Abstract
Several male germ line-specific genes, including MAGE-A1, rely on DNA methylation for their repression in normal somatic tissues. These genes become activated in many types of tumors in the course of the genome-wide demethylation process which often accompanies tumorigenesis. We show that in tumor cells expressing MAGE-A1, the 5′ region is significantly less methylated than the other parts of the gene. The process leading to this site-specific hypomethylation does not appear to be permanent in these tumor cells, since in vitro-methylated MAGE-A1 sequences do not undergo demethylation after being stably transfected. However, in these cells there is a process that inhibits de novo methylation within the 5′ region of MAGE-A1, since unmethylated MAGE-A1 transgenes undergo remethylation at all CpGs except those located within the 5′ region. This local inhibition of methylation appears to depend on promoter activity. We conclude that the site-specific hypomethylation of MAGE-A1 in tumor cells relies on a transient process of demethylation followed by a persistent local inhibition of remethylation due to the presence of transcription factors.
Cytosines in CpG sequences are often methylated in mammalian genomes. This heritable modification of the DNA contributes to gene silencing by preventing the binding of activating transcription factors and by attracting repressor complexes that induce the formation of inactive chromatin structures (3). The repressive role of DNA methylation has been clearly demonstrated in such processes as the inactivation of one of the two X chromosomes in female somatic cells and the monoallelic silencing of parentally imprinted genes (20, 37). The establishment and the maintenance of specific genome methylation patterns are essential for the appropriate pattern of gene expression in the different cell types (24). Lineage-specific DNA methylation patterns appear to be established during embryonic development by a process that involves an early phase of extensive demethylation followed by active de novo methylation (26, 33). The mechanisms directing the methylation to certain regions of the genome and not to others is still unclear. The unmethylated status of certain regions has been ascribed to their association with active promoters (9, 31). Methylation patterns that are established in the embryo are generally maintained in differentiated adult somatic cells, which show little methylation change (36).
Genome methylation patterns are often modified in cancer cells (2). Modifications include both localized de novo methylation and genome-wide hypomethylation, but the net result is usually a decrease in the total methylation level (2, 18, 21). De novo methylation usually affects sequences with a high density of CpGs, termed CpG islands, which are classically unmethylated in all normal tissues (1). The patterns of hypermethylated CpG islands vary according to the histological origin of the tumor (8). Cancer-related de novo methylation appears to contribute to tumor initiation and progression by repressing the expression of tumor suppressor genes (2). DNA hypomethylation in cancers affects most parts of the genome, including both repetitive and single-copy sequences (15, 21). It has been proposed that hypomethylation in cancers contributes to tumor progression by inducing genome instability via the demethylation of transposons and pericentromeric repeats (22, 35, 45). Global genome hypomethylation in tumors was initially expected to induce the expression of many genes, including oncogenes (19, 21). It is only recently, however, that several reports provided clear evidence that hypomethylation in tumors causes the activation of multiple genes, including the MAGE genes, which are silent in the corresponding normal tissues (17).
The MAGE family of genes belongs to a group of germ line-specific genes that become activated in different types of tumors (42). Activation of such “cancer-germ line” genes in tumor cells results in the expression of tumor-specific antigens recognized by cytolytic T lymphocytes. We have studied the mechanisms of activation of the gene MAGE-A1 in tumors. Transient-transfection studies indicated that cells that do not express the gene nevertheless contain transcription factors capable of inducing significant MAGE-A1 promoter activity (11). This implied that the total lack of MAGE-A1 transcription in somatic cells requires a local repression mechanism to prevent activation by these ubiquitous transcription factors. We observed that MAGE-A1 has a CpG-rich promoter, which, unlike classical CpG-rich promoters, is methylated in all normal somatic tissues (13). The gene, which is on chromosome X, is methylated not only on the inactive X chromosome but also on the active X chromosome, since methylation was observed in tissues of male origin and on both alleles in tissues of female origin. In contrast, the promoter region of MAGE-A1 is completely unmethylated in testicular germ cells and in tumor cells that express the gene (13). Moreover, the expression of MAGE-A1 can be induced by treating nonexpressing cells with demethylating agents (12, 43). Conversely, transfection experiments showed that in vitro methylation of MAGE-A1 is sufficient to block its transcription in cells that normally express the gene (13). Altogether, these observations point to DNA methylation as an essential component of the repression of MAGE-A1 in somatic cells. Demethylation, and therefore activation, of MAGE-A1 in tumors appears to be a consequence of the genome-wide demethylation process, since the expression of this gene in tumor cells correlates with a decreased level of overall DNA methylation (12). Several studies suggest that DNA demethylation is also involved in the activation of other cancer-germ line genes in tumors (7, 13, 44).
How DNA demethylation is propagated over the genome of cancer cells is largely unknown. Hypomethylated CpGs in tumor cells may be either randomly scattered over the genome or clustered within specific DNA regions. To clarify this issue, we isolated DNA from several melanoma samples and analyzed the patterns of DNA hypomethylation within a large genomic segment including the gene MAGE-A1. We found that the activation of MAGE-A1 in melanoma is associated with hypomethylation of a restricted region overlapping the promoter of this gene. We proceeded to study the dynamics of demethylation in this region by analyzing the methylation changes occurring within MAGE-A1 sequences that were transfected either methylated or unmethylated into expressing and nonexpressing tumor cells.
MATERIALS AND METHODS
Cell lines and tissue samples.
Tumor cell lines and conditions of cell culture have been described previously (11). The testis sample was obtained from a prostate cancer patient, the sperm sample was obtained from a 33-years-old healthy donor, and the skin sample was obtained from a breast fragment of a patient who had undergone plastic surgery. Melanoma tissues originated from lymph node metastases (LB929 and LB454) or from a cutaneous metastasis (IGR28).
Cosmid and plasmid constructions.
Construction of the cosmid carrying MAGE-A1 has been described elsewhere (10). The sequence of both extremities of the human genomic fragment cloned into this cosmid was obtained and aligned to a human Xq28 contig sequence (GenBank accession number U82670). This revealed that the cosmid insert extends between positions −21710 and +20711 relative to the transcription start site of MAGE-A1.
