Abstract
Bloom's syndrome (BS) is an autosomal-recessive human disorder caused by mutations in the BS RecQ helicase and is associated with loss of genomic integrity and an increased incidence of cancer. We analyzed the mitotic and the meiotic roles of Caenorhabditis elegans him-6, which we show to encode the ortholog of the human BS gene. Mutations in him-6 result in an enhanced irradiation sensitivity, a partially defective S-phase checkpoint, and in reduced levels of DNA-damage induced apoptosis. Furthermore, him-6 mutants exhibit a decreased frequency of meiotic recombination that is probably due to a defect in the progression of crossover recombination. In mitotically proliferating germ cells, our genetic interaction studies, as well as the assessment of the number of double-strand breaks via RAD-51 foci, reveal a complex regulatory network that is different from the situation in yeast. Although the number of double-strand breaks in him-6 and top-3 single mutants is elevated, the combined depletion of him-6 and top-3 leads to mitotic catastrophe concomitant with a massive increase in the level of double-strand breaks, a phenotype that is completely suppressed by rad-51. him-6 and top-3 are thus needed to maintain low levels of double-strand breaks in normally proliferating germ cells, and both act in partial redundant pathways downstream of rad-51 to prevent mitotic catastrophy. Finally, we show that topoisomerase IIIα acts independently during a late stage of meiotic recombination.
Mutations in three genes encoding human RecQ helicases BLM, WRN, and RecQ4 are associated with distinct clinical disorders namely, Bloom's syndrome (BS), Werner's syndrome (WS), and Rothmund-Thomson syndrome (RTS) (13, 31, 68), respectively. BS is characterized by an elevated risk for a wide variety of cancers, immunodeficiency, slow growth, male sterility, and female subfertility (20). Furthermore, BS cells exhibit a hyperrecombination phenotype and greatly enhanced reciprocal exchanges between sister chromatids, as well as chromatid gaps and breaks. These phenotypes are consistent with recent evidence from Drosophila melanogaster that BLM might be required for synthesis-dependent strand annealing (2). WS and RTS are both associated with cancer predisposition, and WS is associated with premature aging (50, 57). The fact that patients suffering from these syndromes are prone to develop cancers shows the importance of the role played by the RecQ family of helicases in the maintenance of genome stability and the prevention of tumorigenesis.
The prototypic RecQ helicase is Escherichia coli RecQ. RecQ is a component of the RecF pathway, which is required for recombinational repair, that is essential for nearly all non-double-strand break recombination in E. coli (41, 42, 52). Because of its helicase activity that can unwind DNA at broken ends, the RecQ protein, together with the RecJ 5′-3′ nuclease, was proposed to act as functional analogs of the RecBCD helicase/nuclease in the RecBCD pathway during recombination repair (21).
In budding yeast, the unique RecQ protein Sgs1p plays a key role in regulating mitotic recombination and functions in the S-phase checkpoint response (15, 60). When expressed in yeast, the human BLM and WRN proteins are able to partially suppress the mitotic recombination defects of sgs1 mutants (66), suggesting that the function of Sgs1p and its human counterparts are evolutionarily conserved. The phenotype of sgs1 mutants includes a slight reduction in growth rate, a 10-fold increase in chromosome missegregation, an elevated mitotic recombination frequency, a high sensitivity to methyl methanesulfonate and hydroxyurea (HU) (15, 66), and premature aging as a consequence of an increased number of extrachromosomal ribosomal DNA circles in the cell (17, 51, 60, 61). Sgs1p acts redundantly with the budding yeast helicase Srs2p, which has been shown to inhibit loading of Rad51p onto single-stranded DNA (ssDNA) in vitro (33, 56). In addition, the sgs1 mutants display some meiotic defects such as delayed meiotic prophase and defective chromosome synapsis (16, 45). In vitro, Sgs1p interacts physically with Top3p (17), and Sgs1p-Top3p, together with Srs2p, suppresses crossovers during double-strand break repair (25). Top3p is a type I topoisomerase that catalyzes passage of two DNA double strands through each other for two consecutive break and rejoining cycles, one single strand at a time (58). In humans, an association between BLM and topoisomerase IIIα was observed in somatic and meiotic cells. The importance of this association was corroborated by their interaction in vitro (26, 64). In budding and fission yeast mutations in sgs1 act as suppressors of the slow-growth phenotype of top3 (17, 34). It has been proposed that Top3p acts on substrates created by Sgs1p. Similarly to Sgs1p, Top3p is involved in regulating the levels of mitotic recombination (17).
To better understand the in vivo function of the BS gene as well as its genetic interactions with top-3 and rad-51 in a multicellular system, we undertook a comprehensive analysis of the mitotic and meiotic roles of the Caenorhabditis elegans ortholog of the human BS RecQ-like helicase. C. elegans provides several advantages to study the function of the BLM ortholog. It is a multicellular animal that circumvents intrinsic complications of mammalian systems, including phenotypes associated with gene disruptions of recombination genes such as early embryonic lethality or programmed cell death at early meiotic stages, both of which confound a detailed cytological analysis in the respective mutants (22, 35, 36, 55, 65, 67). Several features of the adult C. elegans germ line make it a powerful system for the cytological and genetic dissection of meiotic recombination and DNA damage responses. Within the C. elegans gonad cells are organized in a highly polarized way. The distal end of the ovotestes contains a mitotic stem cell compartment that is followed by cells in different stages of meiotic prophase. Within a dissected gonad both mitotic cells, as well as distinct stages of meiotic chromosome pairing, can be observed. In response to DNA damage, checkpoint pathways cause cell cycle arrest of mitotic germ cells and apoptotic death of meiotic pachytene cells (3, 19, 24, 49).
The C. elegans genome encodes four RecQ-like helicases. Here we show that HIM-6 (for high incidence of males) corresponds to the worm ortholog of the human BS protein. Homozygous him-6 loss-of-function mutants are radiation sensitive and are partially defective in S-phase checkpoint function and ionizing radiation-induced apoptotic checkpoint response. him-6 mutants exhibit a decrease in the levels of meiotic crossover recombination and an altered pattern of RAD-51 recombination foci, suggesting defects in the progression of meiotic crossover recombination. him-6 and top-3 are needed to maintain genome stability in mitotic germ cells and, in contrast to the situation in yeast, mutations in both him-6 and top-3 lead to mitotic catastrophy during division of the germ cells. Mitotic catastrophy in top-3(RNAi); him-6 worms depends on rad-51. In addition, we show that topoisomerase IIIα by itself acts during meiotic recombination at a step after the initiation of meiotic recombination.
MATERIALS AND METHODS
Nematode strains and culture conditions.
All nematodes were grown at 20°C under standard conditions (9). The mutations used in the present study were obtained from the Caenorhabditis Genetic Center (University of Minnesota, St. Paul, Minn.). The spo-11(ok79) strains were kindly provided by Anne Villeneuve (Stanford University, Stanford, Calif.): LG I, dpy-5(e61), unc-101(m1), and unc-54(e190); LG III, glp-1(q224ts); LG IV, him-6(e1423), him-6(e1104), spo-11(ok79), and rad-51(lg08701); and LG X, unc-1(e719), dpy-3(e27), lon-2(e678), dpy-7(e88), and unc-3(e151).
