mutant ICR acquired methylation in the ICR and in adjacent regions in cis—the H19 promoter and gene accompanied by down-regulation of H19. This could be the result of a silencing effect of the methylated maternal ICR.
MATERIALS AND METHODS
EMSAs.
Whole-cell extract for the binding assays was made from confluent plates of 13.5-day postcoitum (dpc) normal mouse primary embryo fibroblasts (PEFs) as described previously (49) with minor modifications. Cells were trypsinized and washed with cold Dulbecco’s phosphate-buffered saline without calcium. Aliquots of 10 × 106 cells were suspended in 565 μl of extraction buffer (50 mM Tris HCl [pH 8.0], 500 mM KCl, 0.5 mM EDTA [pH 8.0], 0.5 mM EGTA [pH 8.9], 1% Nonidet P-40, 2 mM dithiothreitol, and 1× protease inhibitor cocktail [P2714; Sigma]) and incubated on ice for 30 min. After centrifugation at 13,000 rpm with a MicroCentaur (MSE) microcentrifuge for 5 min, the supernatant was placed into a new tube, gently mixed with 10% ice-cold glycerol, and snap-frozen in small aliquots on dry ice. The aliquots were kept at −80°C. To prepare the probe for electrophoretic mobility shift assays (EMSAs), 10 pmol of one strand of the oligonucleotide probe was end labeled and annealed to 20 pmol of the other strand and then diluted to 10 fmole/μl (∼30,000 to 60,000 cpm/μl). For the binding reaction mixture, 2 μl of PEF extract was mixed with 2 μl of anti-CTCF antibody (Upstate Biotechnology) or 1 μl of specific competitor in 1× binding buffer (20 mM HEPES [pH 7.6], 60 mM KCl, 2 mM MgCl2, 5% glycerol, 1 mM dithiothreitol, and 1% Nonidet P-40) and incubated on ice for 20 min in the presence of 0.2 μg of sheared herring sperm DNA and 0.25 μg of poly(dI-dC) (Sigma). One microliter (10 fmole) of probe was added, the incubation was continued for another 20 min at room temperature, and the bound and unbound fractions were separated on a prerun 4% polyacrylamide gel in 25 mM Tris-borate-EDTA for 3 h at 200 V. After 1 h of drying at 80°C, the gel was analyzed on a PhosphorImager (Molecular Dynamics). Oligonucleotides used were as follows (upper strands are shown): CTCF1, 5′-CAGTTGTGGGGTTTATACGCGGGAGTTGCCGCGTGGTGGCAGCAAAATCGATTGCGCCAAACCTAAAGAG-3′; CTCF2, 5′-AATCCTTTGTGTGTAAAGACCAGGGTTGCCGCACGGCGGCAGTGAAGTCTCGTACATCGCAGTCCTAAAA-3′; CTCF3, 5′-CTGTTATGTGCAACAAGGGAACGGATGCTACCGCGCGGTGGCAGCATACTCCTATATATCGTGGCCCAAA-3′; CTCF4, 5′-TTGGCTATAGCTAAATGGACAGACGATGCCGCGTGGTGGCAGTACAATACTACATATTGCTCGGCAGACG-3′; CTCF1NdeI, 5′-CAGTTGTGGGGTTTATACGCGGGAGTTACATATGTTTTTCAGCAAAATCGATTGCGCCAAACCTAAAGAG-3′; CTCF2NdeI, 5′-ATCCTTTGTGTGTAAAGACCAGGGTTACATATGTTTTTCAGTGAAGTCTCGTACATCGCAGTCCTAAAAC-3′; CTCF3NdeI, 5′-CTGTTATGTGCAACAAGGGAACGGATGCTACATATGTTTTTCAGCATACTCCTATATATCGTGGCCCAAA-3′; CTCF4NdeI, 5′-TTGGCTATAGCTAAATGGACAGACGATACATATGTTTTTCAGTACAATACTACATATTGCTCGGCAGACG-3′; FII, 5′-CCCAGGGATGTAATTACGTCCCTCCCCCGCTAGGGGGCAGCA-3′; FIIX3′, 5′-CCCAGGGATGTAATTACGTCCCTCCAAATAGCTTTTTCAGCA-3′.
ICR targeting vector.
The same gene targeting vector as previously described was used (41), except that the ICR, defined as a 2.4-kb BglII fragment, was mutagenized at each of the four CTCF binding sites. Nine or 10 bases of the 14-bp consensus sequence were changed, based on the FIIx3′ mutation (31), yielding four identical mutant 14-bp binding sites that included an NdeI restriction site (Fig. 1A). This mutation abolished binding in vitro (Fig. 1B to D). We would also expect this mutation to fully eliminate binding of CTCF to the ICR in vivo; it was shown that changing the mouse CTCF insulator binding site core sequence from CCGC(G/A)(T/C)GG(T/C)GGCAG to CCGCG(G/A)(T/C)ATATGCAG was sufficient to eliminate all CTCF binding to the ICR in vivo at the level of detection of chromatin immunoprecipitation assays (26). In comparison, our mutation involved changing the three invariant nucleotides of these same four nucleotides: three G→T, four other invariant nucleotides, two C→A and two G→T, and two other nucleotides, (G/A)→T and (T/C)→G, to give CATATGTTTTTCAG (Fig. 1A). Further, unlike this previous study, in which only three of the four ICR sites were mutated (26), we introduced these changes to all four sites. On the basis of previous in vivo footprinting studies, CTCF can bind equally well to all four ICR sites (38). Before insertion into the targeting construct, the 2.4-kb BglII fragment was subcloned into the BglII site of pSL1180. Site-directed mutagenesis was done in two cycles using the Transformer site-directed mutagenesis kit (Clontech). Mutagenic primers were the upper-strand EMSA oligonucleotides for the four mutant CTCF binding sites. Selection oligonucleotides were SmaI-AgeI (first cycle), 5′-CATATATAGGGACCGGTTTATAATTAC-3′, and AgeI-SmaI (second cycle), 5′-CATATATAGGGCCCGGGTTATAATTAC-3′. Mutant clones were identified by their NdeI digestion pattern. DNA sequence analysis of the entire 2.4-kb BglII fragment confirmed that no bases other than the ones at the CTCF sites had been altered. In the long arm of the targeting vector, immediately 3′ of the ICR, a tandem duplication of the 193-bp BglII-BamHI fragment was present. However, this small duplication had no effect on the allele-specific expression of Igf2 and H19, or on the extent or pattern of DNA methylation in surrounding sequences. This has been assessed in control mice which carry the duplication but which do not carry the mutant CTCF binding sites in the ICR (data not shown).
