Abstract
PCR-ribotyping, a typing method based on size variation in 16S-23S rRNA intergenic spacer region (ISR), has been used widely for molecular epidemiological investigations of C. difficile infections. In the present study, we describe the sequence diversity of ISRs from 43 C. difficile strains, representing different PCR-ribotypes and suggest homologous recombination as a possible mechanism driving the evolution of 16S-23S rRNA ISRs. ISRs of 45 different lengths (ranging from 185 bp to 564 bp) were found among 458 ISRs. All ISRs could be described with one of the 22 different structural groups defined by the presence or absence of different sequence modules; tRNAAla genes and different combinations of spacers of different lengths (33 bp, 53 bp or 20 bp) and 9 bp direct repeats separating the spacers. The ISR structural group, in most cases, coincided with the sequence length. ISRs that were of the same lengths had also very similar nucleotide sequence, suggesting that ISRs were not suitable for discriminating between different strains based only on the ISR sequence. Despite large variations in the length, the alignment of ISR sequences, based on the primary sequence and secondary structure information, revealed many conserved regions which were mainly involved in maturation of pre-rRNA. Phylogenetic analysis of the ISR alignment yielded strong evidence for intra- and inter-homologous recombination which could be one of the mechanisms driving the evolution of C. difficile 16S-23S ISRs. The modular structure of the ISR, the high sequence similarities of ISRs of the same sizes and the presence of homologous recombination also suggest that different copies of C. difficile 16S-23S rRNA ISR are evolving in concert.
Introduction
Clostridium difficile, an anaerobic, sporogenic bacterium, is one of the most important pathogens causing health care-associated infections. The spectrum of disease ranges from mild diarrhea to colitis and to sometimes fatal pseudomembranous colitis. Although, hospitalization and antibiotic usage are still considered as the main risk factors for C. difficile infection (CDI), affecting mainly elderly patient (>65 years), community-acquired infection is being increasingly reported [1]–[3]. The incidence and severity of CDI has increased dramatically since 2004, partially due to the emergence of more virulent strains (i.e. PCR-ribotypes 027, 078, 017, 053) [4]. Since then, small and large outbreaks are constantly being present in hospital environment [5]–[9].
A diverse set of molecular typing techniques has been used for molecular epidemiological studies of CDI, with PCR-ribotyping being the most popular method. PCR-ribotyping targets the intergenic spacer region (ISR) between the 16S and 23S rRNA genes [10]. Like in many other bacteria, several copies of the rRNA operon are present in C. difficile genome [11]. The 16S-23S rRNA ISRs of C. difficile differ in length and PCR amplification of ISRs with only a single primer pair results in a pattern of bands (ranging from ≈ 200 – 700 bp) which is unique for a specific PCR-ribotype. In C. difficile, the size variability of ISRs seem to be greater than in other bacteria which is reflected in the good discriminatory power of PCR-ribotyping [10], [12].
The rRNA genes in bacterial ribosomal operons are generally organized in the order of 16S-23S-5S rRNA and the individual genes are separated with the intergenic spacer regions (ISR) also called internal transcribed spacers (ITS). The spacer between the 16S and 23S rRNA has been studied the most and, due to various degrees of variability in different species, can be used either for typing, identification or phylogeny studies [13]–[19].
To date, only two groups have studied the variability of 16S-23S rRNA ISR in C. difficile. Sadeghifard et al. showed that ISRs have a mosaic structure and can differ in length as well as sequence [20]. On the other hand, Indra et al. showed that ISRs have a very uniform structure composed of tRNA gene (which is present or not) and different combinations of spacers of variable lengths separated by 9 bp direct repeat. They proposed two mechanisms that could be responsible for the ISR length variations; slipped-strand mispairing and/or homologous recombination [21].
In the present study, we describe the ISR sequence diversity and explore homologous recombination as a possible mechanism driving the evolution of C. difficile 16S-23S rRNA ISRs. Furthermore, we used the ISR sequence information to look for type or lineage specific markers within the ISRs which would be appropriate and have enough discrimination for sequence based typing.
Results and Discussion
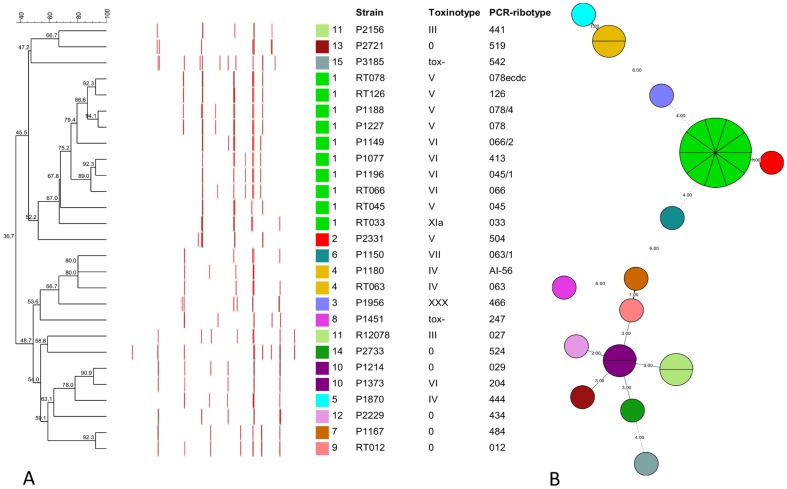
We analyzed 16S-23S rRNA ISR sequences of 43 C. difficile strains (Table 1). Twelve strains were either studied previously or had complete genome available [11], [20]–[23]. Additional thirty-one strains representing 27 different PCR-ribotypes were selected based on the similarity of banding patterns generated by capillary gel electrophoresis-based PCR-ribotyping (Figure 1a, Table S1). After screening the profiles of 136 PCR-ribotypes that were available at the time, we selected a subset of PCR-ribotypes that had similar banding patterns, clustering with PCR-ribotype 078 (these were considered to be more genetically related), and a subset of PCR-ribotypes that had very different banding patterns (considered to be genetically unrelated) (Figure 1a). Genetic relatedness of strains with a similar PCR-ribotyping profile has been suggested previously by Kurka et al. [24]. They compared 14 standard marker genes from 21 different PCR-ribotypes and showed that certain strains (e.g. PCR-ribotypes 078, 126 and 033) always clustered together indicating relatedness. We confirmed genetic relatedness of selected strains with multilocus sequence typing (MLST) and strains that had similar banding patterns by PCR-ribotyping were more likely to also cluster together with MLST (Figure 1b).