Recombination analysis.
Recombination frequencies in the hermaphrodite were measured by scoring the number of recombinant progeny of a cis-heterozygote as described previously (47). The recombination frequency p between two genetic markers was calculated as p = 1 − (1 − 2R)1/2, where R is the number of visible recombinant individuals divided by the total progeny number (9). Since the double homozygote class was not scored because of its reduced viability, the total progeny number was estimated by 4/3 (the number of wild types plus one recombinant class). In some intervals, recovery of only one recombinant class was possible and in these instances, R equals 2X (one recombinant class) divided by the total progeny number. The 95% confidence intervals were estimated by using previously published statistics (11).
Aligning the physical and genetic maps for the him-6 region.
The him-6 gene has been mapped to linkage group IV (23) between unc-22 and unc-31. To map him-6 more precisely, we used deficiencies in the region to do complementation tests. him-6 failed to complement sDf62 but complemented sDf61 (10). This placed him-6 between let-93 and let-99, to the right of unc-22.
Molecular biology.
General molecular manipulations followed standard protocols (48). The sequence of the him-6 genomic region was established by the C. elegans Sequencing Consortium (62). A partial cDNA clone was obtained from Yuji Kohara (National Institute of Genetics, Mishima, Japan). The 5′ end of the cDNA was isolated by reverse transcription-PCR by using the Perkin-Elmer Cetus kit and the conditions described by the manufacturer. The primers used for amplification were SL1 (5′-GGTTTAATTACCCAAGTTTGAG-3′) and RQ-3 (5′-GCTGGCAATTGGTAGCAC-3′). Molecular changes in the him-6 alleles were identified by sequencing PCR-amplified products. All sequence analyses were performed with the GCG sequence analysis software package (Genetic Computer Group, 1991).
Irradiation and HU treatment.
To test for radiation sensitivity, embryonic survival was scored as the percentage of surviving embryos laid after irradiation of mothers with the indicated doses of gamma irradiation. To assay for radiation-induced germ cell apoptosis, worms were irradiated 24 h after the L4 stage with the indicated doses of gamma irradiation, and apoptotic cell corpses were determined 2, 4, and 6 h after irradiation by using Nomarski optics (19). HU-induced cell cycle arrest was determined as described previously (37) (3). Worms at the L4 stage were plated on nematode growth medium plates containing the indicated concentration of HU. After 14 h, cell cycle arrest was determined by scoring for the number of mitotic germ cell nuclei in a volume of 54,000 μm3.
Antibody staining.
For HIM-3 antibody staining of meiotic germ cells, adult hermaphrodites were placed in 0.25 mM levamisole on a charged slide (Menzel Gläser) and cut with a 23-gauge needle below the pharyngeal bulb to expel the gonad arm lying beneath it. Formaldehyde was added onto the slide to a final concentration of 1%. The slide was placed in a humid chamber for 5 min. A coverslip was set on top of the specimens, and the slides were incubated for 10 min on dry ice. After the coverslip was removed, the slides were immersed in 95% ethanol for 1 h. Specimens were washed in phosphate-buffered saline with 0.1% Tween 20 (PBST). Primary antibody incubation was done in PBST at 4°C overnight. After three washes with PBST, specimens were incubated for 1 h at room temperature with fluorescein isothiocyanate-conjugated AffiniPure goat anti-rabbit immunoglobulin G (1:200; Jackson Laboratories), followed by three washes. Stained gonads were then mounted with 2 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml in Vectashield mounting medium (Vector). Specimens were observed by using a Leica Epifluorescence microscope, and images were obtained with a Hamamatsu chilled charge-coupled device camera (C5810). DAPI and immunofluorescence images were processed, merged, and differentiated with false color by using Adobe Photoshop (version 4.0). RAD-51 immunostaining and subsequent immunoflorescence images were obtained as described previously (4). The statistics were done by using the StatView software 4.51 (Abacus Concepts, Inc.), and standard errors of the mean were calculated with 95% confidence intervals.
RNAi.
RNA interference (RNAi) experiments were carried out as described previously (14). Antisense and sense RNA were produced with an in vitro transcription kit (Promega) from a topoisomerase IIIα cDNA clone, which was constructed by reverse transcription-PCR with the primers 5′-GAAAAGAGCCTTATTTGTGGCCG-3′ and 5′-CCCATTTAAAGAAATTACATTTTTCAG-3′and then cloned into a pGEM-T-Easy vector (Promega).
DAPI staining of the gonad.
Adult hermaphrodites were placed in 0.25 mM levamisole on a charged slides (Menzel Gläser) and cut with a 23-gauge needle below the pharyngeal bulb, and the gonads were released. Samples were then fixed in 1% formaldehyde and washed with PBST (PBS with 0.1% Tween 20). Fixed gonads were then mounted with 2 μg of DAPI/ml in Vectashield mounting medium (Vector).
RESULTS
him-6 encodes the C. elegans homolog of the human BS protein.