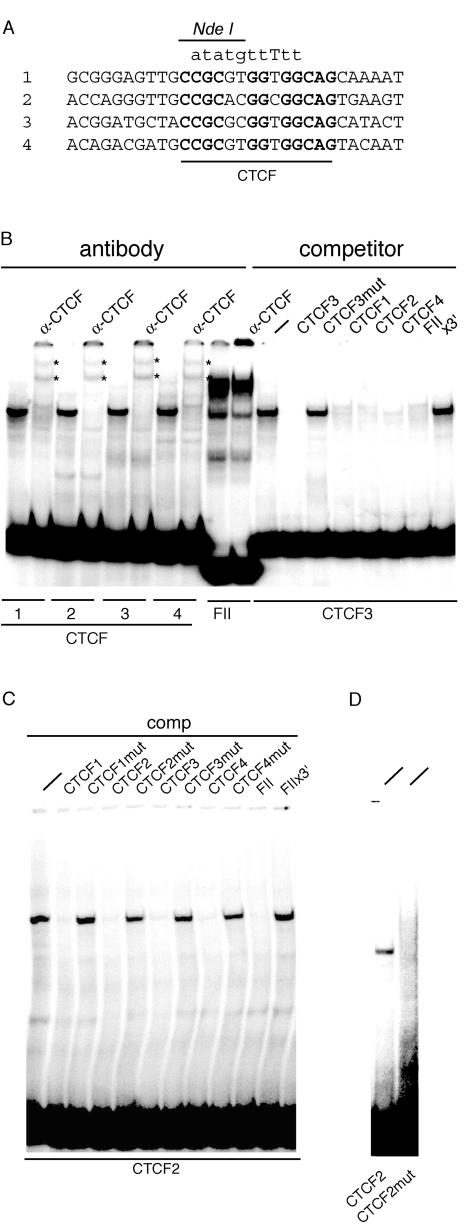
FIG. 1.
EMSA analysis of the CTCF site mutations. Radiolabeled oligonucleotide probes derived from the mouse H19/Igf2 ICR (CTCF 1 to 4) or from the chicken β-globin insulator (FII) were incubated with mouse PEF extract. Unlabeled oligonucleotides were included as competitors at 500-fold excess, or anti-CTCF antibody was given to obtain supershifts. (A) Nature of the CTCF site mutations. The DNA sequence spanning CTCF sites 1 to 4 is aligned. CTCF consensus binding sites are marked in bold. The same 10-bp DNA segment wasreplaced in all four CTCF sites by the sequence shown in lowercase letters on top. The NdeI restriction site created to facilitate screening during site-directed mutagenesis is also indicated. (B) Binding of CTCF complexes to the four ICR CTCF sites (left) and competition of CTCF site 3 (right). (C) Competition of CTCF site 2. (D) Lack of gel shift with the mutant CTCF site 2 oligonucleotide. Mutant oligonucleotide was radiolabeled and run next to the wild-type one after incubation with protein extract.
Production of mice carrying the ICR CTCF binding site mutations.
Gene targeting in mouse embryonic stem (ES) cells was performed using a replacement vector containing a neomycin and diphtheria toxin a-chain positive and negative selection cassette, respectively. The vector was identical to that previously described (41), except for the mutant 2.4-kb BglII ICR. Homologous recombination was detected by PCR amplification across the short arm of the vector to yield a 1.3-kb band with the following primers: upper primer, in the genomic sequence just outside the end of the short arm, 5′-CCTATGCCCATGCCCCATACAAATGACACC-3′; lower primer, in the polyadenylation sequence of the neomycin selection cassette, 5′-GCTGGGGCTCGACTAGAGCTTGCGGAAC-3′. Recombinant clones identified in this PCR assay were examined further by Southern blotting for conservative recombination of the short and long arms as previously described (41) and for retention of all four mutant CTCF binding sites. Excision of the neo cassette was achieved by mating male chimeras with females heterozygous for an X-linked Cre recombinase gene. Eggs of these females excise floxed sequences without mosaicism and regardless of Cre inheritance (42).
Breeding of fetuses carrying the mutation for analysis.
Female mice heterozygous for the mutation and negative for the Cre allele, obtained from mating male chimeras with Cre/0 females (see above), were mated with males homozygous for the Mus musculus castaneus form of distal chromosome 7 as derived from strain CAST/Ei (CS; The Jackson Laboratory, Bar Harbor, Maine). These males were of strain FVB/NJ.CAST/Ei(N7). The use of this mating, and its reciprocal, allowed for allele-specific analysis of expression and methylation. The wild-type allele (+) was derived from strain CS, while the mutant allele (−) was derived from strain 129SI/ImJ (129). Hereafter, heterozygous fetuses maternally and paternally inheriting the mutation are designated −(M)/+ and +/−(P), respectively, except where indicated otherwise.
Gene expression.
Northern blot assays were performed as described elsewhere (23, 37). A 10-μg aliquot of total RNA was loaded per lane. Allele-specific expression was determined using reverse transcription-PCR single-nucleotide primer extension assays as described previously (40, 41). Each assay relies on a single known sequence difference between allelic RNAs. Assays were conducted on the same samples used in Northern blotting. Each sample represented an individual embryo.
DNA methylation in somatic cells.
Southern blotting was performed as described elsewhere (37). For DNA methylation analysis of the H19/Igf2 ICR, DNA was digested with BglII to release the ICR and then with SacI to differentiate between alleles. Digestion with the methylation-sensitive HpaII enzyme was used to test for methylation, while digestion with the methylation-insensitive isoschisomer MspI was used to show the fragment sizes expected in unmethylated DNA. The filter was hybridized to the 2.4-kb ICR BglII fragment (probe a). For methylation around the G-rich repeat element, the DNA was digested with SacI to differentiate between alleles and with HhaI to test for methylation. The P3Q3 PCR fragment was used as probe (P3, 5′-GGTGGGGTTGTATAGCGTGGGTTAT-3′; Q3, 5′-CCCACATCACCCTTGCTATACTCCC-3′) (probe b). To visualize methylation changes localized to the promoter, PvuII restriction was used to release the DNA fragments and K3L3 and M3N3 PCR fragments combined (probe c) were used as probe (K3, 5′-GTATGAGACCCATGCCCTCAAATTCC-3′; L3, 5′-GCCTCACCCCACACCCGCA-3′; M3, 5′-TTTGGAGAATTTCAGGACGGGTGCG-3′; N3, 5′-ACCCCACGACTCTCCTCCAGCTCTC-3′. PvuII restriction was used to release the DNA fragments, and an EcoRI-SalI fragment (probe d) was used for DNA methylation analysis at the body of the H19 gene.