For the construction of pEBFPM1-WT, we cloned a HindIII-EcoRI human MAGE-A1 fragment (−3255 to +4567 relative to the transcription start site) into the pTZ18R plasmid (Pharmacia, Piscataway, N.J.) opened at the same restriction sites. Short insertions were introduced in the MAGE-A1 sequence by introducing oligonucleotide adapters into the SacII site at position +158 (5′-GCTCTAGAGCGC-3′) and into the PmlI site at position +2371 (5′-GGTACATGTACC-3′). This plasmid was digested with BsaI (position +3639 of MAGE-A1), and the resulting ends were blunted by using a DNA polymerase I large Klenow fragment (New England Biolabs, Inc., Beverly, Mass.) in the presence of nucleotides. A 6.9-kb MAGE-A1 fragment was then released following digestion with HindIII (position −3255) and inserted between the HindIII and XmnI sites of a modified version of the pEBFP-N1 plasmid (Clontech, Palo Alto, Calif.), where the CMV-IE promoter had been removed by a digestion with AseI and BamHI and replaced by a short adapter sequence containing a HindIII and a XmnI restriction site (5′-TAAGCTTGAAGGCATTCTG-3′, annealed to 5′-GATCCAGAATGCCTTCAAGCT-3′). For the pEBFPM1-mBB′ construct, we replaced the wild-type promoter sequence of MAGE-A1 in pEBFPM1-WT by a mutated sequence deriving from the previously described 5′Δ-713mB′B plasmid (11).
In vitro methylation of cosmid DNA.
Cosmid DNA was methylated with SssI methylase for 3 h under the conditions recommended by the manufacturer (New England Biolabs) in a volume of 6.25 μl and with 2.5 U of enzyme per μg of DNA. After in vitro methylation, DNA was extracted with phenol-chloroform and precipitated with ethanol. Efficient methylation of the cosmid DNA was confirmed by its resistance to digestion upon incubation with the methylation-sensitive restriction enzyme HpaII.
Transfections.
Cells (6 × 105 to 1 × 106) were inoculated in 75-cm2 flasks 24 h before transfection. Transfections were performed by the calcium phosphate precipitation method as previously described (40). Cosmid and plasmid DNAs were prepared using the EndoFree Plasmid Maxi kit (QIAGEN, Hilden, Germany) prior to transfection. For transfection of the cosmid carrying MAGE-A1, cells received 30 μg of the cosmid, 625 ng of pHMR272 (containing a hygromycin resistance gene), and 10 μg of human genomic DNA as a carrier. Transfected cells were selected in medium containing 180 μg of hygromycin B/ml for 27 to 29 days. Hygromycin-resistant colonies (about 200) were further cultured in the absence of the selective drug. For transfection with pEBFPM1-WT and -mBB′ plasmids, cells received 10 μg of plasmid DNA (carrying a neomycin resistance gene) and 20 μg of EcoRI-digested human genomic DNA as a carrier. Transfectants were selected in medium containing 2 mg of Geneticin/ml for 18 to 21 days (MZ2-MEL3.1 cells) or 13 days (LB23-SAR cells). Geneticin-resistant colonies (300 to 400 for MZ2-MEL3.1 and about 1,000 for LB23-SAR) were further cultured in the absence of the selective drug.
5-azadC treatment of clone TM2.6.
Clone TM2.6 was obtained by limiting dilution of the MZ2-MEL2.2.5 cell population transfected with the in vitro-methylated MAGE-A1 cosmid. For 5-aza-2′-deoxycytidine (5-azadC) treatment, TM2.6 cells were seeded (106 per 75-cm2 flask) in medium containing 2 μM 5-azadC (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). The drug was renewed after 48 h. DNA and RNA were extracted from the cells after 4 days of treatment.
Conventional and quantitative real-time RT-PCR.
Total RNA was extracted using the TriPure isolation reagent (Boehringer Mannheim Corp., Indianapolis, Ind.). Reverse transcription (RT) was carried out on 2 μg of total RNA as described previously (14). Conditions for conventional RT-PCR of MAGE-A1 and β-actin have been described earlier (14). Real-time PCR amplification reactions were prepared using 1/40 of the RT reaction diluted in the qPCR reaction mix (Eurogentec SA, Seraing, Belgium), supplemented with 0.25 U of uracyl-N-glycosylase (Eurogentec), 0.25 μM (each) PCR primers, and 0.1 μM specific 5′-FAM/3′-TAMRA-labeled probe synthesized commercially (Eurogentec). PCR primers were 5′-CATTGACGTGAAGGAAGCA (sense) and 5′-GGTGGTGCAGATGAACTTCA (antisense) for MAGE-A1/EBFP or 5′-GGCATCGTGATGGACTCCG (sense) and 5′-GCTGGAAGGTGGACAGCGA (antisense) for the β-actin gene (β-ACTIN). Probe sequences were 5′-CTCCTATGTCCTTGTCACCTGCCTA for MAGE-A1/EBFP or 5′-TCAAGATCATTGCTCCTCCTGAGCGC for β-ACTIN. Thermal cycling and fluorescence monitoring were performed with an ABI Prism 7700 sequence detector (PE Applied Biosystems, Foster City, Calif.). Thermal conditions were 2 min at 50°C, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 2 min at 60°C. Samples were analyzed in duplicate in three different experiments. Each experiment included a calibration curve of known copy numbers of the template and the cDNA sample derived from MZ2-MEL3.1/pEBFPM1-WT transfectants at day 18 after transfection, which served as 100% reference. MAGE-A1/EBFP expression levels were normalized by the levels of β-ACTIN expression in the corresponding samples. The resulting expression level was divided by the ratio of exogenous over endogenous MAGE-A1 copies in the corresponding group of transfectants, which was determined by the sequencing of amplified MAGE-A1 segments where insertions were introduced in the pEBFPM1-WT and -mBB′ plasmids. MZ2-MEL3.1 and LB23-SAR transfectants contained a mean of 1 or 3 transgenes per cell, respectively.
Bisulfite genomic sequencing.
Genomic DNA was extracted as previously described (12). Twenty micrograms of EcoRI-digested genomic DNA was denatured in 0.4 N NaOH for 10 min in a volume of 80 μl. The denatured DNA was then diluted with 520 μl of a solution of 4.5 M sodium bisulfite and 0.5 mM hydroquinone (pH 5.0), covered with mineral oil, and incubated in a thermal cycler for 16 h at 50°C with 5 min at 95°C every 3 h. The DNA was desalted and concentrated on a silica matrix (GeneClean; BIO101, Vista, Calif.), incubated in 0.3 N NaOH for 10 min at room temperature, neutralized, and precipitated. The bisulfite-modified DNA was resuspended in 100 μl of water. Conditions for PCR amplification of the 5′ region of MAGE-A1 (SF segment from −105 to +225) (see Fig. 1) from bisulfite-modified DNA have been described previously (13). For the other DNA segments (SA to SE and SG to SL) (see Fig. 1 and 2), we performed nested PCR amplifications using 1/500 of the first PCR product in the second PCR. Primer sequences were deduced from the human Xq28 contig sequence (GenBank accession number U82670) and are described in Table 1. PCR mixtures and temperature cycles were identical to those described for the amplification of the MAGE-A1 5′ region. The first and second rounds of PCR were subjected to 36 and 25 cycles of amplification, respectively. Amplification products were purified by using the QiaQuick PCR purification kit (QIAGEN), cloned into the pCR4-TOPO vector using the TOPO TA cloning kit for sequencing, and transformed into electrocompetent Escherichia coli TOP10 (Invitrogen Ltd.; Paisley, United Kingdom). Ampicillin-resistant colonies were subjected to direct PCR amplification in 40-μl reaction mixtures containing 50 mM KCl, 50 mM Tris-HCl [pH 9.0], 2 mM MgCl2, 3 U of Takara Taq polymerase (Biowhittaker Europe, Verviers, Belgium), dATPs and dTTPs at a 72 μM concentration, dCTPs and dGTPs at a 48 μM concentration, and primers (each at 0.125 μM) corresponding to M13 sequences surrounding the cloning site in the pCR4-TOPO vector (5′-CGCTATTACGCCAGCTGGCGA-3′ and 5′-ATGCAGCTGGCACGACAGGT-3′). We used 3 μl of each colony PCR product in a 20-μl sequencing reaction mixture using the ABI Prism BigDye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems) with the −21M13 or M13-Reverse primers. Sequences were resolved on an ABI 3100 genetic analyzer (PE Applied Biosystems) and analyzed with the Sequencher 4.1 sequence analysis program (Gene Codes Corporation, Ann Arbor, Mich.).