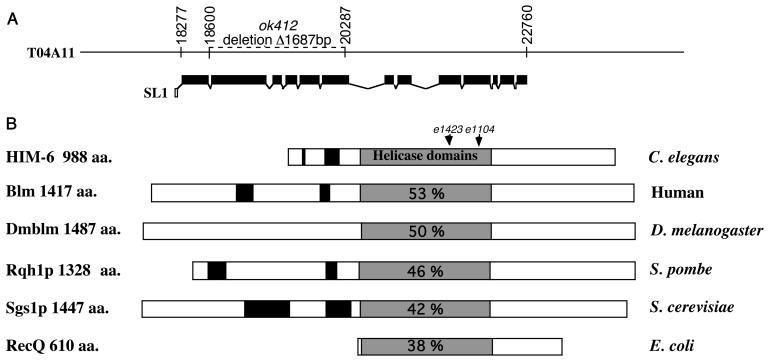
The two alleles of him-6, e1423 and e1104, cause phenotypes including a high percentage of males among the viable progeny (10 and 4.2%, respectively) and reduced viability (47 and 83.4% viable embryos, respectively) (23; the present study). A high incidence of XO males among the self-fertilization progeny of XX hermaphrodites (thus referred to as the Him phenotype) and an increased embryonic lethality can indicate a defect in meiotic chromosome segregation. him-6 maps close to the genomic sequence (T04A11.6) encoding a RecQ-type of DNA helicase. Since RecQ helicases have been implicated in genome stability, we tested whether or not T04A11.6 corresponds to the him-6 locus. Sequence analysis revealed that each of the two alleles e1423 and e1104 had a point mutation within the sequence of T04A11.6 (Fig. 1A). The mutation associated with the strong allele, e1423, is a T-to-A transversion resulting in a premature termination of translation at codon 479, halfway through the helicase domain, whereas the weaker allele, e1104, contains a G-to-A transition causing G-to-Q substitution at codon 561, a highly conserved position in the helicase domain. Furthermore, a 1,687-bp deletion in the him-6 locus (ok412), recovered by the International C. elegans Gene Knockout Consortium (Fig. 1A), removes five exons encoding the entire helicase domain and causes reduced viability and a high incidence of males (54% viable embryos and 11.7% males), comparable to the phenotype of the strong allele, e1423 (47% viable embryos and 10% males). Altogether, these data confirm that T04A11.6 corresponds to him-6. Furthermore, they suggest that the severe alleles e1423 and ok412 are null alleles and that the amino acid substitution in the e1104 mutant leads to a partial loss of function of the him-6 gene. Using RACE (5′ rapid amplification of cDNA ends) and expressed sequence tag sequencing (32), we characterized a 3.1-kb him-6 cDNA trans-spliced to the SL1 splice leader (7) (Fig. 1A). A basic local alignment search tool (BLAST) search with HIM-6 (GenBank accession number AY095296) revealed that its helicase domain is most similar to that of the human and D. melanogaster BS protein, members of the BLM subgroup of RecQ helicases (53 and 51% of identical amino acids, respectively) and is also closely related to other RecQ-like helicases, including the S. cerevisiae Sgs1p, the Schizosaccharomyces pombe Rqh1p and the E. coli RecQ proteins (Fig. 1B). These data were confirmed by the reverse BLAST searches with either human or D. melanogaster BLM protein showing that they are most closely related to him-6. Outside the helicase region, HIM-6 shows little sequence similarity with the BS protein except for conserved features, such as an enrichment in charged and polar amino acids, especially serines, and patches of acidic residues (Fig. 1B) present in the N terminus. Furthermore, domains with low levels of amino acid conservation are found at the C terminus (39), including a putative nuclear localization signal (amino acids [aa] 939 to 954) (27). In summary, the data indicate that him-6 is the C. elegans ortholog of the BS gene.
FIG. 1.
him-6 encodes a RecQ-like helicase. (A) him-6 gene structure. The mutation him-6(ok412) is a 1,687-bp deletion of five exons. The numbers indicate the nucleotide positions in the cosmid T04A11. (B) Schematic representation of RecQ-like DNA helicase; the name of the gene product and its length in amino acids is shown on the left, and the name of the organism is shown on the right. Helicase domains are shown as gray boxes; acidic domains are shown as black boxes. The proteins were arranged by aligning the helicase domains. The percent values indicate the level of conservation throughout the helicase domain between HIM-6 and its counterparts in other organisms. Arrowheads indicate residues mutated in him-6 alleles: K479Stop in e1423 and G561Q in e1104. The HIM-6 GenBank accession number is AY095296.
him-6 is required for normal levels of recombination during meiosis.
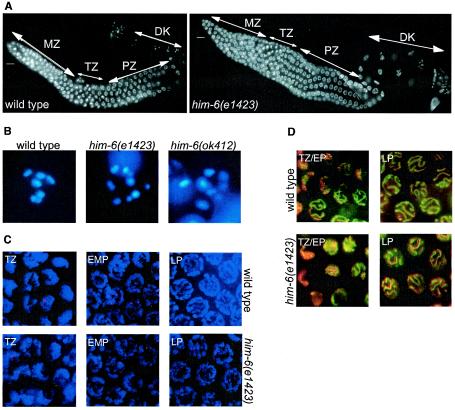
A Him phenotype coupled to reduced viability is characteristic for mutations causing an increased level of chromosome nondisjunction (23). We have examined whether chromosome nondisjunction was due to a decrease in the number of chiasmata, which are the consequences of meiotic recombination. In wild-type worms, germ cells proliferate in the distal portion of the gonad. After passing through a transition zone marking the onset of meiotic prophase, they progress through an extended pachytene region and enter diplotene-diakinesis, staying in diakinesis until oocyte maturation is induced (Fig. 2A). At the final stage of meiotic prophase, six DAPI-stained bodies, corresponding to desynapsed homologous chromosomes physically linked by chiasmata are visible (Fig. 2B). In him-6 mutants the nuclei often contained more than six stained bodies (Fig. 2B). The average numbers of DAPI-stained bodies were 6.01 ± 0.01 (n = 78) in the wild type, 7.36 ± 0.14 (n = 50) in e1423, 7.22 ± 0.12 (n = 60) in ok412, and 6.35 ± 0.09 (n = 45) in e1104. This result indicated that the nuclei contained a mixture of bivalents and achiasmatic chromosomes (univalent). It is unlikely that the smaller stained bodies represented chromosome fragments, since the total number of large stained bodies and small stained bodies always made up for the six pairs of chromosomes. Therefore, we propose that the high percentage of male and nonviable progeny in the him-6 mutant was caused by a reduced number of chiasmata, which are necessary for proper chromosome segregation.
FIG. 2.
Meiotic phenotypes of him-6. (A) Wild-type gonad (left panel) and him-6(e1423) gonad (right panel) stained with DAPI, which show a normal progression of the germ cells through meiotic prophase. (B) Wild-type oocyte at diakinesis stained with DAPI. Six stained bodies can be observed corresponding to the six sets of homologous chromosomes attached by chiasmata, him-6(e1423) oocyte and him-6(ok412) oocyte: more than six structures are present, which correspond to a mix of bivalents and univalents. (C) Meiotic prophase nuclei stained with anti-RAD-51 antibody (red) and DAPI (blue). The RAD-51 immunostaining reveals similar foci pattern in wild-type and him-6(e1423) transition zone and early-mid-pachytene nuclei. Foci are absent from late pachytene nuclei in wild type but persist in the him-6(e1423) mutant. (D) Meiotic prophase nuclei stained with anti-HIM-3 antibody (green) and DAPI (red). The staining revealed no pairing defects in the transition zone and pachytene nuclei of the him-6 mutant. MZ, mitotic zone; TZ, transition zone; EMP, early-mid pachytene; LP, late pachytene; P, pachytene; DK, diakinesis. Scale bars, 10 μm.
The reduced number of chiasmata in him-6 mutants suggested a defect in the meiotic recombination process. Indeed, a general decrease in the level of meiotic recombination was suggested by previous experiments measuring recombination on chromosome I by using the severe allele e1423 (70). To further examine the levels of recombination, we analyzed two intervals on chromosomes I and X by using the e1423 and e1104 alleles (Table 1.) In animals containing the weak allele e1104, crossing over was reduced in only one of the four intervals tested. However, for the strong allele e1423, a 25 to 50% decrease in crossing-over frequency was observed in all intervals tested. The reduction in crossing-over frequency was consistent with the number of univalents observed in the two alleles and with the idea that the loss of function of him-6 causes decreases in the level of meiotic crossing-over.
TABLE 1.