Table 1. Overview of C. difficile strains/genomes included in the analysis.
| Strain | PCR-ribotype | Toxinotype | Host | Remarks |
| P2156 | 441 | III | Human | |
| RT063 | 063 | IV | Human | Reference RT strain1 |
| RT066 | 066 | VI | Human | Reference RT strain1 |
| P2331 | 504 | V | Human | |
| P1956 | 466 | XXX | Human | |
| P1870 | 444 | IV | Human | |
| P1451 | 247 | TOX- | Human | |
| P2229 | 434 | 0 | Human | |
| RT012 | 012 | 0 | Human | Reference RT strain1 |
| RT078 | 078ecdc | V | Human | Reference RT strain1 |
| P2733 | 524 | 0 | Human | |
| P3185 | 542 | TOX- | Human | |
| RT033 | 033 | XIa | Human | Reference RT strain1 |
| RT045 | 045 | V | Human | Reference RT strain1 |
| P1373 | 204 | VI | Human | |
| P1227 | 078 | V | Calf | |
| P1214 | 029 | 0 | Human | |
| RT126 | 126 | V | Human | Reference RT strain1 |
| ZZV08-1263 | 126 | V | Human | |
| P1077 | 413 | VI | Human | |
| P1180 | AI-56 | IV | Human | |
| P2721 | 519 | 0 | Human | |
| P1167 | 484 | 0 | Human | |
| ZZV08-961 | 045 | V | Pig | |
| ZZV08-958 | 045 | V | Pig | |
| SE866 | 045 | V | Human | |
| P1150 | 063/1 | VII | Human | |
| P1188 | 078/4 | V | Human | |
| P1196 | 045/1 | VI | Horse | |
| P1149 | 066/2 | VI | Human | |
| R12078 | 027 | III | Human | |
| CD196 | 027 | III | Human | |
| R20291 | 027 | III | Human | |
| 630 | 012 | 0 | Human |
The 16S -23S rRNA intergenic spacer regions (ISRs) from strains CD196 (NC_013315.1), R20291 (NC_013316.1) and 630 (NC_009089.1) were obtained from published C. difficile genomes. PCR-ribotypes were identified with WEBRIBO (http://webribo.ages.at/). We have included also 78 ISR sequences from 9 different strains (A, B, ATCC43593, 001, 027, AI5, 053, 078 and 176) analyzed in two previous publications [20], [21].
Leiden/Leeds reference PCR-ribotyping strain
Figure 1. Clustering of C. difficile PCR-ribotypes.
(A) Clustering of PRC-ribotypes based on fingerprinting profiles generated by capillary gel electrophoresis-based PCR-ribotyping. Dendrogram is color coded according to MLST type. The exact lengths of the bands, representing the 16S-23S rRNA intergenic spacer regions are given in Table S1. (B) Minimum spanning tree of MLST results showing relatedness of PCR-ribotypes. Each circle represents one sequence type (ST) and is subdivided into sectors corresponding to the number of PCR-ribotypes represented with this ST. The numbers between circles represent number of differing loci between the STs.
Length polymorphism of 16S-23S rRNA ISRs
With cloning and sequencing of amplified ISRs from 31 strains, we obtained 350 sequences. For each fragment (amplified ISR from all 27 PCR-ribotypes) at least one ISR was sequenced (1 – 60 clones per fragment of particular size as seen on banding patterns produced by capillary gel electrophoresis). In addition, we included 30 ISR sequences obtained from published C. difficile genomes of strains CD196 (NC_013315.1), R20291 (NC_013316.1) and 630 (NC_009089.1) and 78 ISRs from 9 different strains from two previous publications [20], [21]. Forty-five different sizes were recognized among the 458 ISR sequences, ranging from 185 to 564 bp.
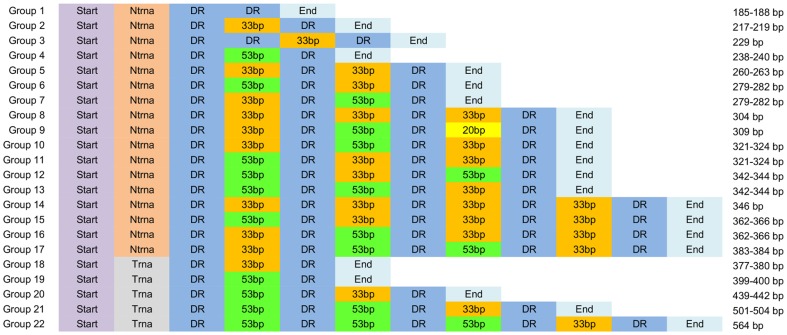
ISR structure - a new spacer
Based on visualization of previously described modules within C. difficile ISRs [21], all ISR sequences were manually grouped into 22 different structural groups (Figure 2), 14 of which have been described previously [21]. In general, the variations in ISRs were due to the different number and organization of building blocks. All ISRs had the same basic structure beginning with a start sequence of 29 bp, followed by either 26 bp (in ISRs without tRNAAla) or 186 bp (in ISRs with tRNAAla). Next was a 9 bp direct repeat followed by different combinations of spacers of three different lengths (33, 53 and 20 bp) where spacers were separated by a 9 bp direct repeat. At the end of the ISR sequence was the last direct repeat followed by a 103–114 bp end sequence (Figure 2). Two of the spacers (33 and 53 bp) have been described already [21], while the third one (20 bp) was found in an ISR of 309 bp in PCR-ribotype 012 (two different isolates, RT012 and CD630). The shortest sequence (185 bp) was composed only of the start sequence (without tRNA gene) followed by two direct repeats and the end sequence (group 1, Figure 2). Only one ISR (229 bp, group 3; found in PCR-ribotype 542) did not follow the general structure and had two direct repeats after the start sequence followed by a spacer of 33 bp and direct repeat before the end sequence.