DNA methylation of the ICR in germ cells. (i) Perinatal germ cells.
Female mice of the homozygous transgenic line, which express the enhanced green fluorescent protein reporter gene specifically in germ cells (39), were mated to males homozygous for the mutant ICR (−/−). From the resulting female and male 17.5-dpc fetuses, germ cells were purified by flow cytometry as described previously (39) using a MoFlo flow cytometer (Cytomation, Fort Collins, Colo.). Bisulfite sequencing was performed on cells embedded into agarose beads (25). DNA from 20,000 and 25,000 cells was amplified using primers described earlier (45). One PCR was performed on DNA from each of two conversion reactions. The products were isolated and subcloned into the pCR2.1-TOPO vector (Invitrogen), and individual clones were sequenced.
(ii) DNA methylation of the ICR on oocytes.
Males homozygous for the mutant ICR (−/−) were mated to normal CF1 females (Charles River), and the resulting adult +/− females were superovulated (11). Oocyte collection was done as published previously (40). Four separate bisulfite reactions were done on pools of 190, 150, 175, and 175 oocytes embedded into agarose beads (25), and each reaction was PCR amplified separately.
RESULTS
The mutant CTCF binding sites are unable to bind CTCF.
In order to inhibit CTCF binding to the ICR completely, we made drastic changes to the consensus sequence of all four CTCF binding sites (Fig. 1A). These mutations were very similar to the mutation (FIIx3′) that inactivated CTCF binding and insulator function of the chicken β-globin CTCF site (FII) (31). Our mutations also included an NdeI restriction site in order to be able to recognize mutants after site-directed mutagenesis and gene targeting in mice. The four wild-type CTCF sites specifically bound CTCF from PEF extracts (Fig. 1B), but the mutant probes were unable to form a specific DNA-protein complex (Fig. 1D; the CTCF 2 mutant is shown). The same gel shift was evident with all four CTCF probes, and this band was supershifted by anti-CTCF antibody. A similar-size gel shift was eliminated from the FII probe by anti-CTCF. CTCF site 3 was competed by CTCF sites 1 to 4, but not by mutant CTCF3 and FIIx3′ (Fig. 1B). CTCF probe 2 was competed by CTCF probes 1 to 4 and FII, but not by mutant CTCF sites 1 to 4 and FIIx3′ (Fig. 1C). With these mutations, we completely eliminated CTCF binding in vitro. This also sacrificed 10 CpGs in the ICR, which slightly reduced its overall CpG density.
Gene targeting.
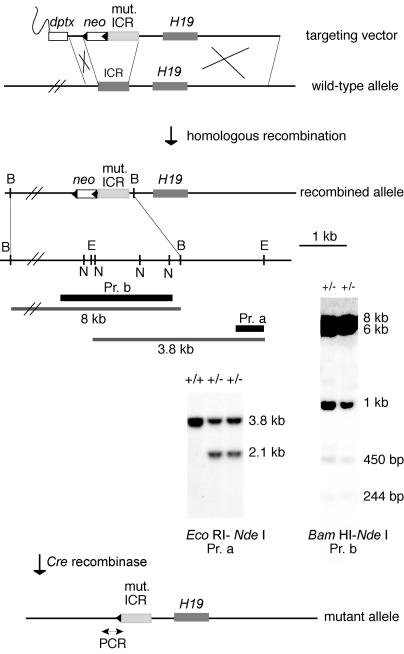
Of 288 G418-resistant clones picked and expanded, 16 underwent homologous recombination as determined by PCR and Southern blotting. Eleven of these clones underwent conservative recombination with respect to the short and long arms and contained the four CTCF binding sites in the ICR (Fig. 2). Recombination in the long arm could have occurred within the ICR, resulting in loss of mutant CTCF sites at the targeted locus; loss of mutant CTCF sites was observed in two clones (data not shown). For each of seven clones, ∼50 blastocysts were injected with ∼15 ES cells and transferred to recipients. Germ line transmission of the ES cell genome was obtained for four clones, and mice derived from clone no. 63 were used in this study.
FIG. 2.
Gene targeting at the H19/Igf2 ICR, as shown by Southern blotting of two ES cell clones that underwent conservative recombination. Dark gray rectangles, wild-type ICR and H19 gene; light gray rectangle, mutant ICR; open rectangles, selection cassettes; black triangles, loxP sites. Probes (black bars): a (Pr. a), 0.6-kb EcoRI-ScaI fragment; b (Pr. b), 2.4-kb BglII ICR fragment. Restriction sites used for analysis: B, BamHI; E, EcoRI; N, NdeI.
Paternal inheritance of the CTCF binding site mutations.
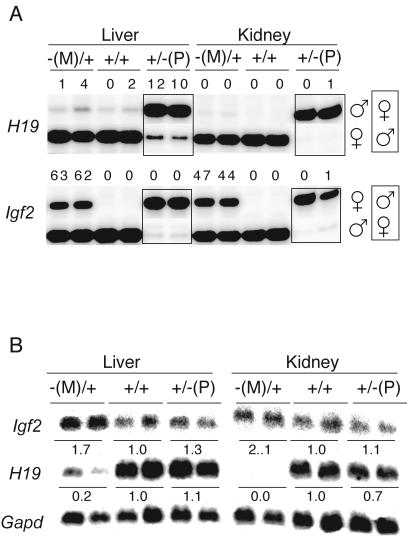
On paternal inheritance, the mutant ICR had little or no effect on the normal allele-specific expression pattern of Igf2 and H19. Both genes were expressed monoallelically in the liver and kidney of 17.5-dpc +/−(P) fetuses (Fig. 3A). The size of 17.5-dpc +/−(P) fetuses was the same as that of their normal littermates (Table 1). The mutant paternal ICR was hypermethylated in somatic tissues of 17.5-dpc +/−(P) fetuses (Fig. 4A; see also Fig. 5B), as is the case in normal fetuses (45, 46). The +/−(P) fetuses showed similar hypermethylation of the paternal chromosome around the G-rich repeats, at the promoter and the body of the H19 gene as found in normal perinatal fetuses (Fig. 4B to D). Thus, the mutant paternal ICR, deficient for CTCF binding, successfully substituted for the imprinting and promoter silencing functions of the normal paternal ICR.