Meiotic recombination in him-6 mutant alleles
| Chromosome and genotype | No. of wild types | No. of recombinant(s)b | Map distance (cM) (CI)a |
|---|---|---|---|
| Chromosome I | |||
| unc-101 unc-54/+ + | 2,321 | 193 Unc | 12.5 (10.5-14.1) |
| unc-101 unc-54/+ +; him-6(e1104)/him-6(e1104) | 1,499 | 152 Unc | 14.9 (12.5-17.2) |
| unc-101 unc-54/+ +; him-6(e1423)/him-6(e1423) | ND | ||
| dpy-5 unc-101/+ + | 1,914 | 130 Unc, 138 Dpy | 10.3 (9.1-11.6) |
| dpy-5 unc-101/+ +; him-6(e1104)/him-6(e1104) | 1,356 | 118 Unc, 94 Dpy | 11.5 (9.9-13.1) |
| dpy-5 unc-101/+ +; him-6(e1423)/him-6(e1423) | 1,746 | 80 Unc, 104 Dpy | 7.8 (6.6-8.9) |
| Chromosome X | |||
| dpy-7 unc-3/+ + | 2,041 | 276 Unc, 246 Dpy | 18.7 (18.0-19.3) |
| dpy-7 unc-3/+ +; him-6(e1104)/him-6(e1104) | 1,533 | 199 Unc, 184 Dpy | 18.3 (15.3-21.2) |
| dpy-7 unc-3/+ +; him-6(e1423)/him-6(e1423) | 1,744 | 130 Unc, 129 Dpy | 10.9 (9.6-12.3) |
| unc-1 dpy-7/+ + | 1,915 | 280 Unc | 21.4 (18.9-23.9) |
| unc-1 dpy-7/+ +; him-6(e1104)/him-6(e1104) | 1,257 | 118 Unc | 13.8 (11.4-16.3) |
| unc-1 dpy-7/+ +; him-6(e1423)/him-6(e1423) | 1,103 | 79 Unc | 10.58 (8.4-13.0) |
CI, confidence interval; cM, centimorgans; ND, not determined.
Unc, uncoordinated; Dpy, dumpy.
In a second approach, dissected gonads were stained with an antibody against RAD-51. RAD-51 is a member of the RecA-strand exchange protein family and catalyzes the invasion of DNA single-strand overhangs into a recipient double-stranded DNA (5, 44, 46, 54). RAD-51 foci are indicative of ongoing meiotic recombination in C. elegans (4). They appear first in the transition zone nuclei and reach their maximal abundance in early pachytene nuclei in wild-type and him-6 animals carrying the mutations e1423 and ok412 (Fig. 2C). In him-6 animals, however, unlike in wild-type worms, RAD-51 foci often persist through the late pachytene and accumulate prominently in him-6 mutants containing two or more univalents in diakinesis (Fig. 2C). To test whether defects in chromosome pairing and SC formation may be responsible for the persisting RAD-51 foci in him-6 mutants, we stained the germ line nuclei with an antibody against HIM-3, a meiotic chromosome core component (69). We observed no obvious abnormalities in the transition zone or pachytene nuclei of the him-6 mutant (Fig. 2D). To distinguish whether the persistent RAD-51 foci observed in late pachytene nuclei of him-6 animals are dependent on the SPO-11 nuclease or whether they were inherited as double-strand breaks generated in premeiotic S phase, we analyzed the distribution of RAD-51 foci in spo-11(ok79) him-6(e1423) worms. In wild-type animals, RAD-51 recruitment depends on meiotic double-strand breaks generated by SPO-11, and virtually no RAD-51 foci are detected during meiosis in spo-11 mutant animals (4). Likewise, in spo-11(ok79) him-6(e1423) double mutants, no RAD-51 staining, even in late-pachytene nuclei, was observed (data not shown), suggesting that RAD-51 foci in him-6 worms are indeed spo-11 dependent and that meiotic double-strand breaks occur in him-6 mutants. Altogether, our observations suggest that him-6 is necessary for normal levels of crossing-over recombination and functions during a late step in this process.
him-6 has defects in response to DNA damage.
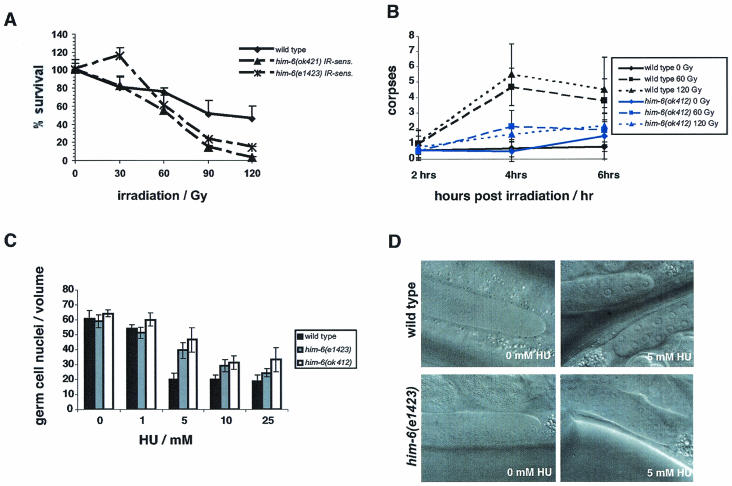
The budding yeast Sgs1p was shown to function during the S-phase checkpoint (15, 40), whereas in fission yeast the Sgs1p homolog, Rqh1p, has no S phase checkpoint function (53). The role of BLM in this process in mammalian cells is still unclear (reviewed in reference 6). To determine whether HIM-6 is needed in response to DNA damage, we exposed L4 larvae to increasing levels of gamma irradiation and scored the survival rate of the progeny as described previously (19). We found that him-6 mutants show an enhanced sensitivity to irradiation (Fig. 3A). In addition, we tested whether germ cell apoptosis (of meiotic pachytene cells) and mitotic germ cell cycle arrest are induced in response to genotoxic stress in him-6 mutants (19). him-6(e1423) and him-6(ok412) animals showed attenuated levels of programmed cell death within 6 h after irradiation (Fig. 3B and data not shown). However, programmed cell death reached almost wild-type levels 36 h after irradiation (data not shown). We did not observe any defects in cell cycle arrest in response to radiation by mitotic germ cells (data not shown). We tested whether him-6 worms were defective in the S-phase checkpoint by growing them on plates containing HU, a drug that leads to the depletion of deoxynucleoside triphosphate pools (12). Cell cycle arrest was measured by counting the average cell number in a defined volume of the germ line, which is indicative of cell cycle progression (wild-type cells transiently stop dividing in response to HU, but S-phase checkpoint-defective cells continue to proliferate) (3, 37). Both him-6(ok412) and him-6(e1423) mutants showed a partially defective cell cycle arrest in response to HU treatment as revealed by the elevated number of mitotic germ cells and their smaller size upon HU (Fig. 3C and D).