Figure 2. Schematic representation of the modular structure of C. difficile 16S-23S rRNA intergenic spacer region.
Start (29 bp) – 5' end of the ISR sequence; Ntrna (26 bp) – part of the ISR without a gene for tRNAAla; Trna (186 bp) – part of the ISR with a gene for tRNAAla; DR – 9 bp long direct repeat; 53 bp, 33 bp, 20 bp – spacers of 53 bp, 33 bp and 20 bp, respectively and End (103–114 bp) is the 3' end of the ISR. The size of each structural group is given on the right. The inverse spacer arrangements can be seen in groups 6 and 7, 10 and 11, 12 and 13 and in groups 15 and 16.
Table 2 lists the number of different sequence variants and consensus sequence for each building block. When looking for sequence variations only the representatives of non-redundant sets (n = 95) were taken into account. All sequence variants of ISR building blocks and sequence logos showing conserved and variable sites are listed in Table S2 and Figure S1.
Table 2. Consensus sequence and number of sequence variants found in ISR building blocks.
| Building block | Sequence length | Nr. of sequences | Nr. of variants | Consensus sequence |
| Start | 29 bp | 95 | 4 | AAGGAGAATTACCTACTGTTTAATTTTGA |
| Ntrna | 26 bp | 72 | 5 | GGGTTCGTTTTTACGAATACTCAAAA |
| Trna | 186 bp | 23 | 11 | AAGTTCTTTACGAACTTTATATATGGGGGTGTAGCTCAGTTGGGA GAGCACTTGCCTTGCAAGCAAGGGGTCAGGAGTTCGACTCTCCTC ATCTCCACCATTTAAGAGTATATTACTTAAATCTTTGATTTACTTAG TAGCCTCTTACAATGCACTTATAGCTTAAATTTATACAAGCTTTGTGCG |
| DR | 9 bp | 335 | 8 | TTAGCACTT |
| 20 bp | 20 bp | 3 | 1 | TAAGCAACGGAATTTATTCG |
| 33 bp | 33 bp | 132 | 42 | TAAGCAACGGAATTTATTCGTTGGCGCTGTGCT |
| 53 bp | 53 bp | 98 | 35 | TAAGCAACAGAATAAACTGAACGCATGTGAAGTTTGTTTGTTG GCGCTGTGCG |
| End | 114 bp | 95 | 33 | TGAAAACTGCATATATATATTTAGTGATATGACATCTAATTTGTA ATATATAAAGCTGATAACTTTTTAAAATTATCGAAGTTGATAGCT TCTAATCTATCAAACCTTTTTAAC |
Only the representatives (n = 95) of non-redundant sets were used to calculate the consensus sequence and to determine the number of sequence variants.
ISR structural group coincide with the sequence length
In the majority of cases, the ISRs of the same length were represented with only one structural group. Four exceptions were found where ISR sequences of the same or similar lengths (±3 bp) were represented by two different structural groups; these included ISRs of 279–282 bp represented by structural groups 6 and 7, ISRs of sizes 321–324 bp represented by structural groups 10 and 11, ISRs of sizes 342 bp and 344 bp represented by groups 12 and 13, and ISRs of 362–366 bp represented by structural groups 15 and 16. The only difference between the two groups with the ISRs of the same lengths was the inverse arrangements of spacers of 33 bp and 53 bp (Figure 2). The inverse arrangement of spacers and their similar sequence can result in incorrect hybridization of different ISRs influencing the correct PCR-ribotype designation when using electrophoresis in non-denaturing conditions for separation of amplified ISRs. This was observed previously for PCR-ribotype AI-5 that was differentiated from PCR-ribotype 001 with classical agarose gel electrophoresis-based PCR-ribotyping by a single band, and had an identical banding pattern with capillary gel electrophoresis-based PCR-ribotyping [21]. Heteroduplex formation in 16S-23S rRNA ISR has also been described for cyanobacteria [25].
Presence of tRNA genes in the ISRs
16S-23S rRNA ISRs can have different tRNA genes. In the majority of bacteria only one or two tRNA genes are present in the ISR [26], but some bacteria can have 3 or even 4 different tRNA genes in a single ISR [27]. In C. difficile only one tRNA gene was present in ISRs and it was always tRNAAla. The occurrence of tRNAAla in C. difficile ISRs has been described by others [20], [21], [26], [27]. The tRNAAla gene was always located at the same site, as a part of the 186 bp sequence located between the start sequence and first direct repeat (Figure 2). When we searched for tRNA genes in published C. difficile genomes, the tRNAAla gene was present only in the ISR and not elsewhere in the genome. This explains why each strain had at least one ISR that contained a tRNAAla gene.
ISR sequence similarities
Single PCR-ribotype yielded from 4 to 11 fragments ranging in size from 234 to 609 bp after separation of the amplified ISRs with capillary gel electrophoresis (Figure 1a, Table S1). Some of the fragments were present only in a single strain (e.g. 609 bp in PCR-ribotype 524 and 279 bp in PCR-ribotype 542) and others in several strains. One fragment of approximately 329 bp (±2 bp) was present in all but one strain (PCR-ribotype 504) included in the analysis (Figure 1a, Table S1).