FIG. 3.
Phenotype of the 17.5-dpc fetuses inheriting the four CTCF site mutations in the H19/Igf2 ICR. (A) Reverse transcription-PCR single-nucleotide primer extension assays. In the −(M)/+ and +/+ lanes, the top row is the presumptive inactive allele. For the +/−(P) lanes (in boxes), these fetuses were obtained from the reciprocal mating, and hence the bottom row is the presumptive inactive allele. Parental origin of alleles is on the right. The value above each band is the amount of RNA contributed by the presumptive inactive allele as a percentage of the total. (B) Northern blotting results. Values under bands are the mean relative amounts of RNA standardized according to Gapd mRNA. Each lane represents an individual embryo. For Igf2 only the shortest transcript is shown, but the value represents the total of all transcripts.
TABLE 1.
Weights of 17.5-dpc fetuses on maternal and paternal inheritance of the CTCF mutations in the ICR
| Alleles | Wt of fetus
|
Wt of placenta
|
||||
|---|---|---|---|---|---|---|
| Mean (g) ± SD (n) | Range | % of +/+ wt | Mean (g) ± SD (n) | Range | % of +/+ wt | |
| −(M)/+a | 1.22 ± 0.09 (13) | 1.03-1.3 | 122 | 99.53 ± 14.25 (13) | 78-126 | 136 |
| +/+a | 1.00 ± 0.09 (20) | 0.84-1.11 | 73.4 ± 10.42 (20) | 53-93 | ||
| +/−(P)b | 1.02 ± 0.09 (15) | 0.91-1.16 | 103 | 71.9 ± 11.41 (15) | 47-88 | 102 |
| +/+b | 0.99 ± 0.08 (18) | 0.87-1.15 | 70.7 ± 11.04 (18) | 56-101 | ||
Siblings from +/−(P) × FVBCSN7 +/+ matings.
Siblings from FBCSN7 +/+ × +/−(P) matings.
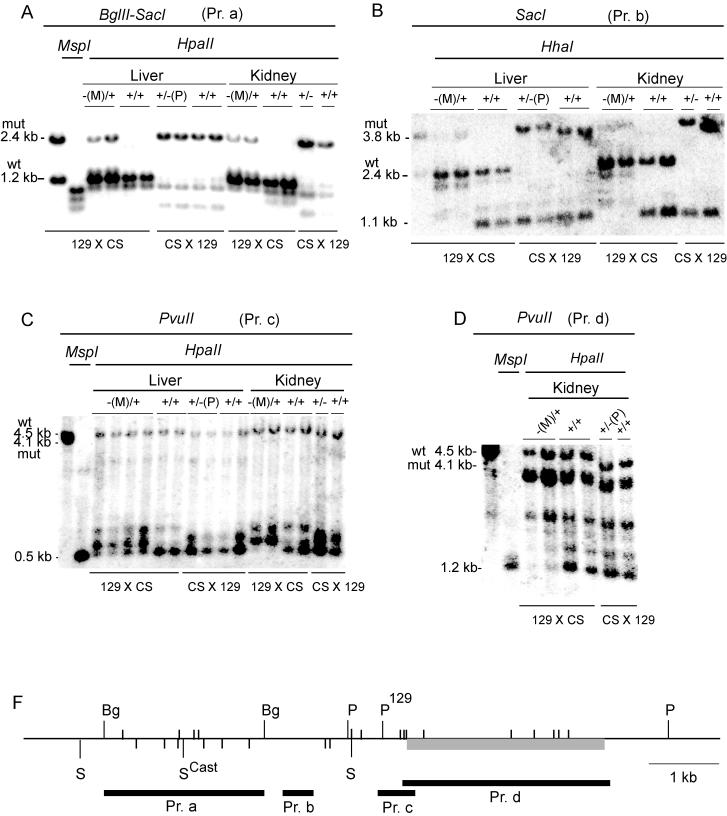
FIG. 4.
Methylation of H19 regions: Southern blots. Tissues were obtained from 17.5-dpc fetuses. Regions analyzed: ICR (A); region surrounding the G-rich repeat elements (B); H19 promoter (C); body of the H19 gene (D). (F) Map of the analyzed regions. Gray rectangle, H19 gene; black rectangles, probes. Restriction sites are SacI (S), BglII (Bg), PvuII (P), and HpaII and HhaI (small vertical lines above and below horizontal line, respectively) (only the sites analyzed are shown). The SacI and PvuII sites which distinguish between the 129 and CS strains (46) are marked.
FIG. 5.