FIG. 3.
him-6 mutants are defective in responding to DNA damage. (A) him-6 germ cells are more sensitive to gamma irradiation. To compare the irradiation sensitivity (IR-sens) curve of the mutant, the survival rates of N2, him-6(e1423), and him-6(ok412) are set to 100 at 0 Gy, although the survival rate of the him-6(e1423) and him-6(ok412) progenies is only 21 and 36%, respectively. (B) him-6(e1423) mutant show decreased germ cell apoptosis. Late-stage L4 hermaphrodites were exposed to 0, 60, and 120 Gy of gamma irradiation and apoptosis was scored after 2, 4 and 6 h later as described previously (19). The y axis indicates the number of dead cells (cell corpses) per gonad arm. (C) him-6 germ lines are sensitive to HU. him-6 and wild-type worms were grown on plates containing 0, 1, 5, 10, or 25 mM HU. Cell cycle arrest was measured by counting the average number of cells in a defined volume of the germ line. The him-6 mutants show a higher number of germ cells upon HU treatment compared to the wild type. (D) Nomarski pictures of wild-type and him-6(e1423) mitotic zones exposed to 0 and 5 mM HU. him-6(e1423) has more mitotic nuclei when exposed to 5 mM HU due to a defect in cell cycle arrest.
To test whether him-6 is needed to maintain genome stability in normally proliferating mitotic germ cells, we scored for the number of RAD-51 foci in untreated animals. The average numbers of RAD-51 foci were 2.3 ± 0.3 in the wild type, 6.6 ± 1.0 in him-6(e1423), and 7.6 ± 0.9 in him-6(ok412) per 100 mitotic germ cells. Thus, RAD-51 is already recruited during the proliferation of the germ line in him-6 mutants, suggesting elevated levels of double-strand breaks or failure to process double-strand breaks. RAD-51 foci are likely to mark sites of ssDNA that have not undergone invasion or sites where ssDNA invasion has occurred but normal disassembly of complexes has not occurred. Taken together, these results indicate that him-6 mutants are partially defective in activating DNA damage checkpoint responses and that him-6 is needed for the efficient maintenance of genome stability.
The combined depletion of HIM-6 and TOP-3 leads to mitotic catastrophy, which is suppressed by the loss of function of rad-51.
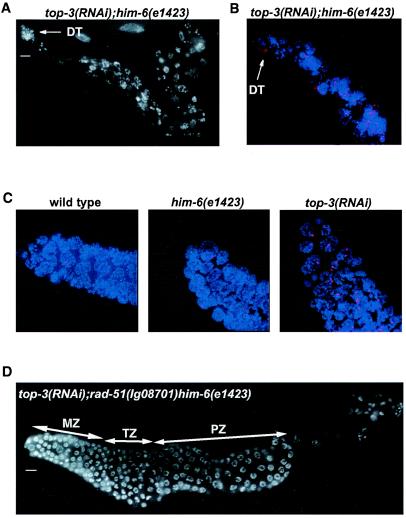
Previously HIM-6 was shown to interact genetically and physically with topoisomerase IIIα (Fig. 4). We have further analyzed this genetic interaction (30). Kim et al. (30) demonstrated that top-3(RNAi); him-6(e1104) germ cells showed severe chromosome abnormalities and were arrested during mitosis. We observed a similar phenotype for top-3(RNAi); him-6(e1423) animals (Fig. 4A). In top-3(RNAi); him-6(e1423), we detected a dramatic increase of RAD-51 foci (136 ± 23.5/100 nuclei), the exact number of which was hard to determine due to nuclear fragmentation (Fig. 4B). The increased number of RAD-51 foci in the mitotic zone of him-6(e1423) (6.6 ± 1.0 foci/100 nuclei) and top-3(RNAi) (47.8 ± 6.7 foci/100 nuclei) worms compared to wild-type animals (2.3 ± 0.3 foci/100 nuclei) already suggested elevated levels of double-strand breaks in mitotic germ cells of the respective single mutants (Fig. 4C). We tested whether the severe chromosome abnormalities of top-3(RNAi); him-6(e1423) germ line nuclei depend on rad-51 by analyzing top-3(RNAi); rad-51(lg08701) him-6(e1423) germ lines. Mutation in rad-51 completely suppressed the top-3(RNAi); him-6(e1423) phenotype (Fig. 4D). top-3(RNAi); rad-51(lg08701) him-6(e1423) germ cell nuclei progress normally through meiotic prophase. The data indicate that the severe defects observed in the top-3(RNAi); him-6(e1423) worms depend on the RAD-51 function. Thus, we propose that RAD-51 might be involved in the generation of toxic recombination intermediates that form in the combined absence of HIM-6 and TOP-3 during the proliferation of mitotic germ cells.
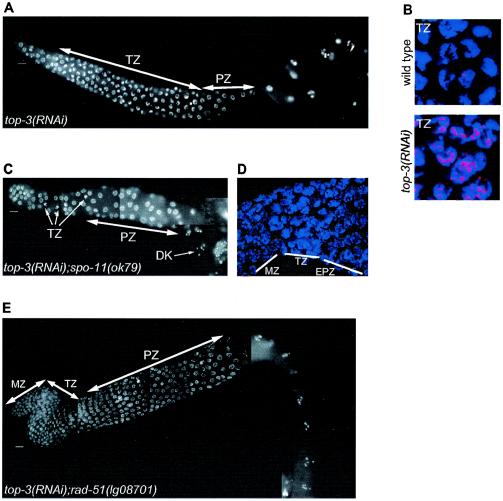
FIG. 4.
top-3(RNAi); him-6(e1423) worms show mitotic catastrophy and massive RAD-51 recruitment in the germ line. The defects are suppressed by mutation in rad-51. (A) top-3(RNAi); him-6(e1423) gonad stained with DAPI, which shows germ line nuclei with severe chromosomal abnormalities; (B) top-3(RNAi); him-6(e1423) gonad stained with DAPI (blue) and anti-RAD-51 antibody (red); (C) mitotic germ nuclei of wild type, him-6(e1423) and top-3(RNAi) worms stained with DAPI (blue) and anti-RAD-51 antibody (red), which revealed a higher number of RAD-51 foci compared to wild type; (D) top-3(RNAi); rad-51(lg08701)him-6(e1423) gonad stained with DAPI, which shows a normal proliferation of the germ line nuclei and a normal progression through meiotic prophase. Scale bars, 10 μm.
Topoisomerase IIIα also acts at a late stage of meiotic recombination.