Sadeghifard et al. concluded that ISRs of similar lengths could have noticeably different sequence, within and between different strains [20]. Furthermore, they suggested that this high degree of diversity is not apparent when just the length polymorphisms are considered. Our data contrast with those results and show that nucleotide sequences of ISRs with the same or similar length (± 3 bp) are very similar (sequence identity > 92 to 100%; only the 95 representatives of non-redundant sets were taken into account). The only exceptions were found in ISRs of 279–282 bp, 321–324 bp, 342–344 bp and 362–366 bp where the similarities were lower than expected; 80,2%, 82,4%, 83,0% and 83,6%, respectively (Supporting information S1). The explanation for this finding is related to the inverse spacer arrangements (Figure 2). ISRs with only one or both variants (inverse arrangement of spacers) were found within a single strain. That the ISR sequences are really very similar is demonstrated also by the unsupervised clustering. All 458 ISRs could be grouped into only 95 non-redundant sets (with 99% sequence identity and 99% alignment coverage).
Some of the ISRs with identical nucleotide sequences differed in length by only 2 bp (e.g. 502 and 504, 280 and 282, 264 and 266…). The length difference in these sequences was always additional or missing di-nucleotide (AT or TA) within the stretch of AT/TA repeats (2–4) at the 3' end of the ISR (Figure S2). Clarke et al. showed that a high error rate at repeat sequence motif could be introduced by PCR when amplifying mono- and di-nucleotide sequence motifs [28]. Whether this 2 bp difference is real and there are two different alleles present in the genome or it is just a PCR artefact needs to be clarified.
Absence of PCR-ribotype specific sequences for possible sequence based typing
The length polymorphism in the C. difficile 16S-23S rRNA ISR has been successfully used for fingerprint based PCR-ribotyping, which has become the method of choice for typing of C. difficile worldwide. However, as with other non-sequence based methods, the disadvantage of the method is still poor inter-laboratory comparability and exchangeability of results. One of the goals of the present work was to develop a sequence based method, utilizing the ISR or parts of the ISR that would enable us to discriminate between strains at least on the level of PCR-ribotyping. But due to the highly uniform ISR sequence structure there were no regions appropriate to position primers which would enable us to specifically amplify a single ISR. Our data also indicate that the nucleotide sequences of ISRs are not variable enough to distinguish between strains of different PCR-ribotypes. ISRs with identical nucleotide sequence were found in related and unrelated PCR-ribotypes. Only a few ISRs were strain specific (in PCR-ribotypes 066/2, 524 and 542) however, it is very likely that these three ISRs are present in some other PCR-ribotypes that were not included in this study.
Despite the lack of PCR-ribotype specific markers, we were able to use the information from the C. difficile ISR sequence to modify the primers currently used for PCR-ribotyping and to describe an approach that enables direct, culture independent PCR-ribotyping [29].
Secondary structure based alignment of ISR
The modular structure and extensive variation in sequence length make the alignment of ISR sequences unreliable. Therefore, we used the primary and secondary structure information of ISRs to align 95 representatives of non-redundant sets (Figure S2). The automatic alignment was then checked manually to verify it and correct misaligned sequences. Secondary structure information can be used to improve the alignment and tree reconstruction of the ribosomal rRNA sequences [19], [30], [31].
Despite large variations in the length of ISRs, the aligned sequences showed parts of highly conserved regions (Figure S2), especially the 5′ end (first 29 bp) and 3′end (last 112 bp) of ISRs that showed very little sequence variation and were highly conserved between all strains. These regions are thought to be involved in maturation of pre-rRNA [26], [32]. To test this we built secondary structure models to illustrate the interactions between these regions. The genomic sequences used for the secondary structure calculations contained parts of approximately 100 bp of the region upstream of 16S rRNA, the 16S-23S ISR, approximately 50 bp at the 5′ end of 23S-5S ISR and tRNAAla when present. Genes coding for 16S and 23S rRNA were excluded from the calculations. Predicted secondary structures were determined for five rRNA operons (two without the tRNAAla gene within the ISR and three with the tRNAAla gene) of C. difficile CD630 strain [11]. In all five rRNA operons, the 5′ end of the ISR was base paired with the region upstream of the 16S, forming a stem that carried the 16S rRNA gene, and the 3′ end of the ISR was paired with the 23S-5S ISR to form a stem that carried the 23S rRNA gene (Figure S3). Direct repeats and spacers, which were identified in the primary structure, did not seem to be conserved in the secondary structure (Figures S2 and S3).
Role of recombination in evolution of C. difficile 16S-23S rRNA ISR
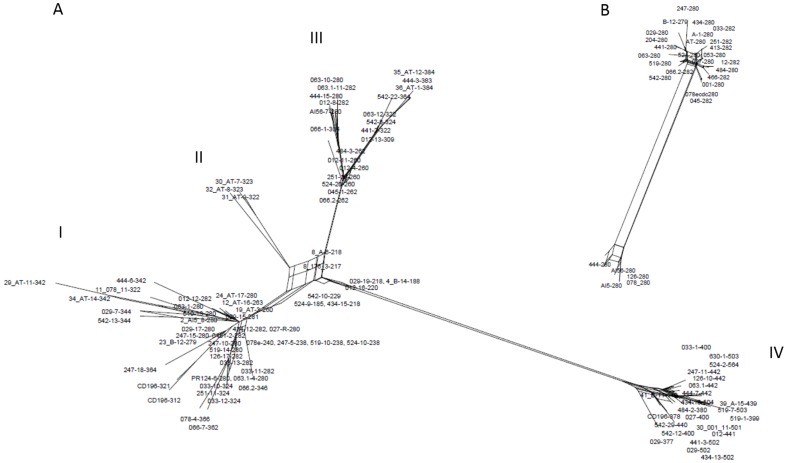
As discussed above, a given ISR is a mosaic of variable blocks and this specific structure of the ISR could be the result of recombination between different rRNA operons. Homologous recombination has been described as one of the possible mechanisms responsible for variation within rRNA operons in other species, such as the exchange of ISRs between rRNA operons detected in Vibrio cholerae [33] and rearrangements of sequence blocks within the ISR as described for Haemophilus parainfluenzae [34]. Other features that suggest homologous recombination as a possible mechanism of variation in the C. difficile 16S-23S rRNA ISR are the absence of strain specific ISRs or sequence blocks, tRNA genes not being present in all ISRs, the inverse spacer arrangements and the observation of identical ISRs in unrelated strains. Two mechanisms that could be responsible for the length variations in C. difficile ISRs have been proposed; slipped-strand mispairing and/or homologous recombination [21]. To further explore the role of homologous recombination in C. difficile ISRs the aligned sequences were analyzed with SplitsTree software. A phylogenetic network was constructed for the 95 representative ISR sequences grouping the sequences into 4 groups (I – IV) (Figure 3A). The ISRs without a gene for tRNAAla clustered in groups I, II and III and the ISRs with a tRNA gene present were in group IV only. The box-like topology of the network (ISRs are connected by several branches), especially within the groups and between groups I, II and III, represents conflicting phylogenetic signals indicating that recombination events might have contributed to the evolution of ISRs (Figure 3A). As conflicting signals can be explained by recombination or homoplasies (parallel mutations) three statistical tests; PHI (pairwise homoplasy index), maximum chi-squared and NSS for detecting the presence of recombination were applied [35]–[37]. All three tests clearly supported recombination (p = 0.0). The phylogenetic network of 29 ISR sequences (279–282 bp) from 29 different strains of C. difficile, only one ISR per strain was included in the analysis (Figure 3B), also showed conflicting signals indicating the possibility of inter-chromosomal recombination (PHI test; p = 0.0013; 55 informative sites).