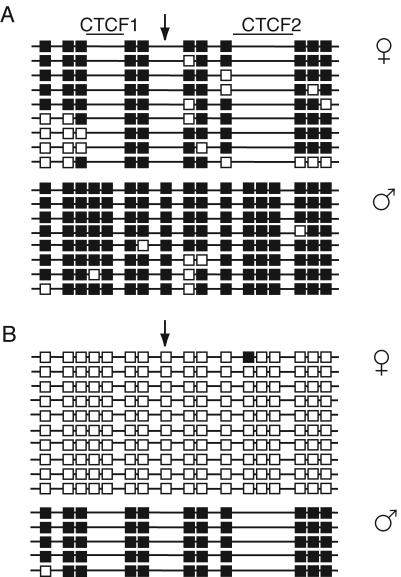
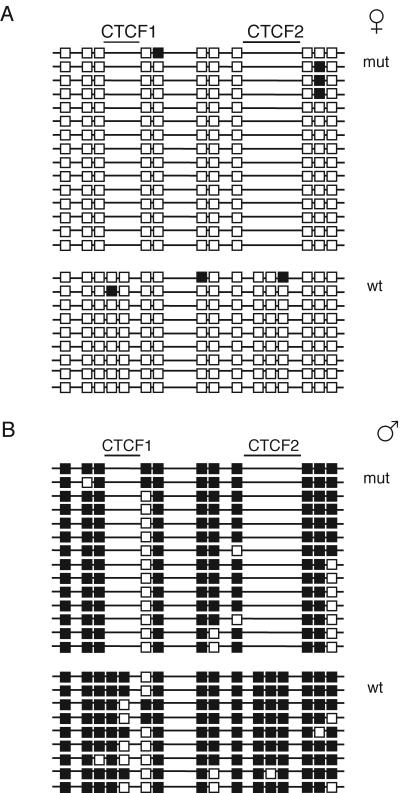
Methylation of the ICR in somatic cells Bisulfite sequencing was performed on DNA isolated from kidneys of 17.5-dpc fetuses derived from reciprocal matings. Data combined from two independent bisulfite conversion reactions are shown. Each horizontal line represents a clone of a separate PCR product. The parental origin of chromosomes is shown on the left. Closed square, methylated CpGs; open squares, unmethylated CpGs. The positions of the five CpGs, removed by mutagenesis, are indicated on the top. Presence of the eighth CpG is specific for CS DNA (arrow). (A) −(M)/+ fetuses; (B) +/−(P) fetuses.
Maternal inheritance of the CTCF binding site mutations.
The 17.5-dpc −(M)/+ fetuses and placentae were 122 and 136% of the weight of +/+ siblings, respectively (Table 1). Eleven of 13 fetuses examined exhibited polydactyly of the forepaws, the defect being essentially the same as that described in mutations involving the insulin-like growth factor receptor gene which result in elevated IGFII activity (13, 21). As expected from the large size of −(M)/+ fetuses, the concentration of Igf2 mRNA in these fetuses was higher than normal (Fig. 3B). In the liver and kidney of −(M)/+ fetuses, Igf2 was biallelically expressed (Fig. 3A). Expression of Igf2 from the maternal allele showed that the CTCF binding-deficient ICR failed to act as a chromatin insulator when maternally derived. H19 mRNA was expressed exclusively from the maternal allele, but the level of expression was much lower than normal in the kidney and liver of 17.5-dpc −(M)/+ fetuses (Fig. 3). These results suggest that the mutant maternal ICR possibly induced postfertilization inactivation of the H19 promoter in cis, as does the normal paternal ICR.
In the absence of CTCF binding, the mutant ICR became hypermethylated in somatic tissues of −(M)/+ mice (Fig. 4A and 5). We observed a low level, but widespread methylation, of the maternal chromosome in 17.5-dpc −(M)/+ fetal tissues that included the region around the G-rich repeats (34) located between the ICR and the promoter, the promoter of H19 itself and the body of the H19 gene (Fig. 4B to D).
ICR DNA methylation in the germ line.
In +/+ mice, differential methylation of the ICR in female and male germ cells is essentially fully established by the perinatal stage, with all copies of the ICR in female and male germ cells being hypo- and hypermethylated, respectively (6, 47). We obtained similar results for the mutant ICR, which was hypo- and hypermethylated in female and male germ cells of 17.5-dpc +/−(P) fetuses, respectively. Further, as we analyzed heterozygous animals, we were able to directly compare the mutant and wild-type ICRs in the same bisulfite sequencing experiments. In female germ cells, the degree of hypomethylation of the mutant ICR was similar to that of the wild-type ICR for all nonmutated CpGs in the region (Fig. 6A). Also, in male germ cells the degree of hypermethylation of the mutant ICR was similar to that of the wild-type ICR for all nonmutated CpGs in the region (Fig. 6B).
FIG. 6.
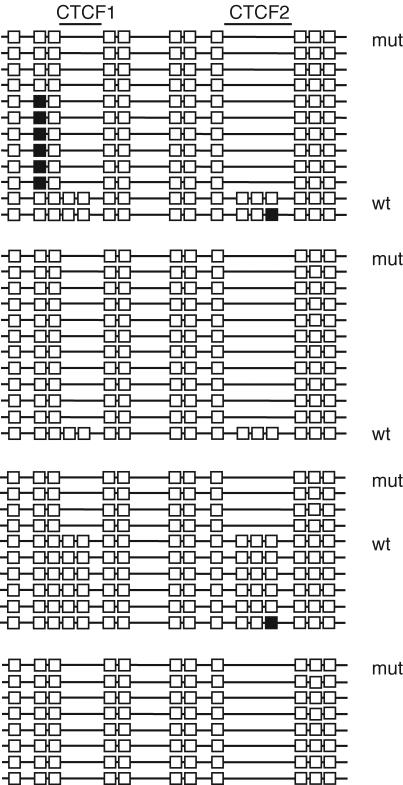
Methylation of the ICR in perinatal germ cells. Bisulfite sequencing was done on pools of 20,000 and 25,000 germ cells. Data combined from two independent bisulfite conversion reactions are shown. Each horizontal line represents a clone of a separate PCR product. (A) Female germ cells; (B) male germ cells.
Hypomethylation of the mutant and wild-type ICRs was also seen in ovulated oocytes of heterozygous +/−(P) females, where an almost complete lack of methylation was observed at all CpGs that could be compared (Fig. 7). Therefore, it is unlikely that the mutant ICR ever attracts methylation during female germ cell development and, therefore, in this regard it appears to be behaving in similar fashion to the wild-type ICR.
FIG. 7.
Methylation of the ICR in oocytes. Bisulfite sequencing was done on four separate pools of oocytes containing 190, 175, 175, and 150 oocytes. Each horizontal line represents a clone of a separate PCR product.
DISCUSSION
We have drastically mutated each of the four CTCF insulator binding sites in the Igf2/H19 ICR by gene targeting in mice. The ICR possesses properties of chromatin insulation on the somatic maternal chromosome, promoter silencing on the somatic paternal chromosome, and imprinting establishment, or differential DNA methylation acquisition, in female and male germ cells. The aim of this study was to determine if these ICR sites play a role in any of these three processes.
Effect of mutant CTCF sites on acquisition of differential methylation or imprinting: CTCF binding sites and imprinting establishment.