In C. elegans TOP-3 is required for fertility. top-3(RNAi) worms had a variable phenotype ranging from normal to completely sterile worms (29). In gonads of top-3(RNAi) animals, the prophase nuclei persist in a transition and/or early pachytene zone-like morphology and only reach full pachytene morphology late during meiotic prophase (Fig. 5A) (29). We studied the RAD-51 staining pattern in nuclei of the transition zone in top-3(RNAi) worms and found an increased number of RAD-51 foci, which also appear larger (Fig. 5B). The increased number of RAD-51 foci in the transition zone indicated that DSBs are accumulating in top-3(RNAi) worms early in meiotic prophase. The data are consistent with the suggestion that top-3(RNAi) worms are defective in processing meiotic recombination intermediates that occur after the formation of RAD-51 bound filaments. This hypothesis is supported by the finding that nuclei in top-3(RNAi); spo-11(ok79) double mutants progress normally through meiotic prophase but in fewer numbers (Fig. 5C). Furthermore, the large amount of RAD-51 foci observed at the transition zone of top-3(RNAi) worms was absent from top-3(RNAi); spo-11(ok79) worms, indicating that the accumulation of double-strand breaks at the transition zone was dependent on spo-11 (Fig. 5D). rad-51 acts after spo-11 in meiotic recombination. In the absence of RAD-51 function top-3(RNAi); rad-51(lg08701) animals had a normal progression through meiotic prophase (Fig. 5E), suggesting that RAD-51 activity is responsible for the top-3 mutant-delayed progression through prophase and that TOP-3 acts after SPO-11 and RAD-51. The reduced number of germ line nuclei in top-3(RNAi); spo-11(ok79) worms may result from a further role for TOP-3 in the (mitotic) proliferation of the germ line (see above).
FIG. 5.
top-3(RNAi) worms show defects, which depend on meiotic recombination. (A) top-3(RNAi) gonad stained with DAPI, which revealed an extended transition zone. (B) Transition zone nuclei stained with an anti-RAD-51 antibody (red) and DAPI (blue). top-3(RNAi) nuclei show an increased number of RAD-51 foci compared to wild type. (C) top-3(RNAi); spo-11(ok79) gonad stained with DAPI, which shows a normal progression of the germ nuclei through meiotic prophase. Arrows point toward nuclei in the transition zone (TZ) and at diakinesis (DK). (D) RAD-51 foci are absent from top-3(RNAi); spo-11(ok79) nuclei in the transition zone and in early pachytene. (E) top-3(RNAi); rad-51(lg08701) gonad stained with DAPI, which shows a normal proliferation of the germ line nuclei and a normal progression through meiotic prophase. Scale bars, 10 μm.
DISCUSSION
In the present study, we show that him-6 encodes for the C. elegans homolog of the mammalian BS protein. him-6 plays a role in DNA damage checkpoint signaling in response to ionizing radiation and partially contributes to the S-phase checkpoint regulation in the germ line. We propose that during mitotic cell divisions him-6 and top-3 act in partial redundant pathways downstream of rad-51 to process lesions generated during normal mitotic germ cell divisions (Fig. 6). During meiotic recombination him-6 and top-3 act downstream of spo-11 and rad-51 (see below) (Fig. 6).
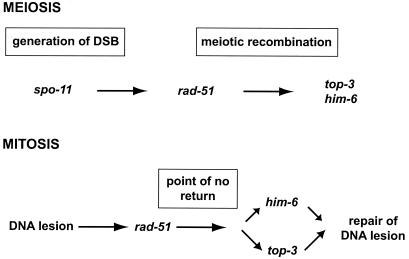
FIG. 6.
him-6 functions during meiosis and mitosis. During meiotic recombination him-6 functions downstream of spo-11 and rad-51. We cannot address the epistatic relationship between top-3 and him-6 during meiotic recombination as top-3; him-6 worms do not enter meiosis. During mitotic proliferation of the germ line, him-6 acts partially redundantly with top-3 on DNA lesions that have been processed by rad-51.
HIM-6 has checkpoint function in the germ line.
him-6 mutant worms show an enhanced sensitivity to ionizing radiation, indicating that it plays a role in the DNA damage response. In addition, we observed a partial checkpoint defect in responding to HU and to ionizing irradiation induced germ cell death in him-6 worms. A function of RecQ helicases in the DNA damage response pathway was already suggested in other organisms but was not observed universally. sgs1-deficient budding yeast cells are partially defective in arresting S-phase progression in response to HU (15, 66). Human BS−/− cells, on the other hand, respond normally to HU (1). S. pombe cells lacking the RecQ-like helicase Rqh1p also react normally to HU but are defective in the recovery from S-phase arrest when exposed to HU (53). It is likely that RecQ-dependent damage checkpoint responses are mediated by functionally redundant or overlapping pathways. We observed only partial checkpoint defects in him-6 C. elegans worms. Similarly, irradiation-induced apoptosis and G2/M cell cycle arrest is only partially compromised in human BS −/− cells (1, 59). It was recently shown that HIM-6 physically interacts with the C. elegans ATR homolog, whose inactivation by RNAi leads to defective checkpoint processes in response to ionizing radiation (8), as well as to HU (A. Gartner, unpublished observation). Therefore, it is conceivable that HIM-6 acts together with the ATM/ATR-like kinases to affect DNA damage checkpoint responses.
TOP-3 and HIM-6 act in partially redundant pathways to prevent mitotic catastrophe in the germ line.
top-3(RNAi); him-6(e1423) germ lines display some profound defects, including a reduced number of germ line nuclei. The chromatin of these nuclei appears to be disorganized and sometimes fragmented, and RAD-51 foci massively accumulate. This strong phenotype is suppressed by the absence of RAD-51. top-3(RNAi); rad-51(lg08701) him-6(e1423) germ line nuclei appear normal and progress normally through meiotic prophase. The results suggest that HIM-6 and TOP-3 act in partially redundant pathways and downstream of RAD-51 to process recombination intermediates that are resulting from spontaneous double-strand breaks in normally proliferating mitotic germ cells (Fig. 6). The presence of an increased number of RAD-51 foci in the mitotic zone of him-6 and top-3 single mutant worms is likely to mark sites of ssDNA that have not undergone invasion or sites where invasion has occurred but normal disassembly of recombination complexes has not occurred. The increased number of RAD-51 foci, as well as the increased radiation sensitivity of him-6 worms, could be either a result of a DNA repair defect or a partial defect during S phase. We consider it likely that in the absence of HIM-6 and TOP-3 recombination intermediates may be formed that cannot be processed and therefore result in massive genomic instability. This could lead to the defects observed in top-3(RNAi); him-6(e1423) germ line nuclei. Once double-strand breaks are generated and processed by RAD-51, cells might pass through a “point of no return” that precludes alternative repair pathways (Fig. 6). Accordingly, in the absence of RAD-51, toxic recombination intermediates cannot be formed and double-strand breaks that form in the absence of RAD-51 may be processed by alternative repair pathways such as, e.g., nonhomologous end joining.
The genetic interactions between him-6, top-3, and rad-51 differ from those observed with the corresponding mutations in S. cerevisiae and S. pombe. In yeast it was shown that Sgs1p, as well as other recombination proteins, suppresses the slow-growth phenotype and the retarded cell cycle progression of top3 mutants (17). It is assumed that Sgs1p generates DNA structures during replication or repair that need TOP-3 or other recombination proteins to be resolved. In contrast, C. elegans top-3(RNAi); him-6(e1423) mutants display synergistic defects. What might be the reason for the differences observed in the yeast and worm system? It is likely that him-6 and top-3 act in partially redundant pathways to prevent the illegitimate accumulation of toxic recombination intermediates that occur as a consequence of mistakes happening during normal S-phase progression. According to this model, inactivation of rad-51 function would bypass the combined defects of top-3; him-6 worms by preventing the accumulation of toxic recombination intermediates (Fig. 6). In yeast inactivation of rad51 can rescue srs2 sgs1 double mutants from cell death, indicating that S. cerevisiae rad51 might prevent srs2 sgs1 double mutants form accumulating recombination intermediates that cannot be processed and lead to a point of no return similarly to the situation in C. elegans him-6 top-3 worms (18).
him-6 is required for normal levels of meiotic recombination.