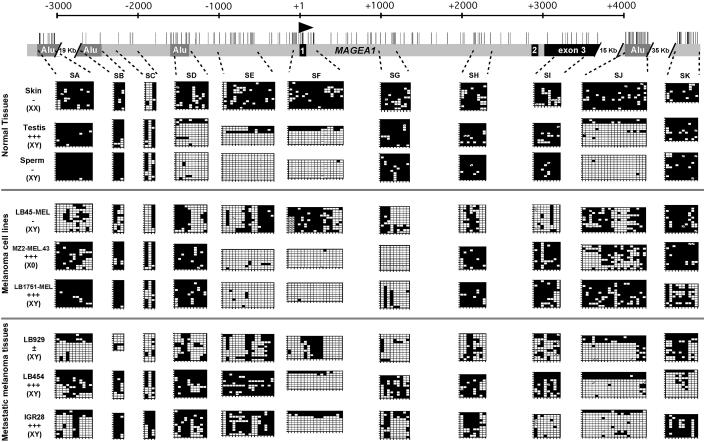
FIG. 1.
Methylation patterns at the MAGE-A1 gene locus in normal tissues, melanoma cell lines, and melanoma tissues. The analyzed human genomic locus is represented at the top with vertical bars at the location of CpG dinucleotides, an arrowhead at the MAGE-A1 transcription start site, black boxes corresponding to the MAGE-A1 exons, and dark-grey boxes representing Alu repeats. The regions corresponding to the DNA segments that were amplified from bisulfite-treated DNA (SA to SK) are indicated by dashed lines. Amplified products were cloned, and several clones (7 to 12) were sequenced. The deduced methylation patterns are represented by the grids, where each line corresponds to one allele and each column represents a CpG site. Black squares correspond to methylated cytosines, and empty squares represent unmethylated cytosines. Hatched squares indicate incomplete sequence information for the corresponding CpG. The expression of MAGE-A1, obtained by RT-PCR, is indicated for each sample: no expression (−), weak expression (±), or strong expression (+++). Since MAGE-A1 is on the X chromosome, the content in sexual chromosomes of the samples is also given.
TABLE 1.
Primers for PCR amplification of bisulfite-modified DNA segments from the human MAGE-A1 gene locusa
| DNA segment | Amplified strand | First PCR
|
Nested PCR
|
||
|---|---|---|---|---|---|
| Primer | 5′ position | Primer | 5′ position | ||
| SA | Lower | CRTCCTATAAATTACTCAATCATA | −20559 | CCCAAACTAAAATACAATAAC | −20476 |
| TYGGTGAAAGGTTAGATGTTAATT | −20182 | GTGGTGGTTTATGTTTGTAAT | −20225 | ||
| SB/SC | Upper | GTAGAAGGTTTTTATAGAGGTAAT | −2777 | GTTTGAAATTAATATATGAGGTTb | −2719 |
| CATATTTTAAAAAACTACCTCAAACTA | −1919 | CCCTCTATCTACCAAACTAAb | −2431 | ||
| GGAGTATAGTATATTTATTGATGAc | −2343 | ||||
| CCATTTTATCAACTACAATCTAAc | −1984 | ||||
| SD | Lower | ACAACTACCTACATATATCCTTAA | −1676 | ACCCACRCTAAAATTCAATAA | −1583 |
| TAGTTAGAGTTATATTTGGTAAAT | −1292 | GTAATATGTTTTATTTTTAGGTA | −1330 | ||
| SE | Upper | GAGATGGTAGAAATAGTATTTTT | −1155 | GGAAGATTTTGGTAATGTTAT | −1010 |
| CTCAATCCTCCCTCAACATA | −507 | TTACCTCCTTACAAAACCTAA | −532 | ||
| SG | Upper | GGTTAGGATAGATGTTTTAGTTGGAT | +846 | TAGGGTAATTTGTAGTTATAGTT | +906 |
| CCCCTAAAACCCTATTCAAA | +1230 | CCTCCATCAACCCTAAATAA | +1203 | ||
| SH | Lower | CCCAAAACCAAAACACTAAAA | +2085 | CCCCAAAAAACATAAACTAAA | +2142 |
| TATGATAAGGAATGGAAGGAT | +2417 | GGGGATAAATATTTGGTTAT | +2395 | ||
| SI | Upper | TTTGGAGGAGGTGTTTATTGT | +3215 | TTAGAGTTTTTAGGGAGTTTT | +3254 |
| CRCCCTCCATTACAATCATAA | +3708 | CCCAACAAACCATCATAAAA | +3640 | ||
| SJ | Lower | CAACCAACCCAACRTAACACTA | +17125 | AAACTACAACATAAAACCRAAAC | +17187 |
| GATGGTATGTAATTTGTGATTTT | +17674 | GAAGGTGTATTTTTTGGTATT | +17570 | ||
| SK | Lower | CTAATTATAAAACCCACTCCAATA | +51865 | CTTAACRCTAAAAACTTTCTCCTAA | +51956 |
| GGGATTTGGGTTGTGTTTATT | +52307 | GAGYGGTTTTGGGTTTTTAT | +52407 | ||
| SL | Upper | GGTTGGTTAAGATAGTATGGATT | −1872 | GTGGGTGAGAAATTGTGTTTT | −1814 |
| CTTACACCTAAAAACRAAATTTAC | −1522 | AAATCRTACCATTAAACTCTAAC | −1555 | ||
Positions are expressed relative to the transcription start site of MAGE-A1.