In the establishment of genomic DNA methylation patterns, some DNA binding proteins might restrict access of methyltransferases to CpGs, while others may target methyltransferases to CpGs (14). Based on these models, in the present study we hypothesized that CTCF—or another protein that can recognize the four ICR sites in question—binds to the ICR in female germ cells and excludes methyltransferases, or binds to the ICR in male germ cells and attracts methyltransferases. Our results clearly showed the hypothesis to be false: we observed no abnormal behavior of the mutant ICR in respect to its acquisition of DNA methylation in the female and male germ lines. We can therefore conclude that the CTCF insulator binding sites are dispensable for the maintenance of ICR hypomethylation in female germ cells and for the acquisition and maintenance of ICR hypermethylation in male germ cells. Thus, the binding of CTCF, or another protein which recognizes the site, e.g., the “brother of the regulator of imprinted sites” (BORIS) protein (20, 26), is not required in either of these processes. With respect to what other ICR sites might be involved, we have identified a set of conserved nuclear hormone receptor-like sites in the ICR which show in vivo footprints in somatic cells and which can bind proteins specifically in male fetal germ cell nuclear extracts in EMSAs (41a. We are currently investigating the role of these sites in imprinting by mutating them in mice.
In a recent study, reduction of CTCF in growing oocytes by RNAi was accompanied by methylation of the ICR, indicating that CTCF binding at the ICR insulator sites inhibits methylation of this region in these cells (7). Given the present results, this interpretation no longer seems plausible, and three other explanations can be put forward: (i) a nonspecific effect of RNAi. Based on estimates of viability, mammalian oocytes and undifferentiated cells appear to lack the “interferon response” to double-stranded RNA exposure (35, 36, 48, 50). However, little else is known regarding possible side effects of this system, and it would be of interest to determine if other double-stranded RNA molecules could induce methylation of the ICR in oocytes. (ii) CTCF binds to other sequences in the ICR—aside from the insulator sites—to inhibit methylation. Further, this mechanism would have to be oocyte specific, as CTCF is unable to prevent postfertilization methylation of maternally inherited ICRs containing mutated insulator binding sites. This possibility seems unlikely and is difficult to test. (iii) CTCF may inhibit methylation of the ICR in oocytes by an indirect mechanism which does not involve binding of the protein to the region. For example, CTCF might sequester a cofactor that is able to direct methylation of the ICR. Also, CTCF is a versatile factor, binding at a wide variety of promoters and exhibiting transcriptional repression and activation functions (24). Loss of CTCF would certainly result in a significant change in gene expression profiles, and one of these changed profiles might result in ICR methylation. It would be of interest to determine if conditional elimination of CTCF in a somatic cell type could result in hypermethylation of the ICR.
CTCF binding sites and imprinting maintenance.
The mutant maternal ICR was unmethylated in oocytes (Fig. 7) but became methylated after fertilization (Fig. 4A and 5); therefore, CTCF binding maintains the hypomethylated state of the ICR in somatic cells, i.e., binding to the maternal ICR is required for excluding methyltransferases from the region. Similarly, the hypomethylated state of CpG islands, which are often coincident with promoters, might result from the exclusion of methyltransferases by bound transcription factors (5, 22, 27). The paternal ICR became methylated in the male germ line, and this methylation persisted into somatic development. This means that the maintenance mechanism of the methylation status was not affected in the mutant paternal ICR.
Effect of mutant CTCF binding sites on chromatin insulation.
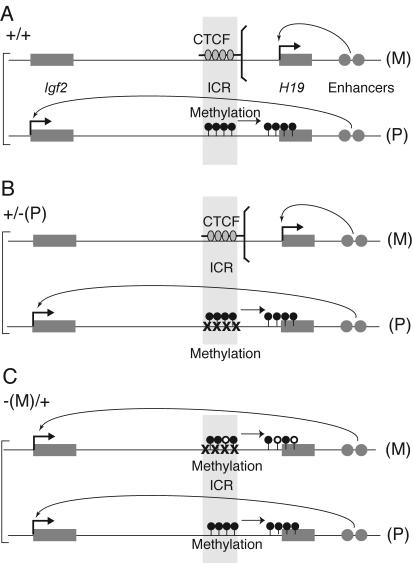
The normal maternal ICR is unmethylated and binds CTCF molecules at four sites in vivo which insulate the effect of the downstream enhancers towards the Igf2 promoters (Fig. 8A). In the present study we showed that insulator function of the maternal ICR was lost when all four CTCF binding sites were inactivated (Fig. 8C), resulting in Igf2 expression from the maternal allele and consequently large fetus size. This provides evidence that maternal Igf2 inactivity is brought about through insulator activity by CTCF binding to ICR sequences in cis.
FIG. 8.
Summary of the results. (A) Wild-type +/+ fetuses; (B) +/−(P) fetuses. Mutation of each of the four CTCF binding sites has no consequences; methylation of the ICR is undisturbed, and allele-specific expression of H19 and Igf2 is the same as in wild type (C) −(M)/+ fetuses. The CTCF mutant ICR does not function as an insulator, the enhancers now activate Igf2 in cis (50% activity), and Igf2 is now biallelically expressed, while H19 expression is dramatically down-regulated (0 to 12%). Methylation accumulates in the maternal ICR, promoter, and body of the H19 gene in somatic cells. M, maternal allele; P, paternal allele. CTCF molecules are indicated by gray ovals.
The normal paternal ICR does not bind CTCF and lacks insulator activity (Fig. 8A). In this respect, the mutations in the paternal ICR did not result in any change: Igf2 was expressed at a normal level from the paternal chromosome (Fig. 8B).
Effect of mutant CTCF binding sites on H19 promoter silencing.
The normal paternal ICR, while possessing no insulator activity, establishes silence of the H19 promoter in cis during early embryogenesis by directing secondary DNA methylation of the H19 promoter (33). The CTCF binding-deficient paternal ICR clearly retained this property, as shown by the analysis of allele-specific expression and H19 promoter methylation in the liver and kidney of 17.5-dpc +/−(P) fetuses.
The normal maternal ICR is unmethylated and does not have a promoter-silencing activity in cis. However, the mutant maternal ICR exhibited a high level of methylation in 17.5-dpc −(M)/+ fetuses compared to +/+ littermates: in kidney DNA, 78% of the CpGs were methylated in the region analyzed with bisulfite sequencing (Fig. 5A). At the same time, H19 RNA levels were greatly reduced upon maternal transmission of the mutation (Fig. 3B), and the maternal H19 promoter was more methylated (Fig. 4C). If methylation of the ICR were the key requirement that directs silencing of the H19 promoter, then it is possible that silencing of the maternal H19 promoter was directed by methylation of the mutant maternal ICR in cis.