What might be the function of HIM-6 during meiotic recombination? HIM-6 could play a role at a late step of recombination. In him-6 mutants the rate of meiotic crossing-over is reduced and RAD-51 foci persist in late pachytene, whereas in wild-type nuclei of the same stage virtually no RAD-51 foci are observed. This phenotype is reminiscent to that of msh-5 mutants. Genetic and cytological evidence indicated that MSH-5 acts after the initiation of recombination (4, 28, 44). Like MSH-5, HIM-6 could act after the initiation of recombination and might be necessary for a late step of recombination (Fig. 6).
RecQ proteins play an important role during meiosis (63). BS is characterized by male sterility and female subfertility and BLM and RAD-51 colocalize in mouse spermatocytes during meiotic prophase (38). However, unlike in the him-6 mutant, the frequency of meiotic crossing-over seems not to be affected in blm knockout mice (36). In S. cerevisiae, the single RecQ homolog (Sgs1p) is not required for the initiation of double-strand breaks but seems to be needed for the processing of recombination intermediates during meiosis. The first meiotic division in sgs1 mutant is delayed, and this delay is alleviated in a spo11-deficient background (16). More recently, Sgs1p was shown to regulate chromosome synapsis and meiotic crossing-over (45). However, Sgs1p seems to function differently from HIM-6, since crossing-over frequency is increased in an sgs1 loss-of-function mutant.
TOP-3 is required for meiotic recombination.
While studying a possible interaction between HIM-6 and TOP-3, we observed that a top-3 single mutant displayed an interesting phenotype distinct from those resulting from the lack of recombination machinery components such as RAD-51 and SPO-11. Absence of TOP-3 leads to an accumulation of nuclei at the so-called transition stage. This defect is dependent on meiotic recombination, since it was suppressed by mutations in spo-11 and rad-51. Furthermore, RAD-51 foci were very abundant in transition zone nuclei of top-3 worms, indicating that an increased number of recombination events were initiated but not resolved. These results are consistent with the notion that TOP-3 might act downstream of RAD-51 during meiotic recombination (Fig. 6). Due to excessive mitotic defects top-3; him-6 worms do not enter meiosis, and we were unable to study the effect of this double mutant on meiotic recombination. The accumulation of recombination intermediates in top-3 worms could cause a severe delay or even an arrest in the progression through the transition zone. It remains to be seen whether the accumulation of transition zone nuclei in top-3 worms depends on the DNA damage checkpoint. These defects and the abundant RAD-51 foci in the transition zone are alleviated by mutation in spo-11, indicating that the top-3 mutant phenotype depends on the generation of double-strand breaks. Indeed, we also observed a similar defect in irradiated rad-51 and mre-11 worms in which massive levels of double-strand breaks are generated but cannot be repaired (A. Gartner and A. Alpi, unpublished observations). Our finding thus generalizes observations made in yeast, suggesting that top-3 might be required during meiosis in a stage acting after the initiation of meiotic double-strand breaks (16).
C. elegans and BS.
BS is mainly associated with somatic phenotypes, such as short stature, skin disorders, and predisposition to cancer, although the germ line is also affected (20). In the present study we have demonstrated a role for him-6 during both mitotic division and meiosis. The him-6 gene is predominantly expressed in the germ line of C. elegans (43), and its loss-of-function has no apparent effect in the soma. However, during C. elegans somatic development cells only go through a very limited number of cell divisions before becoming terminally differentiated. This is in contrast to the massive proliferation needed in mammalian systems to expand rare stem cells into terminally differentiated tissues. Thus, it is not surprising that mechanisms to maintain genome stability are more active in the C. elegans germ line, which is continuously proliferating. This particular developmental setting in C. elegans allows analysis of the combined effect of multiple DNA repair-associated mutations. We discovered new synthetic interactions between C. elegans repair proteins that were not anticipated from previous studies in yeast. Synthetic phenotypes, such as those we uncovered for top-3; him-6 double and top-3; rad-51 him-6 triple mutants, might indeed in the long run be useful for cancer therapy. For example, in instances where single genes such as blm are lost due to genome instability associated with cancer progression, drugs against TOP-3 might selectively target cancer cells.
Acknowledgments
We thank members of the Müller and Gartner laboratories for discussions. We are grateful to David Lydall, Karel Riha, Simon Boulton, and Thomas Caspari for critical reading of the manuscript and to Yolande Molleyres, Björn Schumacher, and Laurence Bulliard for technical support. Some strains were supplied by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
This research was supported by the Swiss National Foundation (grants 31-56953.99 and 3100-040776), by the Sandoz Stiftung für Förderung der Medizinisch-Biologischen Wissenschaften, by the German DFG grant GA703/2, by the Max Planck Society (Erich Nigg), and by an NSERC grant to A.R.