SB segment.
SC segment.
RESULTS
Pattern of unmethylated regions in the MAGE-A1 gene locus.
The human gene MAGE-A1 comprises three exons and is preceded by a promoter whose activity is mainly driven by two contiguous binding sites for transcription factors of the ETS family (10, 11). These sites are located between positions −63 and −46 relative to the transcription start site of MAGE-A1. The methylation status of 17 CpG sites located between positions −81 and +169 has been tested previously by the bisulfite genomic sequencing method (13). These CpGs were found to be heavily methylated in normal somatic tissues and completely unmethylated in male germ cells and in tumor cells that express the gene, as also shown in Fig. 1 (segment SF).
In order to determine whether hypomethylation in expressing cells is restricted to the 5′ region of MAGE-A1 or whether it extends over a larger region, we tested the methylation profile of eight different genomic segments distributed within a 6.5-kb genomic region that contains the MAGE-A1 gene locus (segments SB to SI) (Fig. 1). Most CpGs were heavily methylated in a normal skin sample. Analysis of a testis sample and a sperm sample indicated that the unmethylated region of MAGE-A1 in male germ cells is restricted to a region of less than 3 kb across the promoter of the gene (segments SD to SF) (Fig. 1). Some largely methylated SD, SE, or SF sequences found in the testis sample presumably originated from somatic cells present in the tissue sample.
All melanoma cell lines and melanoma tissues, expressing or not expressing MAGE-A1, were partially unmethylated in different parts of the MAGE-A1 gene locus (Fig. 1). But unlike nonexpressing tumor cells, cells expressing MAGE-A1 contained a region centered on the transcription start site that was nearly completely unmethylated. The boundaries of this hypomethylated region varied among the different tumors. It included segments SE to SG in the MZ2-MEL.43 and LB1751-MEL cell lines and only segment SF in LB454 and IGR28 tumor samples, implying that demethylation of the SF region, which overlaps the transcription start site, is sufficient to allow transcription of MAGE-A1. The presence of some methylated SF sequences in LB454 and IGR28 melanoma samples presumably reflects the presence of noncancerous cells in these tissue samples. The LB929 melanoma sample, which only poorly expressed MAGE-A1, showed a high level of methylation at only five sites within the SF sequence, corresponding to the CpGs at positions −30, +14, +17, +26, and +30.
Altogether, this detailed methylation analysis indicates that while different parts of the MAGE-A1 locus become partially unmethylated in tumors, the activation of MAGE-A1 is associated with increased hypomethylation of a restricted region extending 200 to 1,500 bp both upstream and downstream from the transcription start site of the gene. While demethylation of the region extending between −81 and +169 (segment SF) appears to be sufficient for MAGE-A1 expression, demethylation of the CpG sites located in close proximity to the transcription start site (between positions −30 and +30) seems to be necessary for the transcription of the gene.
We also analyzed the methylation status of several DNA segments located outside the MAGE-A1 gene locus (segments SA, SJ, and SK) (Fig. 1). The results revealed the presence of an AluYb8-type repeated sequence located about 20 kb downstream from the MAGE-A1 start site (segment SJ) that was mostly unmethylated in male germ cells. This confirms a previous report demonstrating that this type of Alu sequence is hypomethylated in the male germ line (23). The same Alu sequence also appeared to be selectively hypomethylated in several tumor samples.
MAGE-A1-expressing cells lack demethylating activity targeted to the 5′ region.
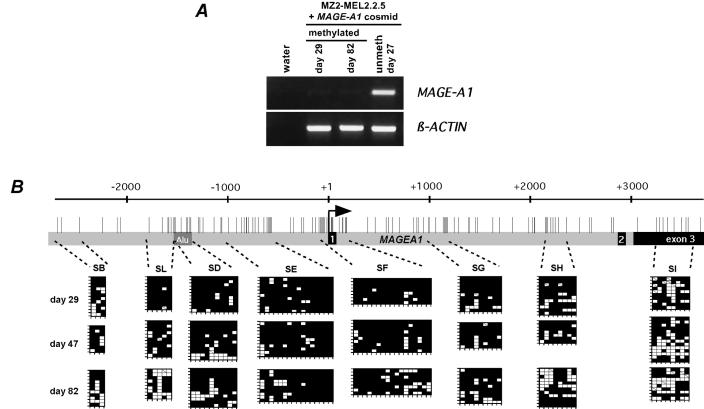
Having shown that in tumor cells expressing MAGE-A1 the 5′ region of the gene is selectively undermethylated, we wondered whether such cells have a permanent demethylating activity targeted to this site. To this end, we tested whether MAGE-A1 sequences that were methylated in vitro undergo targeted demethylation after being transfected into a tumor cell line that can express the gene. As recipient cells we used MZ2-MEL2.2.5 melanoma cells, which derive from the MAGE-A1-positive MZ2-MEL3.1 cell line but have incurred a deletion of the gene. The methylation analysis of a transfected MAGE-A1 fragment therefore cannot be confounded by the presence of an endogenous copy of the gene. Unmethylated MAGE-A1 sequences transfected into the MZ2-MEL2.2.5 cell subline are expressed, indicating that the cells have retained the appropriate transcription factors (13). For the transfection, we used a cosmid carrying MAGE-A1 in the middle of a 42-kb genomic insert to minimize the influence of the vector sequences and of the genomic sequences neighboring the integration sites. The cosmid was treated in vitro with the bacterial methyltransferase SssI, which methylates cytosines in CpG dinucleotides. Incubation of modified cosmid DNA with the methylation-sensitive restriction enzyme HpaII did not result in any detectable fragmentation, indicating that more than 99% of the CpGs were methylated. The methylated MAGE-A1 cosmid was cotransfected with a plasmid conferring resistance to hygromycin into MZ2-MEL2.2.5, and the transfectants were selected with hygromycin. These transfectants had a very low level of MAGE-A1 expression compared to cells transfected with the unmethylated MAGE-A1 cosmid (Fig. 2A). Their DNA was extracted at days 29, 47, and 82 after transfection and was submitted to bisulfite genomic sequencing in order to analyze the methylation status of several DNA segments located within the transfected MAGE-A1 gene (Fig. 2B). Whereas we observed sporadic demethylation throughout the transgene, there was clearly no preferential demethylation in the 5′ region, even after 82 days in culture, which corresponds to about the same number of cell divisions (Fig. 2B). Consistently, the level of MAGE-A1 expression did not increase in these transfectants during culturing (Fig. 2A). Therefore, tumor cells that express MAGE-A1 do not seem to have a permanent process that selectively brings the 5′ region of MAGE-A1 from the methylated state to the unmethylated state.
FIG. 2.