Figure 3. Indication of homologous recombination in C. difficile 16S-23S rRNA intergenic spacer region.
(A) Phylogenetic network constructed for 95 representative ISR sequences from 43 different C. difficile strains. (B) Phylogenetic network of 29 ISR sequences (279–282 bp) from 29 different strains of C. difficile. Only one ISR per strain was included in the analysis. Box-like branches seen on both figures indicate relative support for alternative relationships among ISRs, probably resulting from homologous recombination that was subsequently confirmed by statistical analysis. In groups I, II and III the ISRs without a gene for tRNAAla are clustered and in group IV the ISRs with a tRNA gene.
Since our results suggest that the evolution of C. difficile 16S-23S rRNA ISRs is mainly influenced by recombination, ISRs would not be considered as appropriate marker for study of phylogenetic relationships of C. difficile strains. However, comparison of similarities between PCR-ribotypes showed that PCR-ribotypes that have similar banding patterns are related, as shown by MLST analysis (Figure 1) and the whole genome comparisons shown by others [24], suggesting that despite recombination in ISR PCR-ribotyping is an appropriate method to group C. difficile strains. This could indicate that recombination within ISR regions occurs with slow rate and in parallel to other evolutionary changes within C. difficile species. Furthermore, Valiente et al. recently described 3 different C. difficile PCR-ribotypes that have probably evolved from the 027 lineage and which banding patterns differ from the 027 in just one or two bands (i.e. 176 and 198 differing from 027 by a single band and 244 differing by two bands, one absent and one additional band) [38]. It is therefore possible that gain or loss of ISRs of particular sizes is due to rearrangements of building blocks within the ISRs, or even entire ISRs or rRNA operons, via the process of homologous recombination or possibly also slipped-strand mispairing, another proposed mechanism responsible for size variations in 16S-23S rRNA ISR [21].
All these features described for the 16S-23S rRNA ISRs are consistent with the highly mobile and mosaic C. difficile genome with up to 11% of the genome comprising horizontally acquired genetic elements [11]. Moreover, studies based on whole genome comparisons have shown that beside mobile elements homologous recombination has also played an important role in the evolution of C. difficile [23]. This was recently demonstrated by Brouwer and colleagues [39] who showed that large sequence blocks (up to 270 kbp) can be transferred between C. difficile strains and subsequently being integrated into the recipient genome.
Our results indicate that the evolution of C. difficile 16S-23S rRNA intergenic spacer region (ISR) is mainly influenced by recombination (intra- and inter-genomic). The modular structure of the ISRs, the high sequence similarities of ISRs of the same sizes and evidence of homologous recombination also suggest concerted evolution (homogenization among different loci in multigene families) of 16S-23S rRNA ISRs. Despite the strong evidence of recombination in ISRs our data show that PCR-ribotyping is an appropriate method to group C. difficile strains, as strains with similar banding patterns were related by MLST.
Materials and Methods
Isolates and genomes
Thirty one isolates of 27 different PCR-ribotypes were selected from two strain collections (Table 1). Ten strains were from the National Laboratory for Health, Environment and Food (NLZOH) strain collection and 21 strains were from the Austrian National reference Centre for C. difficile, Vienna, Austria. Three isolates originated from animals, two from piglets from two Slovenian farms and one from a calf from Canada. The remaining 28 isolates were from humans from different geographical locations. ISRs from 3 sequenced C. difficile genomes were also included in the analysis; CD196 (NC_013315.1), R20291 (NC_013316.1) and 630 (NC_009089.1) [11], [22], [23]. For one strain (PCR-ribotype 027) the ISRs were obtained from sequenced genome (CD196) and with cloning of amplified ISRs. We have included also 78 ISR sequences from 9 different strains; A, B, ATCC43593, 001, 027, AI5, 053, 078 and 176 analyzed in two previous publications [20], [21].
Molecular characterization
DNA was extracted using a MagNA Pure Compact instrument (Roche Diagnostics) according to the manufacturer's recommendations.
All strains were characterized by toxinotyping involving amplification and enzymatic restriction of PCR fragment A3 of the tcdA gene and PCR fragment B1 of the tcdB gene as described previously [40]. Absence of the PaLoc was confirmed by PCR amplification of the 115 bp sequence integrated at this site [41].