REFERENCES
- 1.Ababou, M., V. Dumaire, Y. Lécluse, and M. Amor-Guéret. 2002. Bloom's syndrome protein response to ultraviolet-C radiation and hydroxyurea-mediated DNA synthesis inhibition. Oncogene 21:2079-2088. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. D., M. McVey, and J. J. Sekelsky. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299:265-267. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, S., A. Alpi, M. O. Hengartner, and A. Gartner. 2001. Caenorhabditis elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11:1934-1944. [DOI] [PubMed] [Google Scholar]
- 4.Alpi, A., P. Pawel, A. Gartner, and J. Loidl. 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112:6-16. [DOI] [PubMed]
- 5.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of Escherichia coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 6.Bjergbaek, L., J. A. Cobb, and S. M. Gasser. 2002. RecQ helicases and genome stability: lessons from model organisms and human disease. Swiss Med. Wkly. 132:433-442. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. [DOI] [PubMed] [Google Scholar]
- 8.Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson, D. E. Hill, and M. Vidal. 2002. Combined functional genomic maps of the Caenorhabditis elegans DNA damage response. Science 295:127-131. [DOI] [PubMed] [Google Scholar]
- 9.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, D. V., and D. L. Baillie. 1992. Genetic analysis and complementation by germ-line transformation of lethal mutations in the unc-22 IV region of Caenorhabditis elegans. Mol. Gen. Genet. 232:97-105. [DOI] [PubMed] [Google Scholar]
- 11.Crow, E. L., and R. S. Gardner. 1959. Confidence intervals for the expectation of a Poisson variable. Biometrika 46:441-453. [Google Scholar]
- 12.Dasika, G. K., S. C. Lin, S. Zhao, P. Sung, A. Tomkinson, and E. Y. Lee. 1999. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 18:7883-7899. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 14.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 15.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 16.Gangloff, S., B. de Massy, L. Arthur, R. Rothstein, and F. Fabre. 1999. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 18:1701-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genetics 25:192-194. [DOI] [PubMed] [Google Scholar]
- 19.Gartner, A., S. Milstein, S. Ahmed, J. Hodgkin, and M. O. Hengartner. 2000. A conserved checkpoint pathway mediates DNA damaged-induced apoptosis and cell cycle arrest in Caenorhabditis elegans. Mol. Cell 5:435-443. [DOI] [PubMed] [Google Scholar]
- 20.German, J. 1993. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine 72:393-406. [PubMed] [Google Scholar]
- 21.Harmon, F. G., and S. C. Kowalczykowski. 1998. RecQ helicase, in concert with recA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 12:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirao, A., Y.-Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 23.Hodgkin, J., R. Horvitz, and S. Brenner. 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91:67-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann, E. R., S. Milstein, S. J. Boulton, Y. Mianjia, J. J. Hofmann, L. Stergiou, A. Gartner, M. Vidal, and M. O. Hengartner. 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12:1908-1918. [DOI] [PubMed] [Google Scholar]
- 25.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, F. B., D. B. Lombard, N. F. Neff, M.-A. Mastrangelo, W. Dewolf, N. A. Ellis, R. A. Marciniak, Y. Yin, R. Jaenisch, and L. Guarente. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic cells and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 27.Kaneko, H., K. O. Orii, E. Matsui, N. Shimozawa, T. Fukao, T. Matsumoto, A. Shimamoto, Y. Furuichi, S. Hayakawa, K. Kasahara, and N. Kondo. 1997. BLM (the causative gene of Bloom syndrome) protein translocation into the nucleus by a nuclear localization signal. Biochem. Biophys. Res. Commun. 240:348-353. [DOI] [PubMed] [Google Scholar]
- 28.Kelly, K. O., A. Dernburg, F., G. M. Stanfield, and A. Villeneuve, M. 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156:617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, Y. C., J. Lee, and H. S. Koo. 2000. Functional characterization of. Caenorhabditis elegans DNA topoisomerase IIIα. Nucleic Acids Res. 28:2012-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y. C., M. H. Lee, S. S. Ryu, J. H. Kim, and H. S. Koo. 2002. Coaction of DNA topoisomerase IIIα and a RecQ homologue during the germ-line mitosis in Caenorhabditis elegans. Genes Cells 1:19-27. [DOI] [PubMed] [Google Scholar]
- 31.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 32.Kohara, Y. 1996. Large scale analysis of Caenorhabditis elegans cDNA. Tanpakushitsu Kakusan Koso 41:715-720. [PubMed] [Google Scholar]
- 33.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 34.Laursen, L. V., E. Ampatzidou, A. H. Andersen, and J. M. Murray. 2003. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 10:3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim, D.-S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 37.MacQueen, A. J., and A. M. Villeneuve. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moens, P. B., R. Freire, M. Tarsounas, B. Spyropoulos, and S. P. Jackson. 2000. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113:663-672. [DOI] [PubMed] [Google Scholar]
- 39.Morozov, V., A. R. Mushegian, E. V. Koonin, and P. Bork. 1997. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 22:417-418. [DOI] [PubMed] [Google Scholar]
- 40.Myung, K., and R. D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakayama, H., K. Nakayama, R. Nakayama, N. Irino, Y. Nakayama, and P. C. Hanawalt. 1984. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K-12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 195:474-480. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama, K., N. Irino, and H. Nakayama. 1985. The recQ gene of Escherichia coli K-12: molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. 200:266-271. [DOI] [PubMed] [Google Scholar]
- 43.Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren, R. Begley, S. J. Jones, E. B. Davis, S. Scherer, S. Ward, and S. K. Kim. 2000. A global profile of germline gene expression in Caenorhabditis elegans. Mol. Cell 6:605-616. [DOI] [PubMed] [Google Scholar]
- 44.Rinaldo, C., P. Bazzicalupo, S. Ederle, M. Hilliard, and A. La Volpe. 2002. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rockmill, B., J. C. Fung, S. S. Branda, and G. S. Roeder. 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13:1954-1962. [DOI] [PubMed] [Google Scholar]
- 46.Roeder, G. S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600-2621. [DOI] [PubMed] [Google Scholar]
- 47.Rose, A. M., and D. L. Baillie. 1979. Effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92:409-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 49.Schumacher, B., K. Hofmann, S. J. Boulton, and A. Gartner. 2001. The Caenorhabditis elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11:1722-1727. [DOI] [PubMed] [Google Scholar]
- 50.Shen, J.-L., and L. A. Loeb. 2000. The Werner syndrome gene. Trends Genet. 16:213-220. [DOI] [PubMed] [Google Scholar]
- 51.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 52.Smith, G. R. 1991. Conjugational recombination in Escherichia coli: myths and mechanisms. Cell 64:19-27. [DOI] [PubMed] [Google Scholar]
- 53.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takanami, T., S. Sato, T. Ishihara, I. Katsura, H. Takahashi, and A. Higashitani. 1998. Characterization of a Caenorhabditis elegans recA-like gene, Ce-rdh-1, involved in meiotic recombination. DNA Res. 5:373-377. [DOI] [PubMed] [Google Scholar]
- 55.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and T. Morita. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 57.Vennos, E. M., and W. D. James. 1995. Rothmund-Thomson syndrome. Dermat. Clin. 13:143-150. [PubMed] [Google Scholar]
- 58.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 59.Wang, X. W., A. Tseng, N. A. Ellis, E. A. Spillare, S. P. Linke, A. I. Robles, H. Seker, Q. Yang, P. Hu, S. Beresten, N. A. Bemmels, S. Garfield, and C. C. Harris. 2001. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 276:32948-32955. [DOI] [PubMed] [Google Scholar]
- 60.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of Escherichia coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, R., R. Ainscough, K. Anderson, C. Baynes, M. Berks, J. Bonfield, J. Burton, M. Connell, T. Copsey, J. Cooper, et al. 1994. 2.2 Mb of contiguous nucleotide sequence from chromosome III of Caenorhabditis elegans. Nature 368:32-38. [DOI] [PubMed] [Google Scholar]
- 63.Wu, L., and I. D. Hickson. 2001. RecQ helicases and topoisomerases: components of a conserved complex for the regulation of genetic recombination. Cell. Mol. Life Sci. 58:894-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, L., L. D. Sally, S. N. Phillip, H. Goulaouic, J.-F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 65.Xiao, Y., and D. T. Weaver. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25:2985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyper-recombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakasu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18:6619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, C., J. Oshima, Y. Fu, E. M. Wijsman, F. Hisama, R. Alish, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]
- 69.Zetka, M. C., I. Kawasaki, S. Strome, and F. Müller. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13:2258-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zetka, M. C., and A. M. Rose. 1995. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]