Widespread methylation of the mutant maternal allele was induced by the CTCF site mutations. Spreading of methylation could be imagined from two directions: (i) methylation could have spread from the ICR into the promoter. This is likely, as mutations were made in the ICR and methylation levels appeared as a gradient, being highest in the ICR, lower at the promoter, and lowest in the body of the H19 gene (Fig. 4). Furthermore, this possibility is supported by the fact that there is a mechanism in place where methylation of the H19 promoter is directed by the ICR, as it was shown for the normal paternal allele. The methylated paternal ICR is needed in the early embryo to direct inactivation of the paternal H19 promoter, but it is dispensable after the epigenetic changes at the promoter have been made (33). (ii) Methylation could have spread from the promoter to the ICR. In the lack of insulation from the ICR, the H19 and Igf2 promoters would both be accessible to the downstream enhancers and the Igf2 promoter could outcompete the H19 promoter, eventually shutting it down. The downregulated H19 promoter would then accumulate methylation, which could spread into the ICR. This sequence of events seems less likely in the light of other experiments. Upon maternal transmission of the ICR deletion, when enhancer insulation function was missing, the H19 promoter did not become methylated and its activity was 50% of the normal maternal allele (15, 33, 44). This situation is clearly different from our results, when maternal transmission of the CTCF site mutations resulted in increased H19 promoter methylation and a dramatic reduction in H19 expression (0 to 12%). It indicates that something else, e.g., methylation of the remaining sequences of the ICR, other than purely lack of CTCF binding, was necessary to drastically reduce H19 expression upon maternal transmission of the mutant ICR.
On maternal transmission of the mutation, the H19 promoter was more methylated than the promoter of normal siblings but was less methylated than the normal paternal promoter (Fig. 4C). Can this methylation account for the high degree of inactivation of the H19 promoter (Fig. 3B)? It was suggested earlier that de novo methylation may be directed by alterations in the chromatin structure that would reflect differences between transcriptionally active and repressed regions (4). Evidence that such a regulatory pathway exists in mammals was recently published, demonstrating that histone H3-K9 trimethylation is required to direct Dnmt3-dependent DNA methylation at pericentric heterochromatin (17). Similarly, upon maternal transmission of the CTCF site mutations, the methylated mutant maternal ICR could direct inactivation of the H19 promoter, e.g., by posttranslational histone modifications. Similarly, histone modifications rather than promoter methylation could be the primary mechanism by which the methylated normal paternal ICR directs inactivity of the normal paternal H19 promoter, and the role of promoter methylation would be a secondary mechanism to stabilize the inactive state. The normal paternal promoter is indeed packaged into rotationally positioned nucleosomes (38).
In two recent studies, CTCF binding was inactivated in such a way that all CpGs in the ICR remained intact (26, 32). In one study, there was residual CTCF binding activity in vitro (32). In the other study, only three of the four CTCF binding sites were mutated (26), yet all four sites bound CTCF in vivo (38). Our study is in agreement with the results of these two studies with respect to the role of CTCF binding in insulator function and maintenance of the unmethylated state of the ICR in somatic cells. Both groups found activation of maternal Igf2 expression upon maternal transmission of the mutations, indicating that the insulator function was impaired. Both groups found the mutant maternal ICR to be methylated in somatic tissues. Both studies reported down-regulation of H19 expression upon maternal transmission of the CTCF site mutations (26, 32). Schoenherr and colleagues concluded that the drastic down-regulation of H19 expression was caused by lack of activation of the H19 promoter by CTCF in the mutant maternal ICR (32) This explanation does not account for the great difference (50% versus 0 to 12%) between the levels of H19 expression from the mutant maternal alleles when the entire insulator is deleted (15, 33, 44) versus when only the four CTCF sites are inactivated (our quantitative data). Schoenherr and colleagues did not find methylation at the H19 promoter, and they interpreted this finding as indicating that methylation protection by CTCF is specific to the ICR (32). In the other study, methylation was not investigated apart from the ICR. By contrast, our study, in which all CTCF binding sites were drastically mutated, shows a much wider spreading of methylation that includes the promoter and the body of the H19 gene. The differences between the studies could be explained on the basis of the differences in the mutations made. Our data indicate that methylation of the ICR, brought about by lack of CTCF binding, can spread over a wider region. This points to a role in postfertilization inactivation of the promoter by the mutant methylated maternal ICR, possibly through a mechanism similar to that of the paternal ICR (33). Pant and colleagues reported that the mutant paternal ICR induced methylation of the normal maternal ICR in trans (26). Our data do not support such a trans-sensing effect (Fig. 4). Our data (Fig. 7) are in agreement with those of Schoenherr and colleagues in the fact that there is no methylation at the mutant ICR in oocytes (32). In addition we showed that the ICR is hypomethylated in perinatal female germ cells and methylated in male germ cells (Fig. 6) at the time of imprint establishment (6, 47).
The results of these studies together demonstrate that it is possible to separate different properties of the ICR. The property of chromatin insulation and maintenance of the unmethylated state of the maternal ICR depend on CTCF binding and are separable from the properties of establishment of the differential methylation in the Igf2/H19 ICR in the germ line and from postfertilization methylation of the H19 promoter. These results point to the existence of specific sequences in the ICR, independent of CTCF binding sites, which are capable of marking the parental chromosomes in the germ line. Further mutagenesis of the ICR in mice should lead to identification of these sequences.
Acknowledgments
We thank Lucy Brown, Claudio Spalla, and Jim Bolen for flow cytometry and Gerd Pfeifer for his comments on the manuscript.
This work was supported by National Institutes of Health grant GM064378 to J.R.M. and National Science Foundation grant BIR-9220534.
Footnotes
We dedicate this paper to the memory of William Kaplan, pioneer neurogeneticist.