PCR-ribotyping was performed as described before [42]. Amplified fragments were analyzed with a 310 Genetic Analyzer (Applied Biosystems) with a 41 cm capillary in a POP4 gel. A TAMRA ladder 50–625 bp (Chimerx) was used as a size marker. Injection of samples was done at 5 kV over 5 s, and the total running time was 28 min at 15 kV. The size of each peak was determined using Peakscanner software 1.0 (Applied Biosystems). Peaks were counted as bands when they showed at least 10% of the height of the highest peak of each run. PCR-ribotypes were identified with Webribo software (http://webribo.ages.at/). The peaks were also imported into BioNumerics software version 7.1 (Applied Maths) and the Dice coefficient and UPGMA clustering method were used to compare banding patterns. Optimization and position tolerance were set to 0 and 0.5, respectively.
For multilocus sequence typing (MLST) the scheme described by Lemee et al. [43], using seven housekeeping genes (aroE, dutA, gmk, groEL, recA, sodA and tpi), was used. After PCR amplification, products were purified with Exonuclease I and Shrimp alkaline phosphatase (Fermentas), following manufacturer's recommendations. Sequencing reactions were set up using BigDye Terminator v1.1 kit (Applied Biosystems) and PCR products were purified with Centri-Sep columns (Princeton Separations) before sequencing on an ABI 3130 (Applied Biosystems). Sequences with forward and reverse primers for all seven loci were imported into BioNumerics software version 7.1 (Applied Maths) and analyzed with the MLST plugin. Minimum spanning tree (MST) was used to present MLST data.
Amplification of the 16S-23S rRNA intergenic spacer region (ISR)
The 16S-23S rRNA ISRs were amplified with the same primers used for PCR ribotyping; 16S (5′-GTGCGGCTGGATCACCTCCT) and 23S (5′-CCCTGCACCCTTAATAACTTGACC) [10]. PCR was carried out in a final volume of 50 µl containing 25 µl HotStar Taq Master Mix (Qiagen), 50 pmol of each primer and 1.5 µl of DNA. Amplifications conditions were as follows: an initial denaturation step of 15 min at 95°C, followed by 35 cycles of 1 min at 94°C for denaturation, 1 min at 57°C for annealing and 1 min at 72°C for elongation, plus 7 min at 72°C for a final elongation. The resulting amplification products were first checked by electrophoresis in 1.5% agarose gels and then purified with a MinElute PCR purification kit (Qiagen), following manufacturer instructions.
Cloning and sequencing of 16S-23S rRNA ISR
The purified amplification products were cloned into the pDrive vector (PCR Cloning Plus kit; Qiagen) according to the manufactureŕs instructions. Plasmids were isolated from transformed overnight colonies using QIAprep Spin Miniprep kit (Qiagen).
Plasmids (1 µl) were checked for inserts by re-amplification using primers M13r and M13f. Sequencing of the clones with an insert was carried out using Genetic analyser ABI 3130 (Applied Bosystems) and BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing reactions contained 2 µl of template DNA, 2 µl of BigDye Terminator v1.1, 1 µl of sequencing buffer and 1 µl of either primer M13f or M13r in a total volume of 10 µl. After the sequencing PCR, products were purified using Centri-Sep columns (Princeton Separations), according to the manufacturer's instructions.
The lengths of the individual ISRs obtained from direct cloning were compared to the PCR-ribotyping profile (all PCR amplified ISRs from a given genome). If not all ISRs were obtained with direct cloning of PCR products the remaining fragments were excised and purified from the agarose gel using a QIAquick Gel Extraction Kit (Qiagen). The purified products were then cloned and sequenced as described above.
ISR sequence analysis
The sequence of the vector was trimmed from all sequences using the program CLC DNA workbench (CLC bio, Denmark) and BioEdit Sequence Alignment Editor [44]. The same two programs were also used for assembly of the sequences generated with forward and reverse primers. Consensus sequence and sequence logos were generated with Weblogo3 [45]. tRNAscan-SE was used to scan the genomes for tRNA genes [46].
To facilitate computational analysis, sequence clustering using CD-HIT was used to make non-redundant sequence sets with 99% sequence identity and 99% coverage [47]. Sequences of representatives of non-redundant sets are provided in FASTA format (Dataset S1). All representatives in the non-redundant datasets were aligned using LocARNA software using default parameters [48]. LocARNA first calculates the secondary structure for all ISR using RNAfold and then generates a multiple alignment that conserves as many structural features as possible.
The evolutionary relationships among the ISRs were constructed using the Neighbor-Net (part of the SplitsTree4 software) which illustrates the relationships between the ISR by constructing networks, instead of bifurcating phylogenetic trees, taking into account also recombination events that might have occurred during evolution. Three different statistics were used to test for recombination; PHI test (a pairwise homoplasy index) [36], [49], maximum chi-squared [37] and NSS [35], using the PhiPack software (http://www.maths.otago.ac.nz/∼dbryant/).
A secondary structure prediction of the primary rRNA transcript was constructed using the RNAfold web service [50]. To simplify the secondary structure model, the 16S, 23S and 5S rRNAs were deleted from the sequence.
Supporting Information
Sequence logos showing conserved and variable nucleotide sites in the ISR building blocks.
(PDF)
LocARNA alignment of ISRs with consensus sequence and conservation of the sequences. All 95 representatives of non-redundant data set were included in the alignment.
(TIFF)
Proposed secondary structure model of the RNA transcript of C. difficile rRNA operon. The secondary structure shown in (A) represent the rRNA operon with the ISR without the tRNAAla gene and (B) with tRNAAla gene present. The 16S and 23S are represented by triangles. Direct repeats are marked with boxes.
(PDF)
Banding patterns of PCR-ribotypes included in the analysis. Number and sizes of amplified ISRs (including primers) obtained for each PCR-ribotype using the sequencer based PCR-ribotyping.
(PDF)
List of sequence variants of ISR building blocks. Only the 95 representatives of nun-redundant datasets were taken into account when looking at the variations in ISR sequence building blocks.
(PDF)
Representative 16S-23S rRNA ISR sequences of non-redundant sets (99% sequence identity and 99% sequence coverage). Sequences are in Fasta format.