Lack of spontaneous demethylating activity targeted to the 5′ region of MAGE-A1 in cells that can express the gene. A cosmid carrying the MAGE-A1 gene within a 42-kb genomic insert was methylated in vitro prior to transfection into MZ2-MEL2.2.5 cells. (A) Expression of MAGE-A1 in these transfectants was analyzed by RT-PCR at two time points after transfection (days 29 and 82) and compared with that in MZ2-MEL2.2.5 cells transfected with the unmethylated MAGE-A1 cosmid (unmeth). (B) Bisulfite sequencing was used to test the methylation state of the in vitro-methylated MAGE-A1 transgene at different time points after transfection. The MAGE-A1 segments that were analyzed and the bisulfite sequencing results are represented as in Fig. 1.
The MAGE-A1 5′ region is protected against methylation.
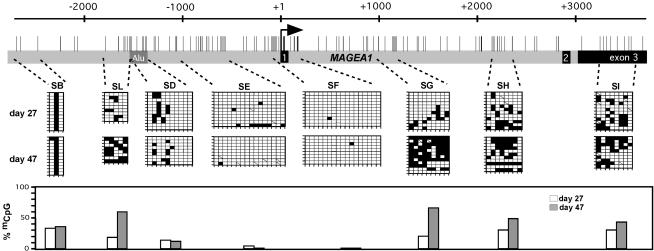
As mentioned above, transfection of the unmethylated MAGE-A1 cosmid into MZ2-MEL2.2.5 leads to expression of the gene. We investigated the methylation status of different segments within the MAGE-A1 locus in these transfectants at days 27 and 47 after transfection. The transgene underwent progressive de novo methylation, but a region including segments SE and SF remained almost completely unmethylated (Fig. 3). The rate of de novo methylation within the transgene appeared to be slow, since only a few sites were methylated at day 27 after transfection. The level of methylation was higher at day 47 (Fig. 3) and did not change significantly at later time points (data not shown). These results indicate that MZ2-MEL2.2.5 cells possess a de novo methylation activity but that this activity is inhibited within the 5′ region of MAGE-A1.
FIG. 3.
The 5′ region of MAGE-A1 is protected against de novo methylation in cells capable of a high-level expression of the gene. The cosmid carrying MAGE-A1 was transfected unmethylated into MZ2-MEL2.2.5 cells. Bisulfite sequencing was used to test the methylation state of the MAGE-A1 transgene at two time points after transfection. The MAGE-A1 segments that were analyzed and the bisulfite sequencing results are represented as in Fig. 1. Histograms represent the overall percentages of methylated CpGs within each segment at the two points after transfection, deduced from the bisulfite sequencing results.
These observations suggested that the process leading to the site-specific hypomethylated state in MAGE-A1 comprises two phases, one transient phase consisting in the DNA demethylation per se, which may not show site specificity, and another permanent phase corresponding to the site-specific maintenance of the unmethylated state within the 5′ region of the gene.
We also examined whether the same region would become selectively hypomethylated upon treatment with 5-azadC, a general inhibitor of DNA methylation. Demethylation by this agent results from irreversible binding of the DNA methyltransferases to 5-azadC and hence cellular depletion of active methylating enzymes. The effect of 5-azadC was investigated in MZ2-MEL.2.2.5 transfectants containing a methylated MAGE-A1 cosmid. In order to avoid cell heterogeneity, we used a clone (TM2.6) isolated from the transfected population. RT-PCR and bisulfite genomic sequencing confirmed that this clone contains a silent and heavily methylated MAGE-A1 transgene (Fig. 4A and B). TM2.6 cells were incubated with 2 μM 5-azadC during 4 days. This resulted in the activation of MAGE-A1 expression (Fig. 4A), although it induced only partial demethylation of the transgene, since the overall number of unmethylated CpGs increased only 2.5-fold after treatment (Fig. 4B). Compared to this overall level of demethylation, segment SF, but not the other regions of the transgene, showed a significantly higher level of demethylation upon 5-azadC treatment (3.9-fold [P ≤ 0.01]), with 4 out of 22 sequences having more than 80% of the CpGs unmethylated (Fig. 4B). This demethylation pattern suggests that the process protecting the 5′ region of MAGE-A1 against remethylation was active during the demethylation induced by 5-azadC and favored the accumulation of unmethylated CpGs in this region. However, we cannot rule out the possibility that the 5-azadC treatment induced the expression of factors that directed site-specific demethylation within the 5′ region of MAGE-A1.
FIG. 4.
Treatment with 5-azadC leads to preferential hypomethylation of the 5′ region of MAGE-A1. (A) MAGE-A1 expression was tested in a clone derived from the MZ2-MEL2.2.5 population transfected with the in vitro-methylated MAGE-A1 cosmid (TM2.6), either before or after treatment with the methylation inhibitor 5-azadC. (B) Methylation analysis of the MAGEA1 transgene in the TM2.6 cell clone either untreated or after 4 days with 5-azadC. The MAGE-A1 segments that were analyzed and the bisulfite sequencing results are represented as in Fig. 1. Histograms compare the level of demethylation upon 5-azadC treatment in each segment with the overall level of demethylation in the entire transgene (dashed line). Only segment SF showed a significantly higher level of demethylation.
Impaired MAGE-A1 promoter activity leads to remethylation of the 5′ region.
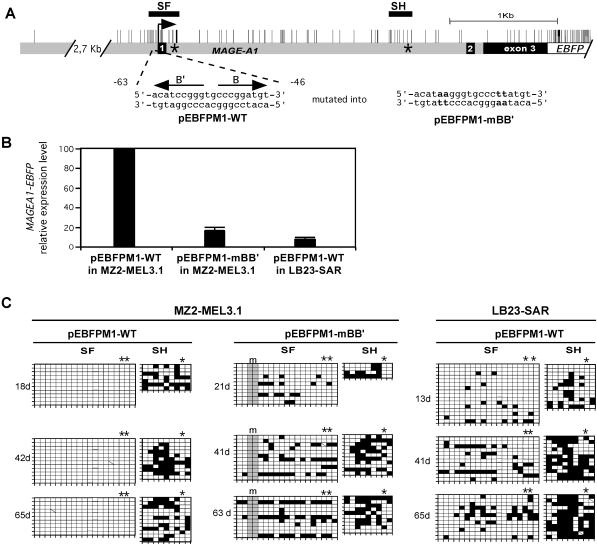
We asked whether the local inhibition of methylation within the 5′ region of MAGE-A1 is related to the transcriptional activity of the promoter. We have shown previously that most of the MAGE-A1 promoter activity is mediated by two elements, termed B and B′, which contain identical but inverted sequences that bind ETS-type transcription factors (CCCGGATGT) (Fig. 5A). Mutations that abolish the binding of ETS factors to the B and B′ sites result in the loss of most of the MAGE-A1 promoter activity (11). We tested the effect of mutating these sites on the remethylation of the 5′ region of MAGE-A1.