REFERENCES
- 1.Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 5.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 6.Davis, T. L., J. M. Trasler, S. B. Moss, G. J. Yang, and M. S. Bartolomei. 1999. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58:18-28. [DOI] [PubMed] [Google Scholar]
- 7.Fedoriw, A. M., P. Stein, P. Svoboda, R. M. Schultz, and M. S. Bartolomei. 2004. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303:238-240. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson-Smith, A. C., H. Sasaki, B. M. Cattanach, and M. A. Surani. 1993. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362:751-755. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson-Smith, A. C., and M. A. Surani. 2001. Imprinting and the epigenetic asymmetry between parental genomes. Science 293:1086-1089. [DOI] [PubMed] [Google Scholar]
- 10.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Holmgren, C., C. Kanduri, G. Dell, A. Ward, R. Mukhopadhya, M. Kanduri, V. Lobanenkov, and R. Ohlsson. 2001. CpG methylation regulates the Igf2/H19 insulator. Curr. Biol. 11:1128-1130. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. R. 1974. Hairpin-tail: a case of post-reductional gene action in the mouse egg. Genetics 76:795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 15.Kaffer, C. R., M. Srivastava, K. Y. Park, E. Ives, S. Hsieh, J. Batlle, A. Grinberg, S. P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14:1908-1919. [PMC free article] [PubMed] [Google Scholar]
- 16.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 17.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone h3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 18.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 19.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9:2079-2089. [DOI] [PubMed] [Google Scholar]
- 20.Loukinov, D. I., E. Pugacheva, S. Vatolin, S. D. Pack, H. Moon, I. Chernukhin, P. Mannan, E. Larsson, C. Kanduri, A. A. Vostrov, H. Cui, E. L. Niemitz, J. E. Rasko, F. M. Docquier, M. Kistler, J. J. Breen, Z. Zhuang, W. W. Quitschke, R. Renkawitz, E. M. Klenova, A. P. Feinberg, R. Ohlsson, H. C. Morse III, and V. V. Lobanenkov. 2002. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. USA 99:6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig, T., J. Eggenschwiler, P. Fisher, A. J. D'Ercole, M. L. Davenport, and A. Efstratiadis. 1996. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev. Biol. 177:517-535. [DOI] [PubMed] [Google Scholar]
- 22.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin, K. J., P. Szabó, H. Haegel, and J. R. Mann. 1996. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development 122:265-270. [DOI] [PubMed] [Google Scholar]
- 24.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 25.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pant, V., P. Mariano, C. Kanduri, A. Mattsson, V. Lobanenkov, R. Heuchel, and R. Ohlsson. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeifer, G. P., R. L. Tanguay, S. D. Steigerwald, and A. D. Riggs. 1990. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 4:1277-1287. [DOI] [PubMed] [Google Scholar]
- 28.Reed, M. R., C. F. Huang, A. D. Riggs, and J. R. Mann. 2001. A complex duplication created by gene targeting at the imprinted H19 locus results in two classes of methylation and correlated Igf2 expression phenotypes. Genomics 74:186-196. [DOI] [PubMed] [Google Scholar]
- 29.Reik, W., and J. Walter. 1998. Imprinting mechanisms in mammals. Curr. Opin. Genet. Dev. 8:154-164. [DOI] [PubMed] [Google Scholar]
- 30.Ripoche, M. A., C. Kress, F. Poirier, and L. Dandolo. 1997. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 11:1596-1604. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava, M., S. Hsieh, A. Grinberg, L. Williams-Simons, S. P. Huang, and K. Pfeifer. 2000. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 14:1186-1195. [PMC free article] [PubMed] [Google Scholar]
- 34.Stadnick, M. P., F. M. Pieracci, M. J. Cranston, E. Taksel, J. L. Thorvaldsen, and M. S. Bartolomei. 1999. Role of a 461-bp G-rich repetitive element in H19 transgene imprinting. Dev. Genes Evol. 209:239-248. [DOI] [PubMed] [Google Scholar]
- 35.Stein, P., P. Svoboda, and R. M. Schultz. 2003. Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev. Biol. 256:187-193. [DOI] [PubMed] [Google Scholar]
- 36.Svoboda, P., P. Stein, H. Hayashi, and R. M. Schultz. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127:4147-4156. [DOI] [PubMed] [Google Scholar]
- 37.Szabó, P., and J. R. Mann. 1994. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120:1651-1660. [DOI] [PubMed] [Google Scholar]
- 38.Szabó, P., S. H. Tang, A. Rentsendorj, G. P. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607-610. [DOI] [PubMed] [Google Scholar]
- 39.Szabó, P. E., K. Hubner, H. Scholer, and J. R. Mann. 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115:157-160. [DOI] [PubMed] [Google Scholar]
- 40.Szabó, P. E., and J. R. Mann. 1995. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 9:1857-1868. [DOI] [PubMed] [Google Scholar]
- 41.Szabó, P. E., S. H. Tang, M. R. Reed, F. J. Silva, W. M. Tsark, and J. R. Mann. 2002. The chicken beta-globin insulator element conveys chromatin boundary activity but not imprinting at the mouse Igf2/H19 domain. Development 129:897-904. [DOI] [PubMed] [Google Scholar]
- 41a.Szabò, P. E., G. P. Pfeifer, and J. R. Mann. 2004. Parent-of-origin-specific binding of nuclear hormone receptor complexes in the H19/Igf2 imprinting control region. Mol. Cell. Biol. 24:4858-4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, S. H., F. J. Silva, W. M. Tsark, and J. R. Mann. 2002. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32:199-202. [DOI] [PubMed] [Google Scholar]
- 43.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsen, J. L., M. R. Mann, O. Nwoko, K. L. Duran, and M. S. Bartolomei. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22:2450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9:407-413. [DOI] [PubMed] [Google Scholar]
- 47.Ueda, T., K. Abe, A. Miura, M. Yuzuriha, M. Zubair, M. Noguchi, K. Niwa, Y. Kawase, T. Kono, Y. Matsuda, H. Fujimoto, H. Shibata, Y. Hayashizaki, and H. Sasaki. 2000. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5:649-659. [DOI] [PubMed] [Google Scholar]
- 48.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 49.Wyszomierski, S. L., J. Yeh, and J. M. Rosen. 1999. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13:330-343. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S., S. Tutton, E. Pierce, and K. Yoon. 2001. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 21:7807-7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zemel, S., M. S. Bartolomei, and S. M. Tilghman. 1992. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 2:61-65. [DOI] [PubMed] [Google Scholar]