(TXT)
Sequence identity matrices of 95 representatives of non-redundant sets.
(PDF)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
SJ was supported by grant 1000-08-310144 from the Slovenian Research Agency. MR and AI were supported by ERA-NET PathoGenoMics CDIFFGEN (grant 3211-09-000141 in Slovenia and 818480 in Austria). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dubberke ER, Reske KA, Yan Y, Olsen MA, McDonald LC, et al. (2007) Clostridium difficile—Associated Disease in a Setting of Endemicity: Identification of Novel Risk Factors. Clin Infect Dis 45: 1543–1549 10.1086/523582 [DOI] [PubMed] [Google Scholar]
- 2. Gupta A, Khanna S (2014) Community-acquired Clostridium difficile infection: an increasing public health threat. Infect Drug Resist 7: 63–72 10.2147/IDR.S46780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7: 526–536 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- 4. Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, et al. (2011) Clostridium difficile infection in Europe: a hospital-based survey. The Lancet 377: 63–73 10.1016/S0140-6736(10)61266-4 [DOI] [PubMed] [Google Scholar]
- 5. Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, et al. (2008) Emergence of Clostridium difficile Infection Due to a New Hypervirulent Strain, Polymerase Chain Reaction Ribotype 078. Clin Infect Dis 47: 1162–1170 10.1086/592257 [DOI] [PubMed] [Google Scholar]
- 6.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, et al. (2008) Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Available: http://www.lenus.ie/hse/handle/10147/104974. Accessed 14 April 2014. [PubMed]
- 7. Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, et al. (2005) A Predominantly Clonal Multi-Institutional Outbreak of Clostridium difficile–Associated Diarrhea with High Morbidity and Mortality. N Engl J Med 353: 2442–2449 10.1056/NEJMoa051639 [DOI] [PubMed] [Google Scholar]
- 8. McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, et al. (2005) An Epidemic, Toxin Gene–Variant Strain of Clostridium difficile . N Engl J Med 353: 2433–2441 10.1056/NEJMoa051590 [DOI] [PubMed] [Google Scholar]
- 9. Pépin J, Valiquette L, Alary M-E, Villemure P, Pelletier A, et al. (2004) Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Can Med Assoc J 171: 466–472 10.1503/cmaj.1041104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC (1999) Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 175: 261–266. [DOI] [PubMed] [Google Scholar]
- 11. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, et al. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38: 779–786 10.1038/ng1830 [DOI] [PubMed] [Google Scholar]
- 12. Janezic S, Rupnik M (2010) Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. Methods Mol Biol Clifton NJ 646: 55–65 10.1007/978-1-60327-365-74 [DOI] [PubMed] [Google Scholar]
- 13. Gianninò V, Rappazzo G, Scuto A, Di Marco O, Privitera A, et al. (2001) rrn operons in Haemophilus parainfluenzae and mosaicism of conserved and species-specific sequences in the 16S-23S rDNA long spacer. Res Microbiol 152: 461–468 10.1016/S0923-2508(01)01219-0 [DOI] [PubMed] [Google Scholar]
- 14. Houpikian P, Raoult D (2001) 16S/23S rRNA Intergenic Spacer Regions for Phylogenetic Analysis, Identification, and Subtyping of Bartonella Species. J Clin Microbiol 39: 2768–2778 10.1128/JCM.39.8.2768-2778.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kostman JR, Edlind TD, LiPuma JJ, Stull TL (1992) Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol 30: 2084–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leblond-Bourget N, Philippe H, Mangin I, Decaris B (1996) 16S rRNA and 16S to 23S Internal Transcribed Spacer Sequence Analyses Reveal Inter- and Intraspecific Bifidobacterium Phylogeny. Int J Syst Bacteriol 46: 102–111 10.1099/00207713-46-1-102 [DOI] [PubMed] [Google Scholar]
- 17. Liguori AP, Warrington SD, Ginther JL, Pearson T, Bowers J, et al. (2011) Diversity of 16S-23S rDNA Internal Transcribed Spacer (ITS) Reveals Phylogenetic Relationships in Burkholderia pseudomallei and Its Near-Neighbors. PLoS ONE 6: e29323 10.1371/journal.pone.0029323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Neill GL, Ogunsola1 FT, Brazier JS, Duerden BI (1996) Modification of a PCR Ribotyping Method for Application as a Routine Typing Scheme for Clostridium difficile . Anaerobe 2: 205–209 10.1006/anae.1996.0028 [DOI] [Google Scholar]
- 19.Siarkou VI, Stamatakis A, Kappas I, Hadweh P, Laroucau K (2011) Evolutionary Relationships among Chlamydophila abortus Variant Strains Inferred by rRNA Secondary Structure-Based Phylogeny. PLoS ONE 6 . Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3101216/. Accessed 14 April 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sadeghifard N, Gürtler V, Beer M, Seviour RJ (2006) The Mosaic Nature of Intergenic 16S-23S rRNA Spacer Regions Suggests rRNA Operon Copy Number Variation in Clostridium difficile Strains. Appl Environ Microbiol 72: 7311–7323 10.1128/AEM.01179-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Indra A, Blaschitz M, Kernbichler S, Reischl U, Wewalka G, et al. (2010) Mechanisms behind variation in the Clostridium difficile 16S–23S rRNA intergenic spacer region. J Med Microbiol 59: 1317–1323 10.1099/jmm.0.020792-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stabler RA, He M, Dawson L, Martin M, Valiente E, et al. (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10: R102 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, et al. (2010) Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci 107: 7527–7532 10.1073/pnas.0914322107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, et al. (2014) Sequence Similarity of Clostridium difficile Strains by Analysis of Conserved Genes and Genome Content Is Reflected by Their Ribotype Affiliation. PLoS ONE 9: e86535 10.1371/journal.pone.0086535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iteman I, Rippka R, Marsac NT de, Herdman M (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology 146: 1275–1286. [DOI] [PubMed] [Google Scholar]