FIG. 5.
Protection of the MAGE-A1 5′ region from remethylation depends on promoter activity. (A) The MAGE-A1 gene fragment cloned in front of the EBFP transcription unit in the pEBFPM1-WT and pEBFPM1-mBB′ constructs is represented as in Fig. 1. Positions of the SF and SH segments are indicated by horizontal bars. Asterisks indicate the positions where short oligonucleotide tags were introduced. The wild-type and mutated MAGE-A1 promoter sequences (present in pEBFPM1-WT and pEBFPM1-mBB′, respectively) are given, with arrows indicating the positions and orientations of B and B′ regulatory elements. (B) MZ2-MEL3.1 and LB23-SAR cells were stably transfected with unmethylated pEBFPM1-WT or pEBFPM1-mBB′ plasmid DNA. Quantitative real-time RT-PCR was used to measure the relative MAGE-A1/EBFP expression levels in the transfectants at the earliest time point after transfection. MZ2-MEL3.1 transfected with pEBFPM1-WT was taken as the 100% reference. Values are the means for three separate RT-PCR experiments. (C) Methylation patterns in the SF and SH segments of the MAGE-A1 transgenes are represented by the grids, as in Fig. 1. Sequences deriving from the exogenous MAGE-A1 transgenes could be distinguished from those deriving from the endogenous MAGE-A1 gene by the presence of sequence tags. Asterisks above the grids indicate CpG sites deriving from these tag sequences. The mutations in the B and B′ promoter elements (m) resulted in the loss of two CpG sites, as indicated by the shading of the corresponding columns.
We constructed the pEBFPM1-WT and pEBFPM1-mBB′ vectors carrying a 6.9-kb fragment of MAGE-A1 (−3255 to +3639) fused to the EBFP transcription unit (Fig. 5A). In pEBFPM1-mBB′, the B and B′ elements contained specific mutations known to abolish their promoter activity (11). In addition, the MAGE-A1 sequence in both constructs contained short insertions immediately downstream of exon 1 and in the 3′ part of intron 1 (Fig. 5A). MAGE-A1 sequences originating from these plasmids could therefore be distinguished from the endogenous MAGE-A1 sequences. This allowed us to use the original MZ2-MEL3.1 cell line as a recipient for transfection, rather than the deletion variant MZ2-MEL2.2.5. The pEBFPM1-WT or pEBFPM1-mBB′ constructs were transfected unmethylated, and the transfectants were selected for their resistance to neomycin, which was conferred by these plasmids. MAGE-A1/EBFP expression levels were evaluated by quantitative real-time RT-PCR in the two transfected cell populations. The level of MAGE-A1/EBFP transcription from the pEBFPM1-WT construct was sixfold higher than that from pEBFPM1-mBB′ (Fig. 5B), confirming the importance of B and B′ ETS binding sites in MAGE-A1 promoter activity. DNA was isolated from the two groups of transfectants at different time points after transfection, and bisulfite genomic sequencing was applied to the analysis of the SF and SH segments located in the 5′ region and in the 3′ part of intron 1 of MAGE-A1, respectively, where the exogenous and endogenous sequences can be distinguished (Fig. 5A). Consistent with the previous result (Fig. 3), the 5′ region of MAGE-A1 carried by the pEBFPM1-WT plasmid remained completely free of methylation, while the sequence at the end of intron 1 was remethylated (Fig. 5C). In contrast, both MAGE-A1 segments showed progressive de novo methylation in the pEBFPM1-mBB′ construct, which contains the mutated B and B′ promoter elements. Therefore, inhibition of methylation within the 5′ region of MAGE-A1 appears to be linked to the activity of the promoter. The level of remethylation in the SF segment of pEBFPM1-mBB′ was lower, however, than in the SH segment. This is probably attributable to the action of transcription factors, other than the ETS factors, that can still bind to the promoter region.
We also tested the methylation of a MAGE-A1 transgene transfected into the LB23-SAR sarcoma cell line, which does not express the gene. We showed previously that although the gene MAGE-A1 is completely repressed in LB23-SAR cells (25), its promoter is significantly activated when transfected unmethylated in these cells (11). The complete repression of MAGE-A1 in LB23-SAR cells was later shown to depend on promoter methylation (13). MAGE-A1 promoter activity in LB23-SAR, however, represented only 15% of that in MZ2-MEL cells, suggesting less-active transcription factors (11). Here, we assessed whether the reduced MAGE-A1 promoter activity in LB23-SAR, relative to that in MZ2-MEL, is associated with a higher degree of remethylation within the 5′ region. The unmethylated pEBFPM1-WT plasmid was stably transfected into the LB23-SAR cell line. Quantitative RT-PCR revealed that the level of MAGE-A1/EBFP expression in the LB23-SAR transfectants represented only 8% of that in MZ2-MEL3.1 cells transfected with the same construct (Fig. 5B), confirming our previous data showing reduced activity of the MAGE-A1 promoter in LB23-SAR cells (11). LB23-SAR transfectants were assessed at different time points for CpG methylation within both the 5′ region and the region at the end of intron 1 of the MAGE-A1 transgene. Unlike MZ2-MEL3.1 cells, LB23-SAR cells induced remethylation in both regions of the MAGE-A1 transgene, although remethylation was induced at a lower level in the 5′ region (Fig. 5C).
DISCUSSION
Genome-wide hypomethylation is a common feature of cancer cells and is accompanied by the ectopic activation of germ line-specific genes, such as gene MAGE-A1, whose repression in normal somatic tissues relies on DNA methylation. We have analyzed the methylation profile within and around the gene MAGE-A1. The expression of this gene is associated with almost complete demethylation of a region which is located around the transcription start site and does not exceed 3,000 bp. This observation suggests that in cancer cells the genome hypomethylation is not homogeneously distributed, nor does it consist of large continuous tracts of unmethylated DNA encompassing several genes.