- 26. Gürtler V (1999) The role of recombination and mutation in 16S–23S rDNA spacer rearrangements. Gene 238: 241–252 10.1016/S0378-1119(99)00224-3 [DOI] [PubMed] [Google Scholar]
- 27. Osorio CR, Collins MD, Romalde JL, Toranzo AE (2005) Variation in 16S-23S rRNA Intergenic Spacer Regions in Photobacterium damselae: a Mosaic-Like Structure. Appl Environ Microbiol 71: 636–645 10.1128/AEM.71.2.636-645.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clarke LA, Rebelo CS, Gonçalves J, Boavida MG, Jordan P (2001) PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol Pathol 54: 351–353 10.1136/mp.54.5.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janežič S, Štrumbelj I, Rupnik M (2011) Use of Modified PCR Ribotyping for Direct Detection of Clostridium difficile Ribotypes in Stool Samples. J Clin Microbiol 49: 3024–3025 10.1128/JCM.01013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letsch HO, Kück P, Stocsits RR, Misof B (2010) The Impact of rRNA Secondary Structure Consideration in Alignment and Tree Reconstruction: Simulated Data and a Case Study on the Phylogeny of Hexapods. Mol Biol Evol 27: 2507–2521 10.1093/molbev/msq140 [DOI] [PubMed] [Google Scholar]
- 31. Rocap G, Distel DL, Waterbury JB, Chisholm SW (2002) Resolution of Prochlorococcus and Synechococcus Ecotypes by Using 16S-23S Ribosomal DNA Internal Transcribed Spacer Sequences. Appl Environ Microbiol 68: 1180–1191 10.1128/AEM.68.3.1180-1191.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Apirion D, Miczak A (1993) RNA processing in prokaryotic cells. BioEssays News Rev Mol Cell Dev Biol 15: 113–120 10.1002/bies.950150207 [DOI] [PubMed] [Google Scholar]
- 33. Lan R, Reeves PR (1998) Recombination between rRNA operons created most of the ribotype variation observed in the seventh pandemic clone of Vibrio cholerae . Microbiology 144: 1213–1221 10.1099/00221287-144-5-1213 [DOI] [PubMed] [Google Scholar]
- 34. Privitera A, Rappazzo G, Sangari P, Gianninò V, Licciardello L, et al. (1998) Cloning and sequencing of a 16S/23S ribosomal spacer from Haemophilus parainfluenzae reveals an invariant, mosaic-like organisation of sequence blocks. FEMS Microbiol Lett 164: 289–294. [DOI] [PubMed] [Google Scholar]
- 35. Jakobsen IB, Easteal S (1996) A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci CABIOS 12: 291–295 10.1093/bioinformatics/12.4.291 [DOI] [PubMed] [Google Scholar]
- 36. Bruen TC, Philippe H, Bryant D (2006) A Simple and Robust Statistical Test for Detecting the Presence of Recombination. Genetics 172: 2665–2681 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34: 126–129 10.1007/BF00182389 [DOI] [PubMed] [Google Scholar]
- 38. Valiente E, Dawson LF, Cairns MD, Stabler RA, Wren BW (2012) Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J Med Microbiol 61: 49–56 10.1099/jmm.0.036194-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brouwer MSM, Roberts AP, Hussain H, Williams RJ, Allan E, et al. (2013) Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4. Available: http://www.nature.com/ncomms/2013/131017/ncomms3601/full/ncomms3601.html. Accessed 14 April 2014. [DOI] [PMC free article] [PubMed]
- 40. Rupnik M, Avesani V, Janc M, Eichel-Streiber C von, Delmée M (1998) A Novel Toxinotyping Scheme and Correlation of Toxinotypes with Serogroups of Clostridium difficileIsolates. J Clin Microbiol 36: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C (1996) Definition of the single integration site of the pathogenicity locus in Clostridium difficile . Gene 181: 29–38. [DOI] [PubMed] [Google Scholar]
- 42. Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, et al. (2008) Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57: 1377–1382 10.1099/jmm.0.47714-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lemée L, Bourgeois I, Ruffin E, Collignon A, Lemeland J-F, et al. (2005) Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile . Microbiology 151: 3171–3180 10.1099/mic.0.28155-0 [DOI] [PubMed] [Google Scholar]
- 44.Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Available: http://brownlab.mbio.ncsu.edu/JWB/papers/1999Hall1.pdf. Accessed 14 April 2014.
- 45. Crooks GE, Hon G, Chandonia J-M, Brenner SE (2004) WebLogo: A Sequence Logo Generator. Genome Res 14: 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 48. Smith C, Heyne S, Richter AS, Will S, Backofen R (2010) Freiburg RNA Tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res 38: W373–W377 10.1093/nar/gkq316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huson DH, Bryant D (2006) Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol 23: 254–267 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 50. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence logos showing conserved and variable nucleotide sites in the ISR building blocks.
(PDF)
LocARNA alignment of ISRs with consensus sequence and conservation of the sequences. All 95 representatives of non-redundant data set were included in the alignment.
(TIFF)
Proposed secondary structure model of the RNA transcript of C. difficile rRNA operon. The secondary structure shown in (A) represent the rRNA operon with the ISR without the tRNAAla gene and (B) with tRNAAla gene present. The 16S and 23S are represented by triangles. Direct repeats are marked with boxes.
(PDF)
Banding patterns of PCR-ribotypes included in the analysis. Number and sizes of amplified ISRs (including primers) obtained for each PCR-ribotype using the sequencer based PCR-ribotyping.
(PDF)
List of sequence variants of ISR building blocks. Only the 95 representatives of nun-redundant datasets were taken into account when looking at the variations in ISR sequence building blocks.
(PDF)
Representative 16S-23S rRNA ISR sequences of non-redundant sets (99% sequence identity and 99% sequence coverage). Sequences are in Fasta format.
(TXT)
Sequence identity matrices of 95 representatives of non-redundant sets.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.