In order to gain more insight into the process that leads to the hypomethylation of the 5′ region of MAGE-A1, we transfected an in vitro-methylated genomic segment containing MAGE-A1 into MZ2-MEL melanoma cells, which are capable of high levels of expression of the gene. This sequence did not become demethylated. We conclude that MAGE-A1-expressing cells do not have a permanent efficient process resulting in the specific demethylation of the gene. We also observed that unmethylated MAGE-A1 genes transfected into these cells underwent remethylation at all CpGs except those located within the 5′ region, indicating that in these cells there is a permanent mechanism that protects the 5′ end of MAGE-A1 against remethylation. These observations suggest that the site-specific hypomethylation in the 5′ region of MAGE-A1 results from a sequence of two distinct processes: a transient DNA demethylation process, followed by persistent site-specific protection of this region against remethylation. The transient demethylation process may not be limited to the region that is permanently hypomethylated but may encompass a larger region, which would later be remethylated, except for the small region that is protected against remethylation. Regarding the timing of the demethylation process, hypomethylation in tumor cells could result from a transient global genome demethylation phase occurring at one stage of the progression of the tumor. Alternatively, there could be a permanent demethylation process of lesser intensity which would be effective for only a few genomic locations at any one time, so that it would be transient for any given genomic region. Another possibility is that MAGE-expressing tumors derive from rare normal precursor stem cells in which these genes are unmethylated and active. The scarcity of these precursor cells would render MAGE expression and hypomethylation undetectable in the normal tissues. However, our observation that the frequency of expression of MAGE genes increases in more-advanced tumor stages strongly suggests that activation of these genes occurs during tumor progression (5, 41).
We examined the possibility that the protection of the 5′ region of MAGE-A1 against remethylation is related to the transcriptional activity of the promoter. The protection was found to be impaired by mutations in two MAGE-A1 promoter elements that bind ETS transcription factors, suggesting that the inhibition of remethylation depends on transcriptional activation. The ability of transcription factors to inhibit methylation has been described previously (4, 27, 32). For instance, ubiquitous transcription factors binding to the Sp1 promoter elements of the Aprt gene appear to maintain a local unmethylated state, allowing permanent expression of the gene in all tissues (4, 32). Transcription factors may inhibit methylation either directly by preventing access of the DNA methyltransferases or indirectly by inducing histone modifications that exclude these methylation enzymes (28, 34, 39).
Previous transfection studies indicated that tumor cell lines, whether or not they express MAGE-A1, contain transcription factors capable of interacting with the unmethylated MAGE-A1 promoter to produce transcriptional activity (11, 13). But the level of transcription of unmethylated MAGE-A1 transgenes varies between different tumor cell lines. The MAGE-A1-negative cell line LB23-SAR, where we observe a lower level of transcription of unmethylated MAGE-A1 transgenes than in MZ2-MEL, also lacks the ability to efficiently protect the 5′ region from remethylation. This reinforces the conclusion that the level of transcriptional activation within the MAGE-A1 promoter is critical for the protection against remethylation.
Altogether our observations imply that the stable activation of MAGE-A1 in tumors depends on two critical processes. First, the genome demethylation process has to take place and should lead to a sufficient loss of cytosine methylation to make the MAGE-A1 promoter become accessible to transcription factors. The second critical point is the presence of transcription factors capable of inhibiting remethylation within the 5′ region of the MAGE-A1 gene. This inhibition may start while the demethylation process is still ongoing and hence accelerate the accumulation of unmethylated CpGs in the 5′ region of the gene. The concentration of the relevant transcription factors may critically determine the probability of evolving from transient demethylation into a permanent hypomethylated state. Low levels of appropriate transcription factors could explain the scarcity of MAGE-A1 expression in some types of tumors, like colon carcinoma, which usually show marked genome hypomethylation.
It is likely that the different cancer-germ line genes respond to different sets of transcription factors. This, and tumor type-specific differences in the concentration of the various transcription factors, may explain why, for instance, MAGE-A genes are expressed more frequently than the TDRD1 cancer-germ line gene in melanomas, whereas the reverse is true in prostate adenocarcinomas (29). Similar cases of preferential activation in certain tumor types have been observed for other cancer-germ line genes (38).
We observed selective hypomethylation of an Alu sequence located about 20 kb downstream from MAGE-A1 in several tumor samples. Interestingly, Alu repeats are known to be the target of several DNA-binding proteins (30), and one of these proteins was shown to be capable of inhibiting DNA methylation at least in vitro (6).
In conclusion, our data strongly suggest that the genome demethylation process in cancer cells evolves into specific hypomethylation patterns according to the set of transcription factors present in the cell. Moreover, the involvement of factors that protect the hypomethylated sequences against remethylation may explain why hypomethylation in tumors persists despite the frequent increase in DNA methyltransferase activity (2) and how hypomethylated and hypermethylated regions can coexist in the genome of cancer cells (16).
Acknowledgments
We are grateful to Adrian Bird, Francis Brasseur, and Etienne De Plaen for advice and comments on the manuscript.
This work was supported by the Belgian Program on Inter-University Poles of Attraction initiated by the Belgian State Prime Minister's Office, Science Policy Programming. A.L. is supported by the Fonds pour la Recherche Scientifique dans l'Industrie et l'Agriculture (Brussels, Belgium).
REFERENCES
- 1.Antequera, F., J. Boyes, and A. Bird. 1990. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62:503-514. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S. B., J. G. Herman, J. R. Graff, P. M. Vertino, and J. P. Issa. 1998. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 72:141-196. [PubMed] [Google Scholar]
- 3.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 4.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 5.Brasseur, F., D. Rimoldi, D. Lienard, B. Lethe, S. Carrel, F. Arienti, L. Suter, R. Vanwijck, A. Bourlond, Y. Humblet, et al. 1995. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int. J. Cancer 63:375-380. [DOI] [PubMed] [Google Scholar]
- 6.Chesnokov, I. N., and C. W. Schmid. 1995. Specific Alu binding protein from human sperm chromatin prevents DNA methylation. J. Biol. Chem. 270:18539-18542. [DOI] [PubMed] [Google Scholar]
- 7.Coral, S., L. Sigalotti, M. Altomonte, A. Engelsberg, F. Colizzi, I. Cattarossi, E. Maraskovsky, E. Jager, B. Seliger, and M. Maio. 2002. 5-aza-2′-deoxycytidine-induced expression of functional cancer testis antigens in human renal cell carcinoma: immunotherapeutic implications. Clin Cancer Res. 8:2690-2695. [PubMed] [Google Scholar]
- 8.Costello, J. F., M. C. Fruhwald, D. J. Smiraglia, L. J. Rush, G. P. Robertson, X. Gao, F. A. Wright, J. D. Feramisco, P. Peltomaki, J. C. Lang, D. E. Schuller, L. Yu, C. D. Bloomfield, M. A. Caligiuri, A. Yates, R. Nishikawa, H. Su Huang, N. J. Petrelli, X. Zhang, M. S. O'Dorisio, W. A. Held, W. K. Cavenee, and C. Plass. 2000. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 24:132-138. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, R., S. Lowell, V. Bolton, and M. Monk. 1997. Transcription of tissue-specific genes in human preimplantation embryos. Hum. Reprod. 12:2251-2256. [DOI] [PubMed] [Google Scholar]
- 10.De Plaen, E., K. Arden, C. Traversari, J. J. Gaforio, J. P. Szikora, C. De Smet, F. Brasseur, P. van der Bruggen, B. Lethe, C. Lurquin, et al. 1994. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics 40:360-369. [DOI] [PubMed] [Google Scholar]
- 11.De Smet, C., S. J. Courtois, I. Faraoni, C. Lurquin, J. P. Szikora, O. De Backer, and T. Boon. 1995. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics 42:282-290. [DOI] [PubMed] [Google Scholar]
- 12.De Smet, C., O. De Backer, I. Faraoni, C. Lurquin, F. Brasseur, and T. Boon. 1996. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc. Natl. Acad. Sci. USA 93:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Smet, C., C. Lurquin, B. Lethe, V. Martelange, and T. Boon. 1999. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 19:7327-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Smet, C., C. Lurquin, P. van der Bruggen, E. De Plaen, F. Brasseur, and T. Boon. 1994. Sequence and expression pattern of the human MAGE2 gene. Immunogenetics 39:121-129. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich, M. 2002. DNA methylation in cancer: too much, but also too little. Oncogene 21:5400-5413. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich, M., G. Jiang, E. Fiala, J. S. Dome, M. C. Yu, T. I. Long, B. Youn, O. S. Sohn, M. Widschwendter, G. E. Tomlinson, M. Chintagumpala, M. Champagne, D. Parham, G. Liang, K. Malik, and P. W. Laird. 2002. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene 21:6694-6702. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg, A. P., and B. Tycko. 2004. The history of cancer epigenetics. Nat. Rev. Cancer 4:143-153. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg, A. P., and B. Vogelstein. 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301:89-92. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 111:47-54. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson-Smith, A. C., and M. A. Surani. 2001. Imprinting and the epigenetic asymmetry between parental genomes. Science 293:1086-1089. [DOI] [PubMed] [Google Scholar]
- 21.Gama-Sosa, M. A., V. A. Slagel, R. W. Trewyn, R. Oxenhandler, K. C. Kuo, C. W. Gehrke, and M. Ehrlich. 1983. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 11:6883-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. [DOI] [PubMed] [Google Scholar]
- 23.Hellmann-Blumberg, U., M. F. Hintz, J. M. Gatewood, and C. W. Schmid. 1993. Developmental differences in methylation of human Alu repeats. Mol. Cell Biol. 13:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 25.Janssen, B. L., L. T. van de Locht, A. Fourkour, C. de Smet, E. J. Mensink, G. N. van Muijen, and T. J. de Vries. 1999. Transcription of the MAGE-1 gene and the methylation status of its Ets binding promoter elements: a quantitative analysis in melanoma cell lines using a real-time polymerase chain reaction technique. Melanoma Res. 9:213-222. [DOI] [PubMed] [Google Scholar]
- 26.Kafri, T., M. Ariel, M. Brandeis, R. Shemer, L. Urven, J. McCarrey, H. Cedar, and A. Razin. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6:705-714. [DOI] [PubMed] [Google Scholar]
- 27.Kirillov, A., B. Kistler, R. Mostoslavsky, H. Cedar, T. Wirth, and Y. Bergman. 1996. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igκ locus. Nat. Genet. 13:435-441. [DOI] [PubMed] [Google Scholar]
- 28.Lin, I. G., T. J. Tomzynski, Q. Ou, and C. L. Hsieh. 2000. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loriot, A., T. Boon, and C. De Smet. 2003. Five new human cancer-germline genes identified among 12 genes expressed in spermatogonia. Int. J. Cancer 105:371-376. [DOI] [PubMed] [Google Scholar]
- 30.Lukyanov, D. V., M. E. Urusova, K. M. Shcherba, and O. I. Podgornaya. 2000. Alu-DNA repeat-binding protein p68 is a part of Alu-RNA containing alpha-RNP. Eur. J. Biochem. 267:2362-2371. [DOI] [PubMed] [Google Scholar]
- 31.Macleod, D., R. R. Ali, and A. Bird. 1998. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: implications for the origin of CpG islands. Mol. Cell. Biol. 18:4433-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 33.Monk, M., M. Boubelik, and S. Lehnert. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99:371-382. [DOI] [PubMed] [Google Scholar]
- 34.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayan, A., W. Ji, X. Y. Zhang, A. Marrogi, J. R. Graff, S. B. Baylin, and M. Ehrlich. 1998. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int. J. Cancer 77:833-838. [DOI] [PubMed] [Google Scholar]
- 36.Pfeifer, G. P., S. D. Steigerwald, R. S. Hansen, S. M. Gartler, and A. D. Riggs. 1990. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl. Acad. Sci. USA 87:8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggs, A. D., and G. P. Pfeifer. 1992. X-chromosome inactivation and cell memory. Trends Genet. 8:169-174. [DOI] [PubMed] [Google Scholar]
- 38.Sahin, U., O. Tureci, Y. T. Chen, G. Seitz, C. Villena-Heinsen, L. J. Old, and M. Pfreundschuh. 1998. Expression of multiple cancer/testis (CT) antigens in breast cancer and melanoma: basis for polyvalent CT vaccine strategies. Int. J. Cancer 78:387-389. [DOI] [PubMed] [Google Scholar]
- 39.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 40.Traversari, C., P. van der Bruggen, B. Van den Eynde, P. Hainaut, C. Lemoine, N. Ohta, L. Old, and T. Boon. 1992. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics 35:145-152. [DOI] [PubMed] [Google Scholar]
- 41.van Baren, N., F. Brasseur, D. Godelaine, G. Hames, A. Ferrant, F. Lehmann, M. Andre, C. Ravoet, C. Doyen, G. C. Spagnoli, M. Bakkus, K. Thielemans, and T. Boon. 1999. Genes encoding tumor-specific antigens are expressed in human myeloma cells. Blood 94:1156-1164. [PubMed] [Google Scholar]
- 42.Van Der Bruggen, P., Y. Zhang, P. Chaux, V. Stroobant, C. Panichelli, E. S. Schultz, J. Chapiro, B. J. Van Den Eynde, F. Brasseur, and T. Boon. 2002. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol. Rev. 188:51-64. [DOI] [PubMed] [Google Scholar]
- 43.Weber, J., M. Salgaller, D. Samid, B. Johnson, M. Herlyn, N. Lassam, J. Treisman, and S. A. Rosenberg. 1994. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 54:1766-1771. [PubMed] [Google Scholar]
- 44.Weiser, T. S., Z. S. Guo, G. A. Ohnmacht, M. L. Parkhurst, P. Tong-On, F. M. Marincola, M. R. Fischette, X. Yu, G. A. Chen, J. A. Hong, J. H. Stewart, D. M. Nguyen, S. A. Rosenberg, and D. S. Schrump. 2001. Sequential 5-Aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J. Immunother. 24:151-161. [DOI] [PubMed] [Google Scholar]
- 45